Abstract
We show that most Salmonella typhimurium mutants resistant to streptomycin, rifampicin, and nalidixic acid are avirulent in mice. Of seven resistant mutants examined, six were avirulent and one was similar to the wild type in competition experiments in mice. The avirulent-resistant mutants rapidly accumulated various types of compensatory mutations that restored virulence without concomitant loss of resistance. Such second-site compensatory mutations were more common then reversion to the sensitive wild type. We infer from these results that a reduction in the use of antibiotics might not result in the disappearance of the resistant bacteria already present in human and environmental reservoirs. Thus, second-site compensatory mutations could increase the fitness of resistant bacteria and allow them to persist and compete successfully with sensitive strains even in an antibiotic-free environment.
During the last decade there has been an alarming increase in the appearance of antibiotic-resistant bacteria as a result of an increased use of antibiotics combined with the exceptional ability of bacteria to develop resistance. One strategy to reverse this development is to decrease the use of antibiotics to promote the disappearance of the resistant bacteria present in human and environmental reservoirs. Implicit in this reasoning is that resistance confers a cost on the bacteria, which results in a counter-selection against resistant strains in an antibiotic-free environment. An associated question and potential problem is whether the supposedly less fit, avirulent-resistant bacteria might accumulate compensatory mutations that restore fitness and virulence without loss of resistance, and thereby stabilize the resistant population.
In spite of the importance of these questions, there are few experiments that explicitly address them under the relevant conditions, i.e., in animal model systems using genetically defined bacterial strains (for a review, see ref. 1). For example, it has been shown in Escherichia coli that carriage of resistance genes on a plasmid is associated with a decreased growth rate, and that these strains can accumulate chromosomal compensatory mutations that, by an unknown mechanism, compensate for the growth rate decrease (2, 3). Likewise, it has been shown that slow-growing streptomycin-resistant mutants of E. coli can accumulate compensatory mutations that restore rapid growth under laboratory conditions without affecting the resistance (4). Interestingly, these compensatory mutations appear to create a genetic background in which the streptomycin-sensitive revertants have a strong selective disadvantage, implying that it would be difficult for an evolved resistant strain to become sensitive even in the absence of the antibiotic (5). There are also animal data which indicate that tetracycline-resistant E. coli persist in pigs long after the antibiotic has been removed, suggesting that the added burden of this particular resistance in vivo is in fact small (6). Finally, studies of human clinical isolates of Mycobacterium tuberculosis, which are isoniazid-resistant because of a defect in the catalase encoded by the katG gene, suggest that they accumulate mutations resulting in increased synthesis of an alkyl hydroxy peroxidase supposedly to compensate for the decrease in virulence caused by the loss of a functional catalase (7). However, this interpretation has been questioned recently (8).
In this study we examined the fitness of antibiotic-resistant S. typhimurium in mice. Our results indicate that most resistant mutants are less virulent than the wild type. However, the avirulent mutants rapidly accumulate various types of compensatory mutations that restore virulence to wild-type levels without loss of high-level resistance.
MATERIALS AND METHODS
Bacterial Strains, Growth Conditions, and Resistance Determinations.
Salmonella typhimurium LT2 were used for all experiments. Bacteria were grown in Luria–Bertani broth (LB) to midlog phase when used for infection of mice. In repeated cycling experiments bacteria were grown by serial transfer of ca. 106 bacteria to 1 ml of fresh LB medium. Bacteria were grown for ca. 12 hr at 37°C before the next transfer. Growth rates were determined by optical density (A600) measurements of bacteria grown in LB medium at 37°C as a function of time. Minimal inhibitory concentrations (MICs) were determined by using the E-test (Biodisk AB, Sweden) as described by the manufacturer. When present, antibiotics were used at standard concentrations (9).
Mouse Experiments.
To assess the virulence of the different mutants we measured their growth rates in the reticuloendothelial system (RES, e.g., liver and spleen). Several studies have shown that the growth rate in the RES after intraperitoneal injection of S. typhimurium correlates well with the virulence as measured by standard LD50 tests (see, for example, ref. 10 and references therein). For competition experiments, wild-type and resistant bacteria were grown separately in LB medium, washed once in PBS, mixed in an approximately 1:1 ratio, and then ca. 105 bacteria were injected intraperitoneally into 6- to 8-week-old female BALB/c mice. Mice were sacrificed at various times, and the spleens were removed and homogenized. Suitable dilutions of the homogenizates were plated to determine the number of sensitive and resistant bacteria present. Mice were taken care of according to the guidelines of the Swedish Institute for Infectious Disease Control under permit N268/96.
Genetic Methods and Sequencing.
Transductions were carried out with P22 HT phage as previously described (9). To demonstrate linkage of the original SmR, RifR, and NalR mutations as well as the compensatory mutations to the rpsL, rpoB, and gyrA genes, respectively, Tn10 transposons were used. Transposons were introduced by transduction from a LT2 wild-type strain, and the loss of the resistance/fast growth was demonstrated in the tetracycline-resistant transductants. We also constructed strains where the respective Tn10 was inserted next to the original resistance mutation and the fast growth mutation and showed that resistance and fast growth could be transferred into the wild-type LT2 strain by cotransduction to the Tn10. The complete rpsD, rpsE, and rpsL genes were PCR amplified with primers 5′-GACTCCGATCCCTCATAACGG-3′ (complementary to the downstream end of the rpsK gene), 5′-GCGTCGAACTCACTTGCTCG-3′ (complementary to the upstream end of the rpoA gene), 5′-GTGTTTCGGCAGACGACC-3′ (complementary to the upstream end of the rpmD gene), 5′-GGTCGTGTCCAGGCACTG-3′ (complementary to the downstream end of the rplR gene), 5′-CGTCCTCATATTGTGTGAGGG-3′ (complementary to a region −70 nt upstream of the rpsL gene start codon), and 5′-CAGGATTGTCCAAAACTCTACG-3′ (complementary to a region +449 nt downstream of the rpsL gene start codon). Sequencing reactions of the PCR fragments were performed by using the Sequenase PCR Product Sequencing Kit (Amersham).
RESULTS
Virulence of Resistant Mutants.
Using the genetically well defined bacterium Salmonella typhimurium we have addressed the following questions. (i) Are bacteria that are antibiotic resistant by virtue of target alterations less virulent than the sensitive variants? (ii) Can compensatory mutations be found, after growth of the bacteria in mice, that restore virulence without loss of resistance? (iii) What is the rate of the appearance of such compensatory mutations? (iv) What is the nature of the compensatory mutations? The antibiotics chosen were streptomycin, rifampicin, and nalidixic acid. High-level resistance in S. typhimurium to these antibiotics is conferred by target alterations as a result of mutations in the rpsL, rpoB, and gyrA genes, respectively (11). The choice of bacterial species and resistances used for these studies was based mainly on the availability of a genetically defined system with an established animal model rather than clinical importance. The streptomycin-resistant mutants have been characterized previously and classified into a restrictive and a nonrestrictive class with respect to their translation fidelity (12). One mutant from the nonrestrictive class and two from the restrictive class were chosen for further experiments. The nonrestrictive mutation rpsL105 alters the AAA codon at position 42 to AGA. The restrictive mutations rpsL106 and rpsL116 alter the AAA codon to ACA and AAC, respectively, at the same position (Table 1). Spontaneous mutants resistant to rifampicin and nalidixic acid were isolated, and transductional mapping showed that the mutations were located in the rpoB and gyrA genes, respectively, as expected. Two mutants resistant to each antibiotic were randomly chosen for further study.
Table 1.
Characteristics of wild-type and antibiotic-resistant S. typhimurium LT2
| Strain | Phenotype/selection | No. of independent isolates | Genotype (mutation) | Generation time, min | MIC
|
||
|---|---|---|---|---|---|---|---|
| Sm | Rif | Nal | |||||
| JB270 | Wild-type | * | rpsL+ (AAA42) | 26 | 2 | 24 | 16 |
| JB271 | Restrictive SmR | * | rpsL106 (ACA42) | 29 | >1,024 | † | † |
| JB272 | Restrictive SmR | * | rpsL116 (AAC42) | 33 | >1,024 | † | † |
| JB273 | Nonrestrictive SmR | * | rpsL105 (AGA42) | 27 | >1,024 | † | † |
| JB274 | Mouse-selected fast-growing suppressor of JB271 | 1 | (AAA42) | 26 | 2 | † | † |
| JB275 | Mouse-selected fast-growing suppressor of JB271 | 2 | (CGT93 → CAT93) | 25 | >1,024 | † | † |
| JB276 | Mouse-selected fast-growing suppressor of JB271 | 7 | (AGA42) | 27 | >1,024 | † | † |
| JB277 and JB290 | Mouse-selected fast-growing suppressor of JB272 | 10 | (AGA42) | 27 | >1,024 | † | † |
| JB278 | LB-selected fast-growing suppressor of JB271 | 4 | (AGA42) | 26 | >1,024 | † | † |
| JB279 | LB-selected fast-growing suppressor of JB272 | 1 | rpsE (GGT108 → AGT108) | 29 | >1,024 | † | † |
| B221 | LB-selected fast-growing suppressor of JB272 | 1 | rpsD (AAG205 → AAC205) | 29 | >1,024 | † | † |
| JB219 | LB-selected fast-growing suppressor of JB272 | 2 | Suppressor mutations extragenic to rpsD, rpsE, and rpsL | 29 | >1,024 | † | † |
| JB280 | RifR | * | rpoB (†) | 44 | † | >256 | † |
| JB281 | RifR | * | rpoB (†) | 58 | † | >256 | † |
| JB282 | Mouse-selected fast-growing suppressor of JB280 | 2 | (†) | 26 | † | 16 | † |
| JB283 | Mouse-selected fast-growing suppressor of JB281 | 1 | (†) | 27 | † | 16 | † |
| JB284 | Mouse-selected fast-growing suppressor of JB281 | 1 | (†) | 32 | † | >256 | † |
| JB285 | NalR | * | gyrA (†) | 25 | † | † | >256 |
| JB286 | NalR | * | gyrA(†) | 31 | † | † | >256 |
| JB288 | Mouse-selected suppressor of JB286 | 1 | (†) | 26 | † | † | >256 |
| JB289 | Mouse-selected suppressor of JB286 | 1 | (†) | 27 | † | † | 8 |
The suppressor mutants were isolated as described in the text. The generation times are averages of at least three measurements in rich medium (Luria–Bertani broth) cultures at 37°C. The variance was approximately ±1 min of the average values. Antibiotic minimal inhibitory concentrations (MIC, μg/ml) were determined using E-tests (Materials and Methods).
*Not applicable.
Not determined.
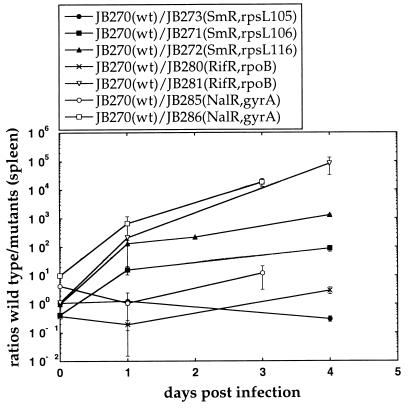
The virulence of the different resistant mutants and the sensitive wild-type bacteria was examined by competition experiments in BALB/c mice after intraperitoneal injection of the bacteria. Bacterial strains were constructed by transductional transfer of the resistance mutations into the S. typhimurium LT2 wild type to ensure isogenicity of the strains used for competitions. Bacteria were recovered from the spleens after various times, and as can be seen in Fig. 1, the wild type outcompeted the restrictive SmR mutants as well as the RifR and NalR mutants in mice. Similar results were also seen when the bacteria were recovered from the livers (data not shown), indicating that these mutants are at a general disadvantage when grown in the reticuloendothelial system. In contrast, the nonrestrictive SmR mutant was similar compared with the wild type, and of the seven resistant mutants examined this was the only mutant that did not show a decreased growth rate in mice. In addition, as seen in Table 1, most of the mutants had a slower growth rate than the wild type in LB medium.
Figure 1.
Competition experiments between wild-type and resistant mutants of S. typhimurium LT2. Each data point represents the average of the viable counts from at least four mice. Error bars represent the standard errors (some of which are smaller than the symbols).
Accumulation of Compensatory Mutations.
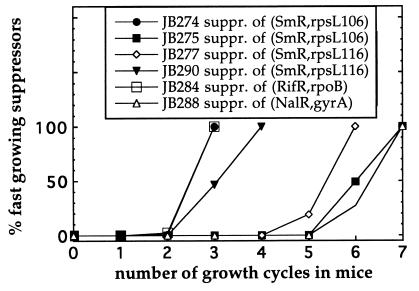
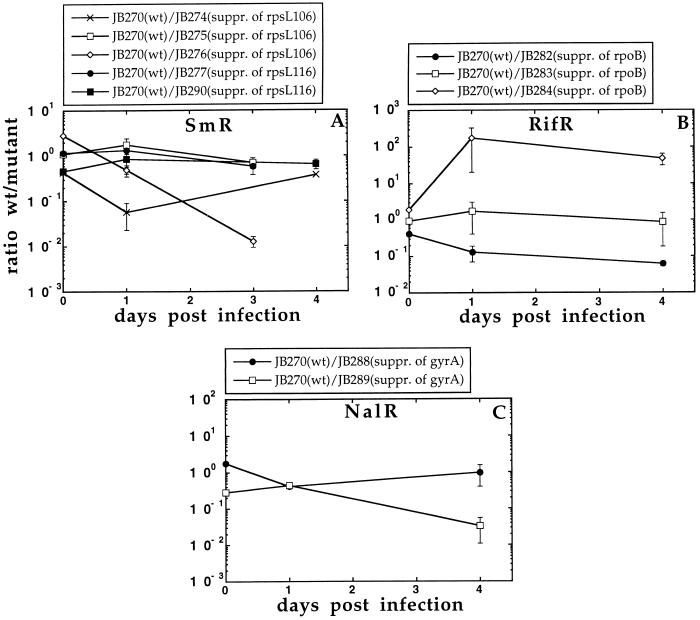
To examine whether the avirulent-resistant mutants could accumulate virulence-restoring compensatory mutations during growth in mice in the absence of antibiotics, 106 resistant bacteria were injected intraperitoneally, recovered from the spleens after 3–5 days of growth, and then 106 bacteria were reinjected into new mice. During each growth cycle the bacterial population was expanded from 106 to 108, corresponding to approximately six generations of growth. This procedure was repeated and the bacteria were examined after each cycle by visual inspection of the colony size of the plated bacteria. Fast-growing variants appeared from the restrictive SmR mutants after three to six growth cycles in mice (Fig. 2), which corresponds to 18–36 generations of growth. Control experiments showed that mutants with a >5% increase in growth rate could be detected by visual inspection of the colonies on plates. Ten independent compensatory mutants were isolated from each of the two restrictive SmR mutants. These 20 mutants were examined for their resistance, and with the exception of 1 mutant they all maintained high-level Sm resistance (Table 1). In addition, when examined for their ability to compete with the wild type in mice as well as their growth rate under laboratory conditions they were similar or better than the wild type (Fig. 3A and Table 1). Similarly, RifR compensatory mutants appeared after three cycles of growth in mice (Fig. 2). These were of two types: mutants that maintained their high-level Rif resistance (e.g., strain JB284) and others that lost their resistance (e.g., strains JB282 and JB283). The mutant strain JB283 showed a partially restored growth rate and virulence (Fig. 3B and Table 1). In contrast, the fast-grower isolated from mutant strain JB280 had a fully restored growth rate and virulence and appeared to be a true revertant as determined by its phenotype. The NalR mutant strain JB286 also accumulated compensatory mutations after six growth cycles in mice. These were either fully resistant to Nal (e.g., strain JB288, Fig. 2) or had lost resistance (e.g., strain JB289). Both strains JB288 and JB289 showed a fully restored virulence and growth rates similar to the wild type (Table 1 and Fig. 3C). Finally, it can be noted that the very fast takeover (<18–36 generations) by the compensatory mutants as shown in Fig. 2 is likely to result from the very slow growth of the resistant mutants. Thus, the wild type showed a much faster growth rate than the resistant mutants (for example, see the JB270/JB273 competition in Fig. 1 where the ratio of wt to mutant is altered from 1:1 to 105:1 in one growth cycle), and rare compensatory mutants could therefore rapidly sweep the whole population in the repeated cycling experiments.
Figure 2.
Rate of appearance of fast-growing compensatory mutants after repeated cycling in BALB/c mice. Each growth cycle represents 3–4 days of growth in a mouse.
Figure 3.
(A–C) Competition experiments between wild-type and compensatory mutants. Each data point represents the average of the viable counts from at least four mice. Error bars represent the standard errors (some of which are smaller than the symbols).
Identification of Compensatory Mutations.
The compensatory mutations were identified for the SmR mutants. Transductional mapping showed that the compensatory mutations were closely linked to the rpsL gene, and sequencing located the mutations in the rpsL gene itself. Ten independent compensatory mutants originating from strains JB271 (SmR, rpsL106) and JB272 (SmR, rpsL116), respectively, were examined. For strain JB271 (SmR, rpsL106), one compensatory mutant found had a reversion of the ACA codon to the wild-type AAA codon at position 42 (strain JB274 in Table 1 and Fig. 3A). Two were mutations that altered the CGT codon to CAT at position 93 while maintaining the original ACA mutation at position 42. These mutants were similar to the wild type in mouse competitions (Fig. 3A). Another class of compensatory mutations (7/10) were intracodonic mutations that altered the ACA codon to AGA. Some of these mutants were similar to the wild type whereas others outcompeted the wild type (e.g., JB276, Table 1 and Fig. 3A), indicating the presence of additional, undefined compensatory mutations. For strain JB272 (SmR, rpsL116), all 10 independent compensatory mutations were unexpectedly alterations of the AAC codon to AGA (e.g., strains JB277 and JB290 in Table 1 and Fig. 3A). The AAC → AGA mutation requires two nucleotide substitutions and ought to be much rarer than the true reversion, which requires only one single nucleotide substitution (AAC → AAA). The frequent finding of the double mutant is intriguing and might either suggest that the mutagenic mechanism promotes the double substitution and/or that the mutant (AGA) has a competitive advantage in mice compared with the wild type (AAA). A slight advantage can actually be seen for the AGA mutant in competitions with the wild type (see JB270/JB273 competition in Fig. 1), but it is unlikely that this small difference can account for this result. This is further supported by the observation that cycling of the wild type (AAA) in mice did not result in selection of the AGA mutant (not shown). Thus, we favor the idea that this particular double mutation is very frequent, rather than the mutant having a competitive advantage over the wild type. Because the number of bacteria in spleen and liver during the cycling experiments does not exceed 108, we conclude that the double mutation has to be unusually frequent in the bacterial population (>10−8/cell). One possible explanation might be that these evolved strains contain mutator genes, which could account for the high mutation rates. However, measurements of the mutation rates to various antibiotic resistances in these strains showed no indication of increased mutation rates (not shown). Previously only mutations in the rpsD or rpsE genes, which encode the ribosomal proteins S4 and S5, have been shown to suppress the growth rate defects of restrictive SmR mutants in E. coli (see, for example, refs. 13–18). Hence, the mutations found in mice in this study are previously undescribed growth rate compensatory mutations of the restrictive SmR mutants.
We also examined linkage of the RifR and NalR virulence compensatory mutations to the original resistance-conferring mutations (rpoB and gyrA, respectively) by transductional analysis. The mapping data indicate that the compensatory mutations are linked to the original rpoB or gyrA mutations, respectively, suggesting that these compensatory mutations are intragenic.
Selections in Laboratory Media.
To examine whether selection for fast-growers in laboratory media would result in different compensatory mutations than selection for virulent mutants in mice, four independent fast-growers were isolated after growth of the restrictive mutant strains JB271 (SmR, rpsL106) and JB272 (SmR, rpsL116) in LB medium. After ca. 250 generations, fast-growing variants appeared that had partly or fully restored growth rates in LB medium and maintained high-level SmR resistance (Table 1). The rpsD, rpsE, and rpsL genes of these mutants were sequenced, one of the JB272 compensatory mutants (JB221) had an rpsD compensatory mutation, and strain JB279 contained an rpsE mutation (Table 1). These mutations have not been described previously in E. coli or S. typhimurium. For the remaining two mutants, no mutations could be found in the rpsD, rpsE, or rpsL genes. The exact location of these mutations remains to be determined. In contrast, strain JB271 compensatory mutants had intragenic mutations that altered the ACA codon to AGA. It is interesting to note that none of the LB-selected mutants contained the double mutation that was commonly found in mouse selections. This might indicate that the types and rates of mutation and/or the type and intensity of the selection pressures in mice and in laboratory media are very different.
DISCUSSION
If the presented results are general and apply to medically relevant pathogens, an implication is that the strategy of getting rid of antibiotic-resistant bacteria by a decreased use of antibiotics can be problematic. This is not to suggest that a prudent use of antibiotics is not very important, but rather that the disappearance of the resistant variants from a population might be slower than anticipated when antibiotics use is decreased. The first problem is that resistance caused by target alterations does not necessarily result in decreased virulence as exemplified by the nonrestrictive SmR mutant. This class of SmR mutant is the most common in S. typhimurium, at least when selected under laboratory conditions (D.H., unpublished data), and our results suggest that this mutant can efficiently compete with the wild type in an antibiotic-free environment. If this type of “no-cost” resistance mutation is prevalent clinically, this might result in stable populations of resistant bacteria even in the absence of antibiotics. A second potential problem is that avirulent-resistant mutants can rapidly accumulate various types of compensatory mutations that partly or fully restore virulence and growth rate without affecting the high-level resistance. It is noticeable that only 4 of the 26 compensatory mutants examined in this study were revertants that regained sensitivity. The remaining 22 contained second-site compensatory mutations that did not decrease resistance. One reason for the bias toward second-site compensatory mutations could be differences in mutational target sizes. Thus, true reversion can occur only by one base substitution whereas second-site suppression is likely to occur at several sites. Thus, for example, the restrictive SmR mutants regained full virulence and growth rates, while retaining high-level resistance, by intragenic mutations as well as undefined extragenic compensatory mutations. Likewise, RifR and NalR mutants obtained compensatory mutations that restored virulence partly or fully without loss of resistance. Interestingly, in LB medium selections, the spectrum of compensatory mutations found for the SmR mutants were different from the ones found in mice, at least for one of the restrictive SmR mutants. The conditional character of the outcome of these selections implies that it is important to use relevant animal models to identify the compensatory mutations, which might be of clinical importance.
These data indicate that knowledge of the cost of resistance and the risk of appearance of compensatory mutations is required for rational use of antibiotics. Thus, the choice of antibiotics used clinically, at least in situations where alternative antibiotics are available, ought to be based not only on the rate of appearance of the initial resistance mutations but also on the possible counter-selection against the resistant variants as well as the rate of virulence-restoring compensatory mutations, which allow resistances to be maintained. In accord with this notion, a theoretical population analysis of resistance shows that the fitness cost of resistance is the primary parameter that determines both the frequency of resistance at any given frequency of antibiotic use and the rate at which that frequency will change with changes in antibiotic use patterns (19). In conclusion, for an ideal antibiotic from the point of view of resistance development, resistance-causing mutations should be rare, the virulence and fitness cost of resistance should be high, and the rate of virulence-restoring second-site compensatory mutations should be low.
Acknowledgments
We thank two reviewers for helpful comments. This work was supported by the Swedish Institute for Infectious Disease Control and the Swedish Natural Science Research Council.
ABBREVIATION
- LB
Luria–Bertani medium
Footnotes
References
- 1.Spratt B G. Curr Biol. 1996;6:1219–1221. doi: 10.1016/s0960-9822(96)00700-2. [DOI] [PubMed] [Google Scholar]
- 2.Bouma J E, Lenski R E. Nature (London) 1988;335:351–352. doi: 10.1038/335351a0. [DOI] [PubMed] [Google Scholar]
- 3.Lenski R E, Simpson S C, Nguyen T T. J Bacteriol. 1994;176:3140–3147. doi: 10.1128/jb.176.11.3140-3147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schrag S J, Perrot V. Nature (London) 1996;381:120–121. doi: 10.1038/381120b0. [DOI] [PubMed] [Google Scholar]
- 5.Schrag S, Perrot V, Levin B R. Proc Royal Acad Sci. 1997;264:1287–1291. doi: 10.1098/rspb.1997.0178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith H W. Nature (London) 1975;258:628–630. doi: 10.1038/258628a0. [DOI] [PubMed] [Google Scholar]
- 7.Sherman D R, Mdluli K, Hickley M J, Arain T M, Morris S L, Barry C E, III, Stover C K. Science. 1996;272:1641–1643. doi: 10.1126/science.272.5268.1641. [DOI] [PubMed] [Google Scholar]
- 8.Heym B, Stavropoulos E, Honore N, Domenech P, Saint-Joanis B, Wilson T M, Collins D M, Colston M J, Cole S T. Infect Immunol. 1997;65:1395–1401. doi: 10.1128/iai.65.4.1395-1401.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davies R W, Botstein D, Roth J R. Advanced Bacterial Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 1980. [Google Scholar]
- 10.Björkman J, Rhen M, Andersson D I. FEMS Microbiol Lett. 1996;139:121–126. doi: 10.1111/j.1574-6968.1996.tb08190.x. [DOI] [PubMed] [Google Scholar]
- 11.LaRossa R A. In: Escherichia coli and Salmonella, Cellular and Molecular Biology. Neidhardt F C, editor. 1996. pp. 2527–2587. [Google Scholar]
- 12.Tubulekas I, Hughes D. Mol Microbiol. 1993;8:761–770. doi: 10.1111/j.1365-2958.1993.tb01619.x. [DOI] [PubMed] [Google Scholar]
- 13.Bjare U, Gorini L. J Mol Biol. 1971;57:423–435. doi: 10.1016/0022-2836(71)90101-x. [DOI] [PubMed] [Google Scholar]
- 14.Donner D, Kurland C G. Mol Gen Genet. 1972;115:49–53. doi: 10.1007/BF00272217. [DOI] [PubMed] [Google Scholar]
- 15.Hasenbank R, Guthrie C, Stöffler G, Wittman H G, Rosen L, Apirion D. Mol Gen Genet. 1973;127:1–18. doi: 10.1007/BF00267778. [DOI] [PubMed] [Google Scholar]
- 16.Olsson M, Isaksson L A. Mol Gen Genet. 1979;169:259–269. doi: 10.1007/BF00382272. [DOI] [PubMed] [Google Scholar]
- 17.Schnier J, Isono S, Cumberlidge A G, Isono K. Mol Gen Genet. 1985;199:265–270. doi: 10.1007/BF00330268. [DOI] [PubMed] [Google Scholar]
- 18.van Acken U. Mol Gen Genet. 1975;140:61–68. doi: 10.1007/BF00268989. [DOI] [PubMed] [Google Scholar]
- 19.Levin B R, Lipsitch M, Perrot V, Schrag S, Antia R. Clin Inf Dis. 1997;24:9–16. doi: 10.1093/clinids/24.supplement_1.s9. [DOI] [PubMed] [Google Scholar]