Abstract
We have examined expression during spermatogenesis in the mouse of three Y-linked genes, 11 X-linked genes and 22 autosomal genes, all previously shown to be germ-cell-specific and expressed in premeiotic spermatogonia, plus another 21 germ-cell-specific autosomal genes that initiate expression in meiotic spermatocytes. Our data demonstrate that, like sex-linked housekeeping genes, germ-cell-specific sex-linked genes are subject to meiotic sex-chromosome inactivation (MSCI). Although all the sex-linked genes we investigated underwent MSCI, 14 of the 22 autosomal genes expressed in spermatogonia showed no decrease in expression in meiotic spermatocytes. This along with our observation that an additional 21 germ-cell-specific autosomal genes initiate or significantly up-regulate expression in spermatocytes confirms that MSCI is indeed a sex-chromosome-specific effect. Our results further demonstrate that the chromosome-wide repression imposed by MSCI is limited to meiotic spermatocytes and that postmeiotic expression of sex-linked genes is variable. Thus, 13 of the 14 sex-linked genes we examined showed some degree of postmeiotic reactivation. The extent of postmeiotic reactivation of germ-cell-specific X-linked genes did not correlate with proximity to the X inactivation center or the Xist gene locus. The implications of these findings are discussed with respect to differential gene regulation and the function of MSCI during spermatogenesis, including epigenetic programming of the future paternal genome during spermatogenesis.
INTRODUCTION
In male mammals, development and differentiation of the germline is a dynamic process (1-5). Primordial germ cells (PGCs) migrate to the developing testis where they form prospermatogonia that subsequently enter mitotic arrest and remain in this state for the duration of fetal development. Shortly after birth in the mouse, these cells resume mitotic activity and a subset of these cells seed basal compartments of the developing seminiferous tubules to form stem spermatogonia. The spermatogonial stem cell population replicates mitotically to both maintain itself and ultimately give rise to differentiating spermatogonia. These cells then enter meiosis as primary spermatocytes that proceed through the first and second meiotic divisions to yield postmeiotic spermatids that differentiate via the process of spermiogenesis to form spermatozoa.
Spermatogenesis is marked by dramatic changes in cellular morphology and cellular contents that are the direct result of dynamic shifts in patterns of gene expression that distinguish premeiotic, meiotic and postmeiotic spermatogenic cell types (6-9). Our previous study to identify germ-cell-specific genes expressed in premeiotic spermatogonia yielded the surprising finding that an abundance of sex-linked, especially X-linked, genes are expressed in premeiotic spermatogonia (10). This was consistent with the evolutionary theories of Rice (11) who proposed that sex-linked genes that influence male-specific processes will be subject to more direct selection and, hence, will tend to propagate more rapidly throughout the population. This also reinforced earlier evidence of many critical and dynamic roles played by sex-linked genes in male germ cell development and differentiation (10,12-20).
The sex chromosomes form a unique cytological structure in meiotic spermatocytes called the ‘XY body’ (21,22) [previously known as the ‘sex vesicle’ (23)]. The XY body is manifest as a non-membrane-bound, darkly staining region of the nucleus in primary spermatocytes. This structure is distinguished on the basis of its relatively condensed chromatin structure compared with that of the autosomes in the same cells. This condensed chromatin appears to be inhibitory to transcriptional activity, resulting in transcriptional repression of sex-linked genes during meiotic stages of spermatogenesis (24-29). This process, known as meiotic sex-chromosome inactivation (MSCI), has previously been characterized primarily through studies of expression of housekeeping genes during spermatogenesis (29-31). Our discovery that a large number of germ-cell-specific sex-linked genes are expressed in spermatogonia (10) has now afforded the opportunity to determine whether these genes are also subject to MSCI. This has also facilitated a direct comparison of the expression patterns of sex-linked and autosomal germ-cell-specific genes during spermatogenesis.
Several questions about MSCI remain to be fully investigated, especially with respect to germ-cell-specific sex-linked genes. These include 1) the extent to which this process affects all sex-linked genes, including both germ-cell-specific and housekeeping genes, 2) the extent to which MSCI is indeed limited to sex-linked genes, 3) the extent to which MSCI is strictly a meiotic phenomenon and/or persists into postmeiotic stages of spermatogenesis and 4) the extent to which chromosomal position of germ-cell-specific X-linked genes, relative to the X-inactivation center, may influence their inactivation during meiosis and/or their reactivation following meiosis. Certain sex-linked genes have been shown to undergo either postmeiotic reactivation of expression following MSCI (31) or de novo initiation of expression in postmeiotic spermatids, but these studies have also been focused primarily on housekeeping genes (32,33). The extent to which an entire set of germ-cell-specific sex-linked genes exhibits postmeiotic reactivation has not previously been examined.
It was recently hypothesized that MSCI may lead directly to repression of genes on the paternal X-chromosome in the trophectoderm, primitive endoderm and early embryo proper following fertilization. Specifically, Huynh and Lee (34) have proposed that proximity to the Xic and/or Xist loci may influence the likelihood that paternally inherited X-linked genes will remain repressed during early embryogenesis in female (XX) embryos, although this remains a point of question (35). This could result from either persistent repression of X-linked genes following MSCI or epigenetic programming of X-linked genes during spermatogenesis resulting in imprinted repression following fertilization. We are now able to test the extent to which germ-cell-specific X-linked genes undergo postmeiotic reactivation and whether or not this is related to their proximity to the Xist/Xci loci.
In this study, we have taken advantage of our ability to recover relatively pure populations of specific premeiotic, meiotic and postmeiotic spermatogenic cell types to examine expression patterns of germ-cell-specific sex-linked and autosomal genes during spermatogenesis in the mouse. Our results indicate that, like sex-linked housekeeping genes, tissue-specific sex-linked genes expressed during spermatogenesis are uniformly subject to MSCI, but many autosomal genes either continue or initiate expression in meiotic spermatocytes. In addition, we find that a significant proportion of germ-cell-specific sex-linked genes show postmeiotic reactivation of expression and that the extent of this reactivation does not correlate with proximity to either the Xic or Xist loci or the pseudoautosomal region.
RESULTS
Meiotic inactivation of germ-cell-specific sex-linked genes during spermatogenesis
We previously used a cDNA subtraction approach to identify three Y-linked genes, 11 X-linked genes and 22 autosomal genes that are expressed in premeiotic spermatogonia, but not in any of a variety of somatic tissues (10). The pattern of expression of these 36 genes throughout spermatogenesis in the mouse is shown in Figure 1. As expected, on the basis of the manner in which these genes were identified, all were expressed in premeiotic spermatogonia, including primitive type A, type A and type B spermatogonia.
Figure 1.
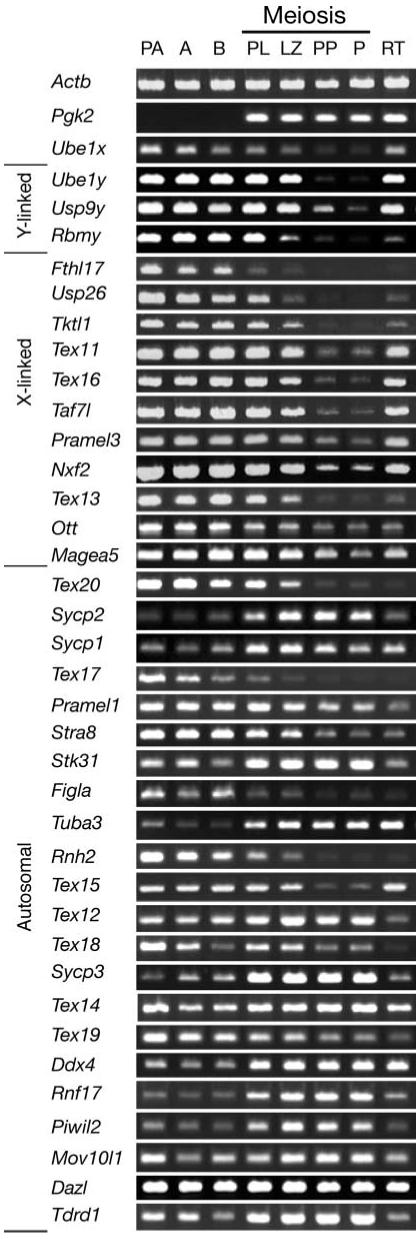
Expression throughout spermatogenesis of 36 germ-cell-specific genes expressed in spermatogonia. Relative levels of transcripts among different spermatogenic cell types were determined by RT-PCR and visualized by gel electrophoresis and ethidium bromide staining. Sex-linked genes are shown in the order of relative map positions on the sex chromosomes (10). Autosomal genes are shown in ascending order on the respective chromosomes to which each maps (10). Levels of Actb mRNA were assessed as a positive control for ubiquitous gene expression, levels of Pgk2 transcripts were determined as a positive control for previously documented autosomal meiotic expression (30) and levels of Ube1x transcripts were included as a positive control for previously documented X-linked gene expression during spermatogenesis (31). Spermatogenic cell types are listed horizontally across the top of the figure. PA: primitive type A spermatogonia; A: type A spermatogonia; B: type B spermatogonia; PL: preleptotene spermatocytes; LZ: leptotene plus zygotene spermatocytes; PP: puberal pachytene spermatocytes; P: adult pachytene spermatocytes; RT: round spermatids. Gene designations are listed vertically and are as described previously (10). Primer sequences and PCR conditions have been deposited in GenBank under accession numbers as described in Materials and Methods (10). Controls without reverse transcriptase were done in parallel and were negative (data not shown).
Importantly, all 14 sex-linked genes showed evidence of MSCI. Among the spermatogenic cell types we were able to assay, repression of sex-linked gene expression was most profound in primary spermatocytes, especially at the pachytene stage of first meiotic prophase, including pachytene spermatocytes isolated from either 18-day-old mice entering puberty or adult mice at 60−70 days of age. Thus, MSCI appears to be a widespread phenomenon affecting the majority, if not all sex-linked genes during each wave of spermatogenesis. For certain sex-linked genes examined, a decline in levels of transcript was evident at earlier stages of first meiotic prophase. For example, the Y-linked Rbmy gene showed an initial decrease in transcript levels in leptotene+zygotene spermatocytes, as did the X-linked Usp26, Tktl1 and Tex13 genes. The X-linked Fthl17 gene showed a noticeable decrease in transcript level in preleptotene spermatocytes. These results are consistent with the suggestion that transcriptional repression associated with MSCI may be initiated at the very beginning of meiotic prophase (29). Presumably the differences in rates of decline of transcript levels among different sex-linked genes are a reflection of differential stabilities of these mRNAs rather than gene-specific differences in the timing of MSCI.
Autosomal germ-cell-specific genes are not uniformly subject to meiotic inactivation
Although the sex-linked genes uniformly showed repression during meiosis, 14 of the 22 autosomal germ-cell-specific genes expressed in spermatogonia showed no decline in transcript levels in spermatocytes (Fig. 1). This supports the notion that, as a chromosome-wide phenomenon, MSCI uniquely affects the sex chromosomes. That eight of these autosomal genes were repressed during meiosis is likely a reflection of the dramatic changes in gene expression patterns that accompany the spermatogonium–spermatocyte transition during spermatogenesis (6,7). Interestingly, of the eight autosomal genes that became repressed during meiosis, only one (Tex15) showed postmeiotic reactivation.
Direct evidence that differential gene expression underlies the spermatogonium–spermatocyte transition is shown in Figure 2. This shows the expression pattern of 21 autosomal germ-cell-specific genes, all but one of which (Prm1) initiate significant expression at the spermatocyte stage, as does the germ-cell-specific autosomal gene, Pgk2, shown as a control in Figure 1. All these genes maintain or increase expression levels in postmeiotic spermatids. Despite the activation of numerous autosomal genes at the onset of meiosis, no study of which we are aware has identified a sex-linked gene that initiates expression in spermatocytes. Thus, repression of sex-linked gene expression during male meiosis is not due to a genome-wide cessation in transcription, but to a specific effect (MSCI) that selectively represses most or all genes on the X and Y chromosomes.
Figure 2.

Expression during spermatogenesis of autosomal germ-cell-specific genes initiating expression in spermatocytes. Relative levels of transcripts were assessed in specific spermatogenic cell preparations as described for Figure 1. Actb serves as a positive control for ubiquitous expression and Prm1 as a control for significant expression initiated in round spermatids. Relative transcript levels for 20 germ-cell-specific genes that initiate significant levels of expression at the onset of meiosis in primary spermatocytes, plus one other germ-cell-specific gene (Prm1) that initiates low-level expression in spermatocytes and higher expression in round spermatids, are shown below these controls. All these genes are autosomal. Primer sequences and PCR conditions have been deposited in GenBank under accession numbers as described in Materials and Methods.
Most germ-cell-specific sex-linked genes undergo postmeiotic reactivation
Of the 14 germ-cell-specific sex-linked genes we examined, all of which underwent MSCI, all but one showed partial or complete reactivation in postmeiotic spermatids (Fig. 1). This demonstrates that MSCI is a meiotic phenomenon that is not maintained on a chromosome-wide basis postmeiotically. Thus it appears that the sex chromosomes return to a transcriptionally potentiated state in postmeiotic spermatids, and the extent to which individual sex-linked genes are reactivated postmeiotically is a function of individual gene regulation and, presumably, the requirement for each particular sex-linked gene product in postmeiotic spermatids or spermatozoa.
Extent of postmeiotic reactivation of germ-cell-specific X-linked genes is not related to their chromosomal position
To determine if the postmeiotic reactivation of germ-cell-specific, X-linked genes is related to their chromosomal position relative to the X inactivation center (Xic) or the Xist gene, we examined the extent of postmeiotic reactivation of each gene as a function of genetic map position on the X chromosome (Table 1). Postmeiotic reactivation was characterized as significant (postmeiotic transcript levels equivalent to premeiotic levels) or none/slight (postmeiotic transcript levels either undetectable or detectable at levels much lower than premeiotic levels). We found no consistent correlation between the map position of X-linked germ-cell-specific genes and the degree of postmeiotic reactivation. Both the most closely linked germ-cell-specific genes on either side of the Xic/Xist loci (Tex11 and Tex16, respectively) showed significant postmeiotic reactivation, although the ubiquitously expressed Pgk1 gene that lies between the Xic/Xist loci and the Tex16 locus shows no postmeiotic reactivation (36). Moving proximally, away from the Xic/Xist loci toward the centromere, Tex11 showed significant reactivation, but the next two genes (Tktl1 and Usp26) displayed little or no reactivation. The ubiquitously expressed Ube1x gene showed significant reactivation, but the X-linked gene that is most distal from the Xic/Xist loci (Fthl17) failed to reactivate post-meiotically. Similarly, there was no obvious pattern with respect to X-linked genes that map distal to the Xic/Xist loci. Tex16, Taf7l, Nxf2 and Pramel3 all showed significant reactivation, but the more distal Tex13 and Ott genes displayed little or no reactivation, and the most distal locus, Magea5 showed significant reactivation.
Table 1.
Map position of X-linked genes and postmeiotic reactivation
| Gene/locus | Position (cm) | Reactivationa |
|---|---|---|
| Centromere | 0 | N/A |
| Fthl17 | 2.6 | None/slight |
| Ube1xb | 5.7 | Significant |
| Usp26 | 16.0 | None/slight |
| Tktl1 | 29.7 | None/slight |
| Tex11 | 40.0 | Significant |
| Xic/Xist | 42.0 | None |
| Pgk1b | 45.0 | None |
| Tex16 | 49.0 | Significant |
| Taf7l | 51.0 | Significant |
| Nxf2 | 53.1 | Significant |
| Pramel3 | 53.2 | Significant |
| Tex13 | 60.5 | None/slight |
| Ott | 62.5 | None/slight |
| Magea5 | 66.1 | Significant |
None, no detectable reactivation; slight, reactivation to transcript level < premeiotic level; strong, reactivation to transcript level > premeiotic level.
Not germ-cell-specific.
The pericentric human X chromosome can be divided into different parts—the short arm (Xp) and the long arm (Xq), or the recently added region (XRA) and the conserved region (XCR) (Fig. 3) (37). In human cells, genes mapping to Xp tend to escape somatic X-chromosome inactivation (SXCI) more often than genes mapping to Xq (38,39), and Xp roughly corresponds to the XRA region of the human X, whereas Xq corresponds to the XCR. Figure 3 shows a comparison of the human and mouse X chromosomes and the relative positions of the germ-cell-specific X-linked genes that can be identified on both homologues. Fthl17 is the only mouse X-linked, germ-cell-specific gene that we examined for which the human homologue both maps to Xp, and lies within the XRA. The human homologue of the mouse Ube1x gene also maps to Xp but lies within the XCR. The eight other germ-cell-specific, X-linked genes we examined all have human homologues that map to Xq (Fig. 3).
Figure 3.

Conservation of germ-cell-specific genes on the mammalian X chromosome. The relative map location of 11 germ-cell-specific genes plus the ubiquitous Ube1x gene is shown on the mouse X chromosome. The location of homologues of 10 of these genes on the human X is also shown. The location of the centromere is shown on each chromosome (arrow), along with the location of the p and q arms and the XRA and XCR regions on the human X chromosome.
DISCUSSION
We have conducted the most extensive study of sex-linked gene expression during spermatogenesis reported so far, and the only such study focused on germ-cell-specific sex-linked genes. Our results confirm that MSCI does occur during mammalian spermatogenesis and demonstrate that it affects germ-cell-specific sex-linked genes in a manner similar to that previously demonstrated for ubiquitously expressed sex-linked genes (18,29-32). These results also show that, as a chromosome-wide repression mechanism, MSCI is limited to the sex chromosomes and does not affect autosomal genes. In addition, these results demonstrate that MSCI is a meiosis-specific phenomenon that does not persist on a panchromosomal basis in postmeiotic spermatogenic cells. Finally, our results indicate no correlation between relative proximity to the Xic/Xist loci and propensity of individual, germ-cell-specific X-linked genes to undergo or avoid post-meiotic reactivation.
Previous studies of sex-linked gene expression during spermatogenesis have been limited to relatively small numbers of genes (≤5 per study) and have been focused predominantly on ubiquitously expressed genes (17,29-33,40-42). These previous studies consistently supported the occurrence of MSCI. Our study demonstrates that germ-cell-specific, sex-linked genes on both the X and Y chromosomes are also subject to MSCI, and thus illustrates the generality of MSCI as a sex-chromosome-wide effect. This is also consistent with a recent genome-wide study to identify the map location of sex-biased genes in which it was found that, as expected, genes subject to selection by MSCI are excluded from the X chromosome (43). In this respect, MSCI appears to be even more comprehensive than SXCI that occurs during preimplantation development of mammalian embryos carrying two or more X chromosomes, as neither we nor anyone else have detected direct evidence for a sex-linked gene that escapes MSCI.
In the case of SXCI, a significant proportion (on the order of 15−25% in humans) of the X-linked genes examined have been found to ‘escape’ inactivation (38,39,44) [actually these genes may undergo initial SXCI but rapidly reactivate (45,46)]. Carrel et al. (38,39) found that a significant majority of X-linked genes known to escape SXCI map to Xp in the human, which largely corresponds to a region (XRA) that is believed to have derived from a previously autosomal region that was more recently added to the mammalian X chromosome (37). Thus genes located within the human XRA tend to escape SXCI more often than genes within the XCR. Although fewer genes have been tested for escape from SXCI in the mouse, a significant minority appear to do so (47-50).
Although we have not tested expression during spermatogenesis of any of the specific genes known to escape SXCI, we have tested X-linked genes located throughout the mouse X chromosome. Fthl17 is the one germ-cell-specific mouse gene we examined for which the human homologue maps well within the XRA (Fig. 3). This gene showed no evidence of postmeiotic reactivation during spermatogenesis. Eight other genes that are X-linked and germ-cell-specific in the mouse (Usp26, Tktl1, Tex11, Tex16, Taf7l, Pramel3, Nxf2, Tex13) have homologues that map to Xq within the XCR region of the human X (Fig. 3). In addition, the mouse Ube1x gene is homologous to the human UBE1X gene that maps to Xp, but still lies within the XCR on the human X. Thus, all the germ-cell-specific X-linked genes which showed some degree of postmeiotic reactivation during spermatogenesis in the mouse have human homologues that map to the XCR region of the human X chromosome. We have not examined expression during spermatogenesis of any genes that map to the pseudoautosomal region of the mouse or human X chromosome.
There are several other distinctions between MSCI and SXCI. SXCI occurs in XX somatic cells and PGCs but not in XX meiotic germ cells. Indeed, female meiosis is characterized by reactivation of the second X chromosome in oocytes (51,52). SXCI affects only one of the two sex chromosomes in each XX cell and achieves dosage compensation between similar cell types in each sex. Finally, SXCI is regulated by the Xist gene (53,54). MSCI, in contrast, occurs exclusively in male meiotic germ cells, affects both sex chromosomes in each cell and is regulated by an Xist-independent mechanism (25,26) that appears to require the Brca1 gene, the kinase ATR and phosphorylation of the histone-variant H2AX (55). Thus MSCI does not lead to dosage compensation in meiotic germ cells, but rather enhances the disparity in sex-linked gene expression between spermatocytes and oocytes.
The function of MSCI in spermatogenic cells remains an enigma. Several possibilities have been suggested, including suppression of X-linked genes, expression of which might otherwise be deleterious to spermatogenesis (56), inhibition of genetic recombination between the X and Y chromosomes especially in the region of the testis-determining gene on the Y (27), exclusion of the poorly paired sex chromosome bivalent from surveillance by the meiotic checkpoint mechanism that otherwise shunts spermatocytes with mispaired homologues into an apoptosis pathway (26,57) and/or a primitive form of dosage compensation (34,57,58) that may still contribute to ‘preinactivation’ of the paternal X chromosome (34). No two of these hypotheses are mutually exclusive.
Recently it was proposed that inheritance of an inactive X chromosome from the sperm, and proximity to the Xic/Xist loci on the paternal X in particular, may contribute to imprinting and/or preinactivation of genes on the paternal X chromosome such that they will be preferentially inactive in the trophectoderm, primitive endoderm and early cells of the embryo proper following fertilization (34), though alternative explanations for non-random X chromosome activity in early female embryos have also been proposed (35). The persistent influence of MSCI on X-chromosome activity following fertilization was proposed with respect to X-linked housekeeping genes (34). Our data clearly show that at least a subset of germ-cell-specific genes reactivate in round spermatids after the completion of MSCI. Thus chromosome-wide inactivation of X-linked genes is not maintained following meiosis. However, most or all of the germ-cell-specific genes we investigated are not active in the preimplantation embryo, so it is not clear that our data bear directly on the preinactivation theory of dosage compensation in mammals (34). It remains possible that MSCI, in particular, or male gametogenesis, in general, contributes to an imprinting mechanism that epigenetically programs postfertilization repression of genes on the paternal X chromosome.
MATERIALS AND METHODS
Isolation of mouse spermatogenic cells
Populations of cells highly enriched for specific spermatogenic cell types were prepared from CD-1 mice (Charles River Laboratories) using the Sta Put method based on sedimentation velocity at unit gravity (59). Primitive type A spermatogonia were prepared from testes of 6-day postpartum (dpp) mice. Types A and B spermatogonia were isolated from 8 dpp mice. Puberal mice at 18 dpp were used for preparation of preleptotene spermatocytes, mixed leptotene plus zygotene spermatocytes and puberal pachytene spermatocytes. Adult mice (60−70 dpp) were used for isolation of adult pachytene spermatocytes and round spermatids. Purities of recovered germ cell populations were assessed on the basis of cellular morphology under phase optics and were >95% for pachytene spermatocytes and round spermatids, respectively, and >85% for all other cell types. Primary contaminants in each population included developmentally adjacent germ cell types in the adult preparations, and somatic Sertoli cells in the prepuberal populations.
Expression analysis
A semi-quantitative RT-PCR technique was used for expression analysis as described (29) with modifications. For puberal–adult germ cell preparations, total RNAs were isolated using TRIzol reagent (Invitrogen) and poly(A)+ RNAs were subsequently isolated using a QuickPrep Micro mRNA purification kit (Amersham Pharmacia Biotech, Piscataway, NJ). Seventy nanograms of poly(A)+ RNAs were reverse-transcribed using oligo(dT)18 as a primer. Bulk cDNAs were diluted to a final volume of 200 μl, and 5 μl was used as template for each 25-μl PCR reaction. To avoid saturation of PCR, products were removed after various cycles (25-35) of PCR and examined by agarose gel electrophoresis. In all cases, products from RT-PCR amplification of actin served as a loading control. Gene-specific PCR primers were designed to avoid cross-amplification from homologues. The primer sequences and PCR conditions have been deposited at GenBank. Gels were stained with ethidium bromide and photographed to visualize the size and intensity of each PCR product. Each reaction was conducted at least twice with similar results to confirm the expression patterns shown.
GenBank accession numbers
Primer sequences and PCR conditions for genes in Figure 1: Actb, BV210360; Pgk2, BV210361; Ube1y, BV210362; Usp9y, BV210363; Rbmy, G65761; Fthl17, G65778; Usp26, G65798; Tktl1, G65797; Tex11, G65787; Tex16, G65792; Taf7l, G65785; Pramel3, G65782; Nxf2, G65780; Tex13, G65789; Ott, BV210394; Magea5, BV210364; Tex20, G65796; Sycp2, BV210365; Sycp1, BV210366; Tex17, G65793; Pramel1, G65762; Stra8, BV210367; Stk31, G65784; Figla, BV210368; Tuba3, BV210369; Rnh2, G65783; Tex15, G65791; Tex12, G65788; Tex18, G65794; Sycp3, BV210370; Tex14, G65790; Tex19, G65795; Ddx4, BV210371; Rnf17, G65763; Piwil2, G65781; Mov10l1, G65779; Dazl, G65760; Tdrd1, G65786.
Primer sequences and PCR conditions for genes in Figure 2: Prm1, BV210372; Ccna1, BV210373; Clgn, BV210374; Hspa2, BV210375; Ldh3, BV210376; Mak, BV210377; Meig1, BV210378; Pdha2, BV210379; Spa17, BV210380; Tenr, BV210381; Tesk1, BV210382; Acrv1, BV210383; Xmr, BV210384; Adam2, BV210385; H1t, BV210386; Mcsp, BV210387; Pabpc2, BV210388; Gapds, BV210389; Gk-rs1, BV210390; Gk2, BV210391; Zfa, BV210392.
Genetic map positions for specific X-linked genes were accessed or estimated from the MGI database (http://www.informatics.jax.org/) and the draft mouse genome sequence in GenBank (39,60).
ACKNOWLEDGEMENTS
This work was supported in part by NIH grants HD 045866 to P.J.W., and HD 046637 and HD 23126 to J.R.M., and support from the Howard Hughes Medical Institute to D.C.P. Funding to pay the Open Access publication charges was provided by the HHMI and NIH.
Footnotes
Conflict of Interest statement. None declared.
REFERENCES
- 1.McCarrey JR. Development of the germ cell. In: Desjardins C, Ewing L, editors. Cell and Molecular Biology of the Testis. V. Oxford University Press; New York: 1993. pp. 58–89. [Google Scholar]
- 2.Eddy EM. Male germ cell gene expression. Recent Prog. Horm. Res. 2002;57:103–128. doi: 10.1210/rp.57.1.103. [DOI] [PubMed] [Google Scholar]
- 3.de Rooij DG, Grootegoed JA. Spermatogonial stem cells. Curr. Opin. Cell Biol. 1998;10:694–701. doi: 10.1016/s0955-0674(98)80109-9. [DOI] [PubMed] [Google Scholar]
- 4.Wiederkehr C, Basavaraj R, Sarrauste de Menthiere C, Hermida L, Koch R, Schlecht U, Amon A, Brachat S, Breitenbach M, Briza P, et al. GermOnline, a cross-species community knowledge base on germ cell differentiation. Nucleic Acids Res. 2004;32(database issue):D560–D567. doi: 10.1093/nar/gkh055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao GQ, Garbers DL. Male germ cell specification and differentiation. Dev. Cell. 2002;2:537–547. doi: 10.1016/s1534-5807(02)00173-9. [DOI] [PubMed] [Google Scholar]
- 6.Shima JE, McLean DJ, McCarrey JR, Griswold MD. The murine testicular transcriptome: characterizing gene expression in the testis during the progression of spermatogenesis. Biol. Reprod. 2004;71:319–330. doi: 10.1095/biolreprod.103.026880. [DOI] [PubMed] [Google Scholar]
- 7.Schultz N, Hamra FK, Garbers DL. A multitude of genes expressed solely in meiotic or postmeiotic spermatogenic cells offers a myriad of contraceptive targets. Proc. Natl Acad. Sci. USA. 2003;100:12201–12206. doi: 10.1073/pnas.1635054100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anway MD, Li Y, Ravindranath N, Dym M, Griswold MD. Expression of testicular germ cell genes identified by differential display analysis. J. Androl. 2003;24:173–184. doi: 10.1002/j.1939-4640.2003.tb02660.x. [DOI] [PubMed] [Google Scholar]
- 9.Pang AL, Taylor HC, Johnson W, Alexander S, Chen Y, Su YA, Li X, Ravindranath N, Dym M, Rennert OM, et al. Identification of differentially expressed genes in mouse spermatogenesis. J. Androl. 2003;24:899–911. doi: 10.1002/j.1939-4640.2003.tb03142.x. [DOI] [PubMed] [Google Scholar]
- 10.Wang PJ, McCarrey JR, Yang F, Page DC. An abundance of X-linked genes expressed in spermatogonia. Nat. Genet. 2001;27:422–426. doi: 10.1038/86927. [DOI] [PubMed] [Google Scholar]
- 11.Rice WR. Sex-chromosomes and the evolution of sexual dimorphism. Evolution. 1984;38:735–742. doi: 10.1111/j.1558-5646.1984.tb00346.x. [DOI] [PubMed] [Google Scholar]
- 12.Mazeyrat S, Saut N, Grigoriev V, Mahadevaiah SK, Ojarikre OA, Rattigan A, Bishop C, Eicher EM, Mitchell MJ, Burgoyne PS. A Y-encoded subunit of the translation initiation factor Eif2 is essential for mouse spermatogenesis. Nat. Genet. 2001;29:49–53. doi: 10.1038/ng717. [DOI] [PubMed] [Google Scholar]
- 13.Oates RD, Silber S, Brown LG, Page DC. Clinical characterization of 42 oligospermic or azoospermic men with microdeletion of the AZFc region of the Y chromosome, and of 18 children conceived via ICSI. Hum. Reprod. 2002;17:2813–2824. doi: 10.1093/humrep/17.11.2813. [DOI] [PubMed] [Google Scholar]
- 14.Burgoyne PS. The role of Y-encoded genes in mammalian spermatogenesis. Semin. Cell Dev. Biol. 1998;9:423–432. doi: 10.1006/scdb.1998.0228. [DOI] [PubMed] [Google Scholar]
- 15.Kay GF, Ashworth A, Penny GD, Dunlop M, Swift S, Brockdorff N, Rastan S. A candidate spermatogenesis gene on the mouse Y chromosome is homologous to ubiquitin-activating enzyme E1. Nature. 1991;354:486–489. doi: 10.1038/354486a0. [DOI] [PubMed] [Google Scholar]
- 16.Brown GM, Furlong RA, Sargent CA, Erickson RP, Longepied G, Mitchell M, Jones MH, Hargreave TB, Cooke HJ, Affara NA. Characterisation of the coding sequence and fine mapping of the human DFFRY gene and comparative expression analysis and mapping to the Sxrb interval of the mouse Y chromosome of the Dffry gene. Hum. Mol. Genet. 1998;7:97–107. doi: 10.1093/hmg/7.1.97. [DOI] [PubMed] [Google Scholar]
- 17.Elliott DJ, Ma K, Kerr SM, Thakrar R, Speed R, Chandley AC, Cooke H. An RBM homologue maps to the mouse Y chromosome and is expressed in germ cells. Hum. Mol. Genet. 1996;5:869–874. doi: 10.1093/hmg/5.7.869. [DOI] [PubMed] [Google Scholar]
- 18.Mahadevaiah SK, Odorisio T, Elliott DJ, Rattigan A, Szot M, Laval SH, Washburn LL, McCarrey JR, Cattanach BM, Lovell-Badge R, et al. Mouse homologues of the human AZF candidate gene RBM are expressed in spermatogonia and spermatids, and map to a Y chromosome deletion interval associated with a high incidence of sperm abnormalities. Hum. Mol. Genet. 1998;7:715–727. doi: 10.1093/hmg/7.4.715. [DOI] [PubMed] [Google Scholar]
- 19.Lyon MF. Mechanisms and evolutionary origins of variable X-chromosome activity in mammals. Proc. R. Soc. Lond. B Biol. Sci. 1974;187:243–268. doi: 10.1098/rspb.1974.0073. [DOI] [PubMed] [Google Scholar]
- 20.Hunt PA, Worthman C, Levinson H, Stallings J, LeMaire R, Mroz K, Park C, Handel MA. Germ cell loss in the XXY male mouse: altered X-chromosome dosage affects prenatal development. Mol. Reprod. Dev. 1998;49:101–111. doi: 10.1002/(SICI)1098-2795(199802)49:2<101::AID-MRD1>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 21.Handel MA, Park C, Kot M. Genetic control of sex-chromosome inactivation during male meiosis. Cytogenet. Cell Genet. 1994;66:83–88. doi: 10.1159/000133672. [DOI] [PubMed] [Google Scholar]
- 22.Handel MA. The XY body: a specialized meiotic chromatin domain. Exp. Cell Res. 2004;296:57–63. doi: 10.1016/j.yexcr.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 23.Solari AJ. The behavior of the XY pair in mammals. Int. Rev. Cytol. 1974;38:273–317. doi: 10.1016/s0074-7696(08)60928-6. [DOI] [PubMed] [Google Scholar]
- 24.Lifschytz E, Lindsley DL. The role of X-chromosome inactivation during spermatogenesis (Drosophila–allocycly–chromosome evolution–male sterility–dosage compensation) Proc. Natl Acad. Sci. USA. 1972;69:182–186. doi: 10.1073/pnas.69.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turner JM, Mahadevaiah SK, Elliott DJ, Garchon HJ, Pehrson JR, Jaenisch R, Burgoyne PS. Meiotic sex chromosome inactivation in male mice with targeted disruptions of Xist. J. Cell Sci. 2002;115:4097–4105. doi: 10.1242/jcs.00111. [DOI] [PubMed] [Google Scholar]
- 26.McCarrey JR, Watson C, Atencio J, Ostermeier GC, Marahrens Y, Jaenisch R, Krawetz SA. X-chromosome inactivation during spermatogenesis is regulated by an Xist/Tsix-independent mechanism in the mouse. Genesis. 2002;34:257–266. doi: 10.1002/gene.10163. [DOI] [PubMed] [Google Scholar]
- 27.McKee BD, Handel MA. Sex chromosomes, recombination, and chromatin conformation. Chromosoma. 1993;102:71–80. doi: 10.1007/BF00356023. [DOI] [PubMed] [Google Scholar]
- 28.Richler C, Ast G, Goitein R, Wahrman J, Sperling R, Sperling J. Splicing components are excluded from the transcriptionally inactive XY body in male meiotic nuclei. Mol. Biol. Cell. 1994;5:1341–1352. doi: 10.1091/mbc.5.12.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCarrey JR, Dilworth DD, Sharp RM. Semiquantitative analysis of X-linked gene expression during spermatogenesis in the mouse: ethidium-bromide staining of RT-PCR products. Genet. Anal. Tech. Appl. 1992;9:117–123. doi: 10.1016/1050-3862(92)90051-6. [DOI] [PubMed] [Google Scholar]
- 30.McCarrey JR, Berg WM, Paragioudakis SJ, Zhang PL, Dilworth DD, Arnold BL, Rossi JJ. Differential transcription of Pgk genes during spermatogenesis in the mouse. Dev. Biol. 1992;154:160–168. doi: 10.1016/0012-1606(92)90056-m. [DOI] [PubMed] [Google Scholar]
- 31.Odorisio T, Mahadevaiah SK, McCarrey JR, Burgoyne PS. Transcriptional analysis of the candidate spermatogenesis gene Ube1y and of the closely related Ube1x shows that they are coexpressed in spermatogonia and spermatids but are repressed in pachytene spermatocytes. Dev. Biol. 1996;180:336–343. doi: 10.1006/dbio.1996.0305. [DOI] [PubMed] [Google Scholar]
- 32.Hendriksen PJ, Hoogerbrugge JW, Themmen AP, Koken MH, Hoeijmakers JH, Oostra BA, van der Lende T, Grootegoed JA. Postmeiotic transcription of X and Y chromosomal genes during spermatogenesis in the mouse. Dev. Biol. 1995;170:730–733. doi: 10.1006/dbio.1995.1252. [DOI] [PubMed] [Google Scholar]
- 33.Turner RM, Johnson LR, Haig-Ladewig L, Gerton GL, Moss SB. An X-linked gene encodes a major human sperm fibrous sheath protein, hAKAP82: genomic organization, protein kinase A-RII binding, and distribution of the precursor in the sperm tail. J. Biol. Chem. 1998;273:32135–32141. doi: 10.1074/jbc.273.48.32135. [DOI] [PubMed] [Google Scholar]
- 34.Huynh KD, Lee JT. Inheritance of a pre-inactivated paternal X chromosome in early mouse embryos. Nature. 2003;426:857–862. doi: 10.1038/nature02222. [DOI] [PubMed] [Google Scholar]
- 35.Okamoto I, Otte AP, Allis CD, Reinberg D, Heard E. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science. 2004;303:644–649. doi: 10.1126/science.1092727. [DOI] [PubMed] [Google Scholar]
- 36.Kumari M, Stroud JC, Anji A, McCarrey JR. Differential appearance of DNase I-hypersensitive sites correlates with differential transcription of Pgk genes during spermatogenesis in the mouse. J. Biol. Chem. 1996;271:14390–14397. doi: 10.1074/jbc.271.24.14390. [DOI] [PubMed] [Google Scholar]
- 37.Wilcox SA, Watson JM, Spencer JA, Graves JA. Comparative mapping identifies the fusion point of an ancient mammalian X-autosomal rearrangement. Genomics. 1996;35:66–70. doi: 10.1006/geno.1996.0323. [DOI] [PubMed] [Google Scholar]
- 38.Carrel L, Cottle AA, Goglin KC, Willard HF. A first-generation X-inactivation profile of the human X chromosome. Proc. Natl Acad. Sci. USA. 1999;96:14440–14444. doi: 10.1073/pnas.96.25.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- 40.Shannon M, Handel MA. Expression of the Hprt gene during spermatogenesis: implications for sex-chromosome inactivation. Biol. Reprod. 1993;49:770–778. doi: 10.1095/biolreprod49.4.770. [DOI] [PubMed] [Google Scholar]
- 41.Singer-Sam J, Robinson MO, Bellve AR, Simon MI, Riggs AD. Measurement by quantitative PCR of changes in HPRT, PGK-1, PGK-2, APRT, MTase, and Zfy gene transcripts during mouse spermatogenesis. Nucleic Acids Res. 1990;18:1255–1259. doi: 10.1093/nar/18.5.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kerr SM, Taggart MH, Lee M, Cooke HJ. Ott, a mouse X-linked multigene family expressed specifically during meiosis. Hum. Mol. Genet. 1996;5:1139–1148. doi: 10.1093/hmg/5.8.1139. [DOI] [PubMed] [Google Scholar]
- 43.Khil PP, Smirnova NA, Romanienko PJ, Camerini-Otero RD. The mouse X chromosome is enriched for sex-biased genes not subject to selection by meiotic sex chromosome inactivation. Nat. Genet. 2004;36:642–646. doi: 10.1038/ng1368. [DOI] [PubMed] [Google Scholar]
- 44.Lingenfelter PA, Adler DA, Poslinski D, Thomas S, Elliott RW, Chapman VM, Disteche CM. Escape from X inactivation of Smcx is preceded by silencing during mouse development. Nat. Genet. 1998;18:212–213. doi: 10.1038/ng0398-212. [DOI] [PubMed] [Google Scholar]
- 45.Disteche CM, Filippova GN, Tsuchiya KD. Escape from X inactivation. Cytogenet. Genome Res. 2002;99:36–43. doi: 10.1159/000071572. [DOI] [PubMed] [Google Scholar]
- 46.Carrel L, Hunt PA, Willard HF. Tissue and lineage-specific variation in inactive X chromosome expression of the murine Smcx gene. Hum. Mol. Genet. 1996;5:1361–1366. doi: 10.1093/hmg/5.9.1361. [DOI] [PubMed] [Google Scholar]
- 47.Dal Zotto L, Quaderi NA, Elliott R, Lingerfelter PA, Carrel L, Valsecchi V, Montini E, Yen CH, Chapman V, Kalcheva I, et al. The mouse Mid1 gene: implications for the pathogenesis of Opitz syndrome and the evolution of the mammalian pseudoautosomal region. Hum. Mol. Genet. 1998;7:489–499. doi: 10.1093/hmg/7.3.489. [DOI] [PubMed] [Google Scholar]
- 48.Greenfield A, Carrel L, Pennisi D, Philippe C, Quaderi N, Siggers P, Steiner K, Tam PP, Monaco AP, Willard HF, et al. The UTX gene escapes X inactivation in mice and humans. Hum. Mol. Genet. 1998;7:737–742. doi: 10.1093/hmg/7.4.737. [DOI] [PubMed] [Google Scholar]
- 49.Salido EC, Li XM, Yen PH, Martin N, Mohandas TK, Shapiro LJ. Cloning and expression of the mouse pseudoautosomal steroid sulphatase gene (Sts) Nat. Genet. 1996;13:83–86. doi: 10.1038/ng0596-83. [DOI] [PubMed] [Google Scholar]
- 50.Mroz K, Carrel L, Hunt PA. Germ cell development in the XXY mouse: evidence that X chromosome reactivation is independent of sexual differentiation. Dev. Biol. 1999;207:229–238. doi: 10.1006/dbio.1998.9160. [DOI] [PubMed] [Google Scholar]
- 51.Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- 52.Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 1997;11:156–166. doi: 10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- 53.Turner JM, Aprelikova O, Xu X, Wang R, Kim S, Chandramouli GV, Barrett JC, Burgoyne PS, Deng CX. BRCA1, histone H2AX phosphorylation, and male meiotic sex chromosome inactivation. Curr. Biol. 2004;14:2135–2142. doi: 10.1016/j.cub.2004.11.032. [DOI] [PubMed] [Google Scholar]
- 54.Heard E, Clerc P, Avner P. X-chromosome inactivation in mammals. Annu. Rev. Genet. 1997;31:571–610. doi: 10.1146/annurev.genet.31.1.571. [DOI] [PubMed] [Google Scholar]
- 55.Epstein CJ. The Consequences of Chromosome Imbalance. Cambridge University Press; Cambridge: 1986. [Google Scholar]
- 56.Forejt J. X–Y involvement in male sterility caused by autosome translocations—a hypothesis. In: Crosignani PG, Rubin BL, editors. Genetic Control of Gamete Production and Function. Academic Press; New York: 1982. pp. 135–151. [Google Scholar]
- 57.McCarrey JR. X-chromosome inactivation during spermatogenesis: the original dosage compensation mechanism in mammals? In: Xue G, Xue Y, Xu Z, Holmes R, Hammond GL, Lim HA, editors. Gene Families, Studies of DNA, RNA, Enzymes and Proteins. World Scientific; Singapore: 2001. pp. 59–72. [Google Scholar]
- 58.Cooper DW. Directed genetic change model for X chromosome inactivation in eutherian mammals. Nature. 1971;230:292–294. doi: 10.1038/230292a0. [DOI] [PubMed] [Google Scholar]
- 59.Bellve AR. Purification, culture, and fractionation of spermatogenic cells. Meth. Enzymol. 1993;225:84–113. doi: 10.1016/0076-6879(93)25009-q. [DOI] [PubMed] [Google Scholar]
- 60.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]


