Abstract
Background
In the course of inflammatory bowel diseases (IBD) and acute murine ileitis following peroral Toxoplasma gondii infection, commensal Escherichia coli accumulate at inflamed mucosal sites and aggravate small intestinal immunopathology.
Aim
To unravel the molecular mechanisms by which commensal E coli exacerbate ileitis.
Methods
Ileitis was investigated in mice that lack Toll‐like receptors (TLR) 2 or 4, specific for bacterial lipoproteins (LP) or lipopolysaccharide (LPS), respectively. Gnotobiotic mice, in which any cultivable gut bacteria were eradicated by antibiotic treatment, were used to study the role of LPS in ileitis.
Results
Microbiological analyses revealed that E coli increase in the inflamed ileum. TLR4−/−, but not TLR2−/−, mice displayed reduced mortality and small intestinal immunopathology. Decreased interferon (IFN)‐γ and nitric oxide (NO) levels in the inflamed terminal ileum of TLR4−/− mice indicated that TLR4 signalling aggravates ileitis via local mediator release from immune cells. E coli strains isolated from the inflamed ileum activated cultured mouse macrophages and induced TLR4‐dependent nuclear factor κB activation and NO production in human embryonic kidney 293 cells and in peritoneal macrophages, respectively. Most strikingly, in contrast with wild‐type mice, gnotobiotic TLR4−/− mice were protected from induction of ileitis by treatment with purified E coli lipid A or colonisation with live E coli. Finally, prophylactic treatment with the LPS scavenger polymyxin B ameliorated T gondii‐induced ileitis.
Conclusion
These findings highlight the innate immune system as a key player in T gondii‐induced ileal immunopathology. Treatment with LPS or TLR4 antagonists may represent a novel strategy for prophylaxis and/or therapy of small intestinal inflammation in IBD.
Inflammatory bowel diseases (IBD) such as Crohn's disease (CD) and ulcerative colitis are characterised by chronic intestinal inflammation with acute episodes.1 Commensal gut bacteria aggravate IBD, and the disturbance of mucosal barrier functions results in increased immunoreactivity against bacterial antigens.2,3,4 In patients with active intestinal inflammation, Gram‐negative bacteria (such as Escherichia coli or Bacteroides spp, etc) accumulate at the inflamed tissue sites and potentiate immunopathology by translocation via microlesions and ulcerations.5,6,7 These bacterial groups are also suspected to trigger intestinal inflammation in the course of acute graft‐versus‐host disease after bone marrow transplantation.8,9 Although the role of commensal gut bacteria in colitis has been studied in a number of experimental models,10,11,12 our knowledge on their contribution to ileitis is still limited.13 Recently, we have described that acute murine ileitis induced by peroral infection with the parasite Toxoplasma gondii is accompanied by a rigorous E coli overgrowth in the terminal ileum.14,15 Within 8 days after infection, commensal E coli increase by 8–11 orders of magnitude reaching levels of up to 1011,12 bacteria per gram ileum content.14,15 Most importantly, the presence of commensal E coli in the ileum was found to be essential for the induction and progression of T gondii‐induced ileal immunopathology. Worsening of ileal histopathology as well as levels of interferon (IFN)‐γ and nitric oxide (NO)16 were suppressed by antibiotic treatment.15 Thus, T gondii‐induced ileitis resembles some important microbiological aspects of human IBD, as it displays both ileal bacterial overgrowth and bacteria‐driven Th1‐type inflammation.14 To date, mechanisms by which bacteria aggravate T gondii‐induced ileal inflammation have not been investigated in detail. The high numbers of E coli in the inflamed ileum point towards an important role of bacterial lipoproteins (LP) or lipopolysaccharides (LPS) in the exacerbation of ileal immunopathology.15 LP and LPS are specifically sensed by the innate immune system via Toll‐like receptors (TLRs) 2 and 4, respectively.17 TLR4 has been shown to be of primary importance for elicitation of cytokine responses to whole Gram‐negative bacteria, whereas TLR2 is the main sensor of Gram‐positive pathogens. Both receptors are expressed in the murine intestinal mucosa and, similar to human IBD, their expression is upregulated in the inflamed colon.18,19,20 In experimental colitis, TLR‐mediated sensing of gut bacteria has been suggested to play a role in intestinal homeostasis and to limit bacterial translocation.21,22,23 It was furthermore demonstrated that the TLR/IL‐1‐signalling adapter protein MyD88 is essential for limiting T gondii dissemination and that TLR9 plays an essential role in the development of T gondii‐induced ileitis.24 Further results suggest that TLRs 2 and 4 could interfere with systemic parasite‐related immune responses.25,26,27 To unravel the mechanism by which Gram‐negative commensal gut bacteria trigger inflammation in the small intestine and to determine contributions of gut bacterial products to ileitis, we investigated small intestinal inflammation and gut microflora changes in mice lacking TLRs 2 and 4, which did not show altered T gondii dissemination in mice.25 To further elucidate sensing of bacterial agents via these TLRs, we re‐investigated the role of bacterial LPS and E coli in TLR‐mediated immunopathology by using gnotobiotic mice, lacking any cultivable gut microflora due to quintuple antibiotic treatment.15 Finally, we examined whether ileitis could be prevented or treated with polymyxin B, a known LPS scavenger.28
Materials and methods
Mice, parasites and ileitis induction
C57BL/10ScSn (wild type),29,30,31,32 TLR2−/− (Tularic, San Francisco, California, USA,30 backcrossed six times to C57BL/10ScSn mice31,32), TLR4−/− (C57BL/10ScN, carrying a deletion of the TLR4 gene29,31), TLR2−/−/TLR4−/− (TLR2−/− backcrossed six times to TLR4−/− C57BL/10ScN mice32), C57BL/6 and NMRI mice were bred in the Forschungsinstitut für Experimentelle Medizin (Berlin, Germany). Mice were infected perorally with 100 T gondii cysts (strain ME49) from homogenised brains of intraperitoneally infected NMRI mice in a volume of 0.3 ml phosphate‐buffered saline (PBS, pH 7.4) by gavage, as described previously.15
Generation of gnotobiotic mice
The cultivable gut flora of 8‐week‐old mice was eradicated by quintuple antibiotic treatment as described previously.15 Resulting animals were designated “gnotobiotic” according to the term “gnotos bios” ( = defined life) in the sense that live bacteria are absent in the gut of these mice.
Defined colonisation and LPS treatment of gnotobiotic mice
E coli strain M (an isolate from the ileum of a diseased mouse) was given to gnotobiotic mice by gavage for three consecutive days starting 6 days before T gondii infection as described previously.15 Gnotobiotic mice received 15 μg/ml of sterile purified E coli serotype R515 lipid A (Axxora Life Sciences, Grünberg, Germany) in autoclaved drinking water until day 9 postinfection (pi) starting 6 days before T gondii infection. This highly purified lipid A preparation was tested by the supplier for the absence of any contaminant interacting with innate immunity receptors other than TLR4.
Prophylactic and therapeutic treatment of ileitis with polymyxin B
Mice received 50 mg/kg/day polymyxin B sulphate (EURO OTC Pharma, Kamen, Germany) in PBS perorally by gavage twice daily. To avoid any interference of polymyxin B with T gondii, the antibiotic was withheld 24 h before and after infection. In prophylactic and therapeutic treatment regimens, the administration of antibiotic was started 5 days before and after T gondii infection, respectively.
Sampling procedures, histological scoring, parasite loads and intestinal length
Mice were killed with Halothan® (Eurim‐Pharm, Mülheim, Germany). Samples from liver, spleen, mesenteric lymph nodes (MLN) and terminal ileum were removed under sterile conditions. Bacterial translocation was determined by the cultivation of tissue homogenates. Histological scores and parasite loads were determined as described15 in formalin‐fixed and paraffin‐wax‐embedded tissue sections taken from the terminal ileum. The relative shortening of the small intestine was calculated by dividing the difference in the mean length of small intestines in naive control mice and the respective length of small intestine at day 8 pi multiplied by 100 with the mean length of small intestines in naïve control mice (relative shortening in length = (mean day 0−day 8 pi)×100/mean day 0). Results were expressed as percentage shortage.
Cultural and molecular microflora analysis
Culture‐based and growth‐independent analyses of the gut flora were performed as described.15 For molecular analyses, luminal content of the terminal ileum was suspended in PBS and subjected to PCR amplification of bacterial 16S rRNA genes. The amplicons were analysed by denaturing gradient gel electrophoresis (DGGE) as described previously.15
Determination of IFN‐γ and NO concentrations
Ileum samples (∼1 cm2, 50–100 μg each) were removed and cultured in 24‐well flat‐bottom culture plates (Nunc, Wiesbaden, Germany) containing 500 μl serum‐free RPMI (Rosewell Park Memorial Institute) medium containing penicillin/streptomycin for 18 h at 37°C. IFN‐γ and NO concentrations in supernatants were determined by ELISA (BD Biosciences, Heidelberg, Germany) and by Griess reaction, respectively, as described previously.15
Interaction of isolated E coli strains with TLR4 and TLR2 in vitro
The ability of E coli M to activate TLR4 or TLR2 signalling was determined in transfected human embryonic kidney cells (HEK293) transiently expressing human TLR4 or TLR2 from plasmids.33 In brief, transfection of cell layers displaying 50–80% confluence with plasmids carrying human TLR4 or TLR2 cDNAs was performed with the FuGENE 6 Transfection Reagent (Roche Diagnostics, Mannheim, Germany) following the manufacturer's instructions. Cells were co‐transfected with plasmids containing a luciferase reporter gene under the control of a nuclear factor κB (NFκB)‐activated promoter (120 ng) and the β‐galactosidase gene under the control of the respiratory syncytical virus promoter (40 ng), to correct for differences in transfection efficiency. Transfected cells were incubated with heat‐inactivated bacteria (see below) or purified TLR2 and TLR4 ligands, Pam2Cys (EMC Microcollections, Tübingen, Germany) and LPS (Sigma, Munich, Germany), at concentrations of 100 ng/ml each, respectively. After 20 h, cell extracts were prepared for determination of luciferase and β‐galactosidase activity using the Luciferase and β‐Gal Reporter Gene Assay (Roche, Mannheim, Germany). For heat inactivation, bacteria grown in appropriate liquid media were harvested by centrifugation, washed with PBS and suspended in PBS to an optical density of 1 at 600 nm (1×109 bacteria/ml). These suspensions were incubated at 95°C for 30 min and 100 μl of the suspension was added to transfected cells in 400 μl Dulbecco's modified Eagle's medium culture (PAA Laboratories, Cölbe, Germany). E coli M isolated from inflamed mouse ileum and the E coli reference strain ATCC 29522 were used in experiments.
Interactions of intestinal bacteria with mouse macrophages
E coli strain M, E coli reference strain ATCC29522 and reference strains of Lactobacillus johnsonii (DSM10533) and L acidophilus (DSM20079) were used for coculture experiments. Peritoneal macrophages (PMs) were prepared and cultured in RPMI (PAA Laboratories, Germany) medium with 5% fetal calf serum (Biochrom, Berlin, Germany) as described previously.26 For stimulation assays, 10 μl of the suspensions with heat‐inactivated bacteria (see above) were added to cells in 90 μl RPMI medium with 5% fetal calf serum. After 24 h the supernatants were removed and analysed for NO concentration by the Griess reaction (see above). RAW264.7 macrophages were grown under standard conditions in RPMI (PAA laboratories) with 2% fetal calf serum. Control cells were incubated with Pam2Cys (EMC Microcollections) and LPS (Sigma).
Statistical analysis
Levels of significance were determined using appropriate tests. p Values of ⩽0.05 were considered significant.
Results
TLR4 signalling increases ileal immunopathology
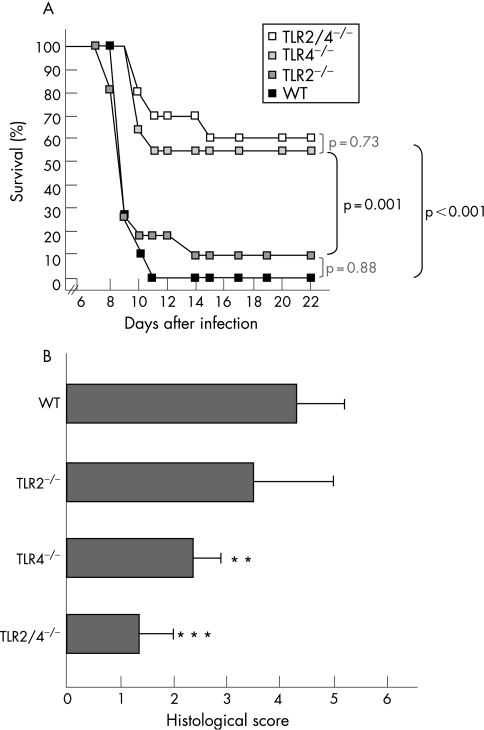
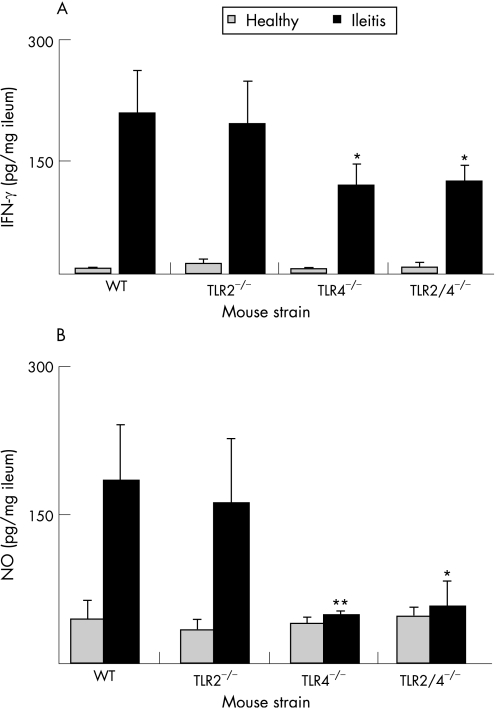
Commensal E coli increase strongly during T gondii‐induced ileitis and aggravate small intestinal inflammation.15 To determine whether bacterial TLR2 or TLR4 ligands may be involved in triggering ileitis, we investigated ileal inflammation in C57BL/10 wild‐type mice and in mice lacking TLR2, TLR4 or both (fig 1) at day 9 T gondii pi. Kinetics of mortality (fig 1A) and histopathology (fig 1B) in C57BL/10 WT mice indicated that the severity of ileitis at day 9 pi was comparable to that observed in C57BL/6 animals at day 8 pi for which the model was established originally.14,15 Most strikingly, the TLR4−/− and TLR2/4−/− animals displayed reduced mortality (fig 1A) and ileal immunopathology (fig 1B) as compared with wild‐type and TLR2−/− mice. Reduced immunopathology in TLR4‐deficient mice was associated with decreased levels of IFN‐γ (fig 2A) and NO (fig 2B), both mediating tissue damage in the ileum.16 Most importantly, parasite dissemination in the ileum was affected by neither TLR2 nor TLR4 deficiency, since tachyzoite numbers in small intestinal sections did not differ between experimental groups (not shown).
Figure 1 Survival rates and ileal immunopathology in TLR2−/−, TLR4−/− or TLR2/4−/− mice. (A) Mortality was monitored daily in T gondii‐infected wild‐type mice (WT, black squares, n = 10) and in mice lacking TLR2 (TLR2−/−, dark grey squares, n = 11), TLR4 (TLR4−/−, light grey squares, n = 11) or both (TLR2/4−/−, white squares, n = 8). Data are from at least two independent experiments. Survival was monitored until day 22 postinfection. Significance levels were determined by Kaplan–Meier analysis. p Values for significant and non‐significant differences (determined by log rank test) are shown in black and light grey, respectively). (B) Histopathology of the terminal ileum in WT mice (n = 5) and in mice lacking TLR2 (TLR2−/−, n = 5), TLR4 (TLR4−/−, n = 5) or both (TLR2/4−/−, n = 5) at day 9 pi SDs and significance levels (as compared with WT animals) determined by Student's t test are indicated (**p<0.01; ***p<0.001).
Figure 2 Proinflammatory mediator concentrations in the ileum of wild‐type TLR2−/−, TLR4−/− and TLR2/4−/− mice. (A) Interferon gamma (IFN‐γ) concentrations in supernatants of ileum cultures from healthy (white bars) and diseased (black bars) wild‐type (WT, n = 5), TLR2−/− (n = 5), TLR4−/− (n = 5) and TLR2/4−/− (n = 3) mice. Ilea from mice with ileitis were analysed at day 9 postinfection SD and significance levels (as compared with WT) determined by Student's t test are indicated (*p<0.05). (B) Nitric oxide (NO) concentrations in supernatants of ileum cultures from healthy (white bars) and diseased (black bars) WT (n = 5), TLR2−/− (n = 5), TLR4−/− (n = 5) and TLR2/4−/− (n = 3). SD and levels of significance (as compared with WT animals with ileitis) determined by Student's t test are indicated (*p<0.05; **p<0.01).
Composition of ileum microflora in mice lacking TLR2 and/or TLR4
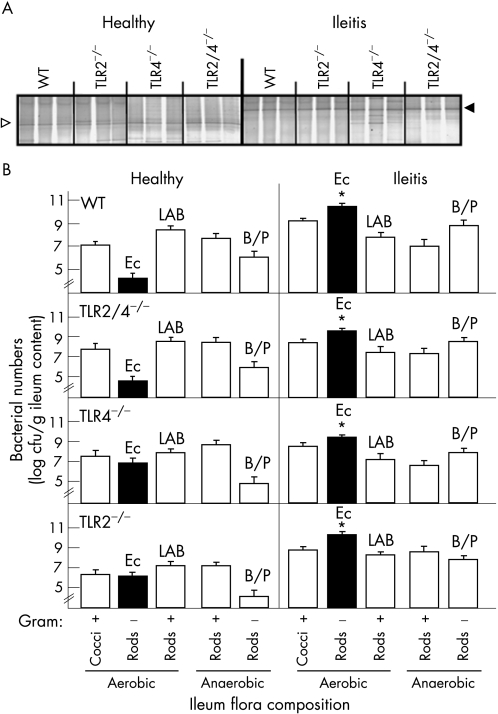
Having shown that E coli overgrowth in the terminal ileum profoundly contributes to the severity of T gondii‐induced ileal inflammation,15 we compared the composition of ileum microflora in healthy and diseased wild‐type mice and in mice lacking TLR2, TLR4 or both. DGGE profiling showed that ileal inflammation was accompanied by a profound shift in flora towards enterobacteria, replacing lactobacilli, usually predominant in the healthy ileum (fig 3A). A detailed culture analysis demonstrated that concentrations of the major gut bacterial groups did not differ between wild‐type and TLR‐deficient mice, irrespective of the degree of inflammation (fig 3B). Wild‐type mice and mice lacking TLR2, TLR4 or both developed a profound E coli overgrowth during ileitis (fig 3B). Genetic fingerprints generated by RAPD‐PCR revealed that the E coli strains accumulating in the inflamed mouse ileum originated from the normal gut flora of healthy mice (data not shown). Gram‐positive rods (including lactobacilli and clostridia) were slightly reduced but did not change significantly during inflammation (fig 3B). Levels of Enterococcus spp (E faecalis, E faecium and E gallinarum) and Bacteroides/Prevotella spp (B ovatus, B merdae, B uniformis, B vulgatus and B thetaiotaomicron, P oralis and P buccae) tended to increase in the inflamed ileum (not significant; fig 3B).
Figure 3 Changes in the composition of gut flora in the course of ileal inflammation. (A) Detection of predominant bacterial communities in the healthy and inflamed ileum by molecular analysis. Genetic fingerprints of the ileal flora were generated by denaturing gradient gel electrophoresis (DGGE) analysis of bacterial 16S rRNA genes amplified by PCR from the total DNA of ileal gut content taken from healthy mice (n = 3) and from mice with ileitis (n = 5) at day 9 postinfection (as indicated above the lanes). Results are shown for wild‐type (WT) mice and for mice lacking TLR2, TLR4 or both as indicated above the lanes. Sequence analysis revealed that DNA bands disappearing (open arrow) and appearing (black arrow) during ileal inflammation refer to Lactobacillus spp and Enterobacteriaceae, respectively. The DGGE profiles are representative for at least three mice per group and experiment. Results were reproduced in two independent experiments. (B) Composition of the gut flora in healthy and inflamed ileum. Bacterial counts (cfu, colony‐forming units) in ileal content from healthy animals (n = 6) and from mice with severe ileitis at day 9 pi (n = 9). E coli, lactic acid bacteria (LAB) (mainly lactobacilli) and Bacteroides/Prevotella spp (B/P) were identified by biochemical analysis and by sequence analysis of the 16S rRNA genes. Results are shown for WT and for mice lacking TLR2, TLR4 or both (TLR2/4−/−). SD and significance levels (as compared to the healthy condition) determined by Student's t test are indicated (*p<0.05).
TLRs 2 and 4 are not required for limiting bacterial translocation
The final stages of T gondii‐induced ileitis are accompanied by bacterial translocation, which is caused by bacterial colonisation of microlesions in the inflamed epithelium.15 As TLR4 has been suggested to be required for limiting bacterial translocation in experimental colitis,23 we determined the concentration of live E coli in MLNs, spleen, liver and cardiac blood taken from wild‐type mice and from TLR2−/−, TLR4−/− and TLR2/4−/− animals with ileitis (at day 9 pi). Commensal E coli were found in MLNs, spleen and liver. However, neither TLR2 nor TLR4 were required for limiting bacterial translocation, as animals carried comparable E coli concentrations in the respective organs (102–106 bacteria/g, not shown). Live bacteria were never detected in cardiac blood.
Murine intestinal E coli strains initiate immune responses via TLR4 in vitro
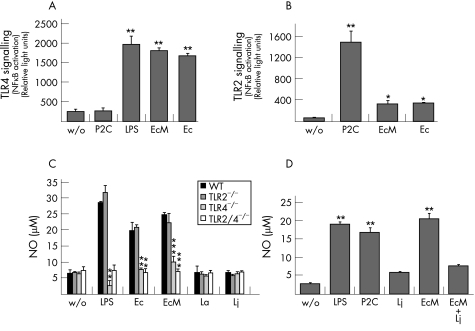
To further investigate whether commensal mouse E coli strains initiate inflammatory responses via TLRs 2 or 4, we analysed the capacity of E coli strain M, an isolate from the inflamed mouse ileum, to activate NFκB in HEK293 cells expressing human TLR2 or TLR4 in vitro. The analysis of TLR‐dependent NFκB activation revealed that E coli M interacts with both TLRs (fig 4). The TLR‐mediated responses were similar to the reference strain E coli ATCC29522. When compared with the natural TLR4 and 2 ligands, LPS and Pam2Cys, respectively (fig 4A and B), both E coli strains mediated strong TLR4‐ (fig 4A) but weak TLR2‐signalling (fig 4B). The inflammatory potential of E coli strains was further examined in PMs isolated from wild‐type, TLR2−/−, TLR4−/− and TLR2/4−/− mice. E coli‐mediated NO production by PMs was strictly TLR4‐dependent. PMs isolated from TLR4−/− and TLR2/4−/− mice did not respond to E coli M (fig 4C). The fact that the reference strains of lactobacilli (L acidophilus and L johnsonii) failed to induce NO release in PMs (fig 4C) strongly suggests that E coli LPS could be involved in triggering inflammation. As the Lactobacillus species dominate the gut flora in healthy mice (fig 3B), we studied the influence of E coli M and L johnsonii on the activation of cultivated RAW264.7 macrophages, responding to both TLR2 and TLR4 ligands (fig 4D). Although E coli M, LPS and Pam2Cys induced strong NO production at similar levels, the response to L johnsonii was at the detection limit (fig 4D). Most interestingly, E coli M‐induced NO release was reduced by heat‐inactivated L johnsonii lysate (fig 4D) in excess (10 times more than E coli M lysate). These results suggest that the composition of gut flora in the healthy ileum provides an environment that downregulates immune responses induced by Gram‐negative bacteria.
Figure 4 In vitro initiation of inflammatory responses via Toll‐like receptors (TLRs) by E coli isolates from the inflamed mouse ileum. (A) Mouse commensal E coli increase nuclear factor κB (NFκB) activity via TLR4. Human embryonic kidney cells (HEK293) transiently transfected with human TLR4 were incubated with purified liposaccharide (LPS), with heat‐inactivated E coli M (EcM, isolated from inflamed mouse ileum) or the E coli reference strain ATCC29522 (Ec29522). Each bar represents mean values of two individual measurements. SD and significance levels (as compared with control cells without stimulation, w/o) determined by Student's t test are indicated (**p<0.01). Similar results were obtained for repeated analyses of four additional E coli strains isolated from the mouse ileum (not shown). (B) E coli‐mediated NFκB activation via TLR2. HEK293 cells transiently transfected with human TLR2 were incubated with the purified TLR2 ligand Pam2Cys (P2C) or with bacterial cell lysates from EcM (isolated from inflamed mouse ileum) or from the E coli reference strain ATCC29522 (Ec29522). Each bar represents mean values of two separate measurements. SDs and significance levels (as compared with control cells without bacterial lysates) determined by Student's t test are indicated (*p<0.05; **p<0.01). Similar results were obtained for repeated analyses of four additional E coli strains isolated from the mouse ileum (not shown). (C) TLR‐dependent activation of mouse PMs by commensal bacteria. PMs isolated from wild‐type mice (WT) and from mice lacking TLR2 (TLR2−/−), TLR4 (TLR4−/−) or both (TLR2/4−/−) (n = 3 each group) were incubated with purified LPS or heat‐inactivated bacteria as indicated (EcM isolated from inflamed mouse ileum; E coli reference strain ATCC29522, Ec29522; L johnsonii, Lj; L acidophilus, La). Cells without LPS treatment or bacterial stimulation served as controls (w/o). Each bar represents mean values of duplicate measurements. SDs and significance levels (if compared to WT) determined by Student's t test are indicated (*p<0.05; **p<0.01; ***p<0.001). Results are representative for two independent experiments. (D) Nitric oxide (NO) release in cultivated RAW264.7 mouse macrophages. Cells were incubated with P2C, LPS or lysates from E coli (Ec), Lj or both (EcM + Lj). Each bar represents mean values of duplicate measurements. SDs and significance levels (if compared with the non‐stimulated control (w/o)) determined by Student's t test are indicated (**p<0.01).
E coli and purified LPS aggravate ileitis via TLR4 in gnotobiotic mice
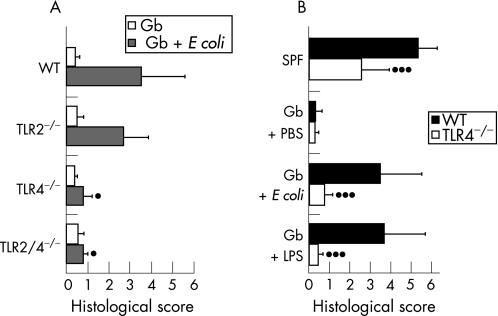
The role of E coli LPS in the exacerbation of ileal inflammation was further investigated in gnotobiotic mice generated by quintuple antibiotic treatment15 as described in the Materials and methods section. T gondii‐infected gnotobiotic mice did not develop ileitis. Most strikingly, mono‐association with E coli M aggravated ileitis in gnotobiotic wild‐type mice and in mice lacking TLR2, whereas TLR4−/− and TLR2/4−/− mice displayed significantly less ileal inflammation, as indicated by lower histopathological scores (fig 5A). E coli counts in the ileum (109–1010 per gram gut content) did not differ significantly within animal groups irrespective of their TLR status. In the ileum of wild‐type mice without T gondii‐infection mono‐association with E coli did not induce immunopathology (not shown). As TLR4 is highly specific for the conserved lipid A component of bacterial LPS, gnotobiotic wild‐type mice and mice lacking TLR4 were treated with purified E coli lipid A (starting 6 days before T gondii infection until day 9 pi). The comparison of histological scores displayed by LPS‐treated mice, gnotobiotic animals mono‐associated with E coli and SPF mice (fig 5B) revealed that in wild‐type mice oral administration of purified lipid A aggravated immunopathology, as did mono‐colonisation of gnotobiotic mice with live E coli (fig 5A, B). However, immunopathology displayed by gnotobiotic LPS‐treated or E coli‐colonised TLR4−/− mice was significantly lower, indicating that TLR4 is essential for LPS‐mediated small intestinal immunopathology.
Figure 5 Induction of ileitis in gnotobiotic mice (Gb) by mono‐association with E coli or treatment with purified lipid A. (A) Immunopathology of the ileum in gnotobiotic mice Gb (white bars) and in Gb mice colonised with E coli (grey bars) at day 9 postinfection. Results for wild‐type (WT) mice (n = 3) and for mice lacking TLR2 (TLR2−/−, n = 3), TLR4 (TLR4−/−, n = 3) or both TLRs (TLR2/4−/−, n = 3) are presented as mean values. SDs and levels of significance (as compared with WT animals and mice lacking TLR2) were determined by Student's t test as indicated (*p<0.05). (B) Influence of oral lipid A administration on ileal immunopathology in Gb WT (black bars; n = 4) and in Gb mice lacking TLR4 (TLR4−/−, white bars, n = 4). Gb mice treated with PBS (WT, n = 22; TLR4−/−, n = 7), colonised with E coli (WT, n = 16; TLR4−/−, n = 7), or SPF animals (WT, n = 22; TLR4−/−, n = 9) served as controls. The severity of ileitis was determined at day 9 postinfection. Results from at least two experiments are presented as mean values. SDs and levels of significance (as compared to WT animals) were determined by Student's t test as indicated (***p<0.001).
Amelioration of ileitis by peroral antibiotic treatment with polymyxin B
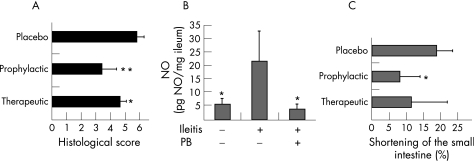
To determine whether TLR4‐mediated sensing of E coli LPS could be used as target for the amelioration of ileitis, we treated T gondii‐infected C57BL/6 mice, displaying slightly increased mortality due to ileitis than C57BL/10 mice, with the LPS antagonist polymyxin B.28 Survival was determined for animals treated with placebo (n = 15) or with polymyxin B starting 5 days before (n = 19) or after (n = 25) T gondii infection. Both regimens ameliorated ileitis, as treated animals displayed higher survival rates than untreated mice. At day 8 pi, when all control mice had died, 63.2% and 48% of the prophylactically and therapeutically treated mice, respectively, were still alive. From the respective treatment groups, 5.3% and 4% of animals survived until day 28 pi (not shown). At day 8 pi, significantly lower immunopathology scores were observed in the ileum of prophylactically treated animals (fig 6A) and this was paralleled by decreased concentrations of NO in the ileum (fig 6B). As ileal inflammation is accompanied by a significant shortening of the upper intestinal tract,15 we determined the lengths of the small intestine (fig 6C). In mice with severe ileitis at day 8 after T gondii infection, the lengths of the small intestines were reduced by 19.0% (4.8%) compared to healthy controls (fig 6C). Animals treated prophylactically with polymyxin B displayed significantly less shortening of the upper intestinal tract. Microbiological analysis in polymyxin B‐treated mice at day 8 pi confirmed that E coli was selectively eradicated (table 1).
Figure 6 Effect of polymyxin B (PB) treatment on ileitis. (A) Reduced histopathology in polymyxin B‐treated C57BL/6 wild‐type mice. The severity of ileitis was determined by histological examination in placebo‐treated mice and in mice receiving polymyxin B either prophylactically or therapeutically (n = 5, each group). The results presented as mean values are representative of at least two independent experiments. SDs and levels of significance determined by Student's t test are indicated (*p<0.05; **p<0.01). (B) Reduced nitric oxide (NO) levels in mice treated with polymyxin B. NO concentrations were determined in supernatants of ileum cultures from animals treated with placebo (n = 5) or prophylactically with polymyxin B (PB, n = 4) as described in the Materials and methods section. Results are presented as mean values. SDs and levels of significance (as compared with placebo‐treated animals) determined by Student's t test are indicated (*p<0.05). (C) Less shortening of the small intestine in animals treated with polymyxin B. The length of the entire small intestine was determined in mice treated with placebo or with polymyxin B either prophylactically or therapeutically (n = 5 each group), as described earlier.15 SDs and levels of significance (as compared to the placebo‐treated animals) determined by Student's t test are indicated (*p<0.05).
Table 1 Bacterial counts (cfu) in the ilea of mice after antibiotic treatment with polymyxin B.
| Treatment group (PB) | Aerobic gram– rods | Aerobic gram+ rods | Aerobic gram+ cocci | Anaerobic gram– rods | Anaerobic gram+ rods |
|---|---|---|---|---|---|
| Without treatment | |||||
| Naive* | 0.7 (1.1) E5 | 5.7 (7.4) E8 | 5.9 (9.8) E6 | <1 E3 | 1.1 (1.0) E9 |
| Ileitis† | 1.2 (1.0) E11 | 5.5 (4.6) E5 | 6.8 (7.3) E8 | 1.1 (2.0) E11 | <1 E3 |
| Prophylactic treatment (PB) ileitis | <1 E3 | 1.1 (1.4) E8 | 2.9 (1.9) E9 | 2.5 (1.4) E9 | <1 E3 |
| Therapeutic treatment (PB) ileitis | <1 E3 | 1.4 (1.7) E7 | 8.5 (9.1) E8 | 2.0 (1.7) E8 | <1 E3 |
PB, polymyxin B.
*Naive mice (uninfected, untreated).
†At day 8 after T gondii infection.
1E3 (below the detection limit of 1 E3 cfu/g ileal content).
Values are mean (SD).
Discussion
Murine small intestinal inflammation following peroral T gondii infection is a suitable model for studying mechanisms by which bacteria aggravate small intestinal inflammation. Similar to human IBD, it displays ileal overgrowth, bacterial translocation and Th1‐type ileal inflammation. The fact that E coli, but not lactobacilli, had a strong inflammatory potential in T gondii‐infected gnotobiotic mice provided strong evidence that LPS or LPs from Gram‐negative bacteria trigger ileitis.15 The finding that TLR4‐, but not TLR2‐signalling potentiated ileitis in the present study underlines the role of E coli LPS in the induction and aggravation of small intestinal inflammation. This was corroborated by treating gnotobiotic animals with purified lipid A, which induced inflammation at levels comparable with E coli colonisation. In addition, E coli strains isolated from the mouse gut initiated strong TLR4‐dependent immune responses in vitro. The fact that at day 9 pi WT and TLR‐deficient mice carried live E coli in MLN, spleen and liver at comparable concentrations indicates that neither TLR2 nor TLR4 influences translocation of bacteria from the small intestine, which results in the direct contact of E coli compounds with immune cells. In the context of decreased IFN‐γ and NO levels observed in the ileum of TLR4‐deficient mice, these results demonstrate that translocating E coli potentiate tissue destruction and intestinal inflammation via the LPS‐mediated stimulation of mediator release from professional phagocytes required for second‐line responses by T‐cells. As a result, specific modulation of the intestinal flora and blockage of LPS‐mediated signalling might open new ways for the treatment or prevention of small intestinal inflammation. Such strategies are strongly supported by the successful treatment of ileitis with the narrow‐spectrum antibiotic polymyxin B (table 1).28 In addition, we found that C57BL/6 mice, impaired in production of the innate immunity adapter protein MyD88, essential for TLR‐signalling, displayed significantly reduced mortality due to ileitis (MMH, SB and UBG, unpublished observations). Furthermore, our results are in line with similar observations in models of experimental colitis. It has been reported earlier that colitis was associated with increased bacterial counts of enterobacteria and that LPS‐hyporesponsive mice develop less colonic inflammation.34
In addition, colonic inflammation was effectively suppressed by TLR4 antagonists.35 The potentiation of intestinal inflammation by innate immune receptors was further underlined by the finding that bacterial DNA triggered colitis via TLR9.36 However, in different models of colitis, the roles of bacterial compounds and innate immunity in aggravating inflammation remain controversial, as neither clinical parameters nor epithelial damage differed between TLR4−/− and WT mice.37 In addition, components of innate immunity were recently suggested to be required for regenerative functions. Mice lacking MyD88, TLR2 or TLR4 developed more severe colitis as a result of defective tissue repair and healing.21,22 Moreover, the role of TLR4‐mediated LPS sensing in human IBD remains unclear. Our results support the view that LPS hyporesponsiveness would rather suppress than aggravate small intestinal inflammation. By contrast, single‐nucleotide polymorphisms (SNPs) within the TLR family exist and may support an involvement of the TLR system in IBD. According to our results, the frequency of functionally relevant TLR4 SNPs should be decreased rather than increased in patients with CD having IBD manifestations in the small intestine. Results from several studies focusing on TLR4 SNPs and IBD are inconsistent,38,39,40,41 as associations of known TLR4 mutations with (1) both CD and UC,39 (2) CD but not with UC40 or (3) UC but not with CD41 were found. Despite these discrepancies, these genetic studies clearly support the role of TLRs in IBD.
In summary, the present study demonstrates that intestinal inflammation is greatly influenced by the composition of the intestinal flora and that variations therein may strongly contribute to ileal inflammation. This opens new potential strategies for prevention of IBD by modulation of the intestinal flora—for example, by antibiotics, prebiotics or by the disruption of mucosal inflammation by anti‐TLR strategies. This assumption is further supported by the finding that ileitis was ameliorated by treatment with the LPS scavenger polymyxin B. However, as polymyxin B treatment targets both E coli (table 1) and the LPS ligand, thus neutralising TLR4 signalling,28 further studies are warranted to explore the beneficial effects of anti‐TLR strategies in small intestinal inflammation. Finally, the aggravation of ileal inflammation in situations of E coli overgrowth furthermore demonstrates the necessity of a detailed analysis of the intestinal flora, in order to determine the contribution of individual bacterial groups or species to disease.
Acknowledgements
AF and MMH equally contributed in this study. AF is supported by a grant from the Sonnenfeld Foundation Berlin, Germany. We thank Jutta Imlau, Michaela Wattrodt, Nadja Goos, Diana Woellner and Gernot Reifenberger for excellent technical assistance. We also thank Dr Jutta Wagner and Dr Matthias Heimesaat for critical discussions. Jana Eckert provided excellent scientific expert advice.
Abbreviations
CD - Crohn's disease
cfu - colony‐forming units
DGGE - denaturing gradient gel electrophoresis
HEK - human embryonic kidney cells
IBD - inflammatory bowel disease
LP - lipoprotein
LPS - lipopolysaccharide
IFN‐γ - interferon gamma
MLN - mesenteric lymph nodes
NFκB - nuclear factor κB
NO - nitric oxide
pi - postinfection
RPMI - Rosewell Park Memorial Institute
PBS - phosphate‐buffered saline
PM - peritoneal macrophage
SNP - single‐nucleotide polymorphisms
TLR - Toll‐like receptor
Footnotes
Funding: This work was supported by grants from the Deutsche Forschungsgemeinschaft to UBG, OL, RRS (KFO104/project 6; SFB633/projects A7, B6) and MF (DFG‐SPP1110/project Fr448/4‐3).
Competing interests: None.
References
- 1.Podolsky D K. Inflammatory bowel disease. N Engl J Med 2002347417–429. [DOI] [PubMed] [Google Scholar]
- 2.Basset C, Holton J. Inflammatory bowel disease: is the intestine a Trojan horse? Sci Prog 20028533–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Y, van Kruiningen H J, West A B.et al Immunocytochemical evidence of Listeria, Escherichia coli, and Streptococcus antigens in Crohn's disease. Gastroenterology 19951081396–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lodes M J, Cong Y, Elson C O.et al Bacterial flagellin is a dominant antigen in Crohn disease. J Clin Invest 20041131296–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Swidsinski A, Ladhoff A, Pernthaler A.et al Mucosal flora in inflammatory bowel disease. Gastroenterology 200212244–54. [DOI] [PubMed] [Google Scholar]
- 6.Swidsinski A, Weber J, Loening‐Baucke V.et al Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J Clin Microbiol 2005433380–3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Darfeuille‐Michaud A, Boudeau J, Bulois P.et al High prevalence of adherent‐invasive Escherichia coli associated with ileal mucosa in Crohn's disease. Gastroenterology 2004127412–421. [DOI] [PubMed] [Google Scholar]
- 8.Beelen D W, Elmaagacli A, Muller K D.et al Influence of intestinal bacterial decontamination using metronidazole and ciprofloxacin or ciprofloxacin alone on the development of acute graft‐versus‐host disease after bone marrow transplantation in patients with hematologic malignancies: final results and long‐term follow‐up of an open‐label prospective randomized trial. Blood 1999933267–3275. [PubMed] [Google Scholar]
- 9.Heidt P J, Vossen J M. Experimental and clinical gnotobiotics: influence of the microflora on graft‐versus‐host disease after allogeneic bone marrow transplantation. J Exp Med 199223161–173. [PubMed] [Google Scholar]
- 10.Sartor R B. Review article: how relevant to human inflammatory bowel disease are current animal models of intestinal inflammation? Aliment Pharmacol Ther 199711(Suppl 3)89–97. [DOI] [PubMed] [Google Scholar]
- 11.Blumberg R S, Saubermann L J, Strober W. Animal models of mucosal inflammation and their relation to human inflammatory bowel disease. Curr Opin Immunol 199911648–656. [DOI] [PubMed] [Google Scholar]
- 12.Strober W, Fuss I J, Blumberg R S. The immunology of mucosal models of inflammation. Annu Rev Immunol 200220495–549. [DOI] [PubMed] [Google Scholar]
- 13.Pizarro T T, Arseneau K O, Bamias G.et al Mouse models for the study of Crohn's disease. Trends Mol Med 20039218–222. [DOI] [PubMed] [Google Scholar]
- 14.Liesenfeld O. Oral infection of C57BL/6 mice with Toxoplasma gondii: a new model of inflammatory bowel disease? J Infect Dis 2002185(Suppl 1)S96–101. [DOI] [PubMed] [Google Scholar]
- 15.Heimesaat M M, Bereswill S, Fischer A.et al Gram‐negative bacteria aggravate murine small intestinal Th1‐type immunopathology following peroral infection with Toxoplasma gondii. J Immunol 20061778785–8795. [DOI] [PubMed] [Google Scholar]
- 16.Liesenfeld O, Kang H, Park D.et al TNF‐α, nitric oxide and IFN‐γ are all critical for development of necrosis in the small intestine and early mortality in genetically susceptible mice infected perorally with Toxoplasma gondii. Parasite Immunol 199921365–376. [DOI] [PubMed] [Google Scholar]
- 17.Kaisho T, Akira S. Pleiotropic function of Toll‐like receptors. Microbes Infect 200461388–1394. [DOI] [PubMed] [Google Scholar]
- 18.Ortega‐Cava C F, Ishihara S, Rumi M A.et al Strategic compartmentalization of Toll‐like receptor 4 in the mouse gut. J Immunol 20031703977–3985. [DOI] [PubMed] [Google Scholar]
- 19.Hart A L, Al‐Hassi H O, Rigby R J.et al Characteristics of intestinal dendritic cells in inflammatory bowel diseases. Gastroenterology 200512950–65. [DOI] [PubMed] [Google Scholar]
- 20.Cario E, Podolsky D K. Differential alteration in intestinal epithelial cell expression of Toll‐like receptor 3 (TLR3) and TLR4 in inflammatory bowel disease. Infect Immun 2000687010–7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rakoff‐Nahoum S, Paglino J, Eslami‐Varzaneh F.et al Recognition of commensal microflora by Toll‐like receptors is required for intestinal homeostasis. Cell 2004118229–241. [DOI] [PubMed] [Google Scholar]
- 22.Araki A, Kanai T, Ishikura T.et al MyD88‐deficient mice develop severe intestinal inflammation in dextran sodium sulfate colitis. J Gastroenterol 20054016–23. [DOI] [PubMed] [Google Scholar]
- 23.Fukata M, Michelsen K S, Eri R.et al Toll‐like receptor‐4 is required for intestinal response to epithelial injury and limiting bacterial translocation in a murine model of acute colitis. Am J Physiol Gastrointest Liver Physiol 2005288G1055–G1065. [DOI] [PubMed] [Google Scholar]
- 24.Minns L A, Menard L C, Foureau D M.et al TLR9 is required for the gut‐associated lymphoid tissue response following oral infection of Toxoplasma gondii. J Immunol 20061767589–7597. [DOI] [PubMed] [Google Scholar]
- 25.Hitziger N, Dellacasa I, Albiger B.et al Dissemination of Toxoplasma gondii to immunoprivileged organs and role of Toll/interleukin‐1 receptor signaling for host resistance assessed by in vivo bioluminescence imaging. Cell Microbiol 20057837–848. [DOI] [PubMed] [Google Scholar]
- 26.Mun H S, Aosai F, Norose K.et al Toll‐like receptor 4 mediates tolerance in macrophages stimulated with Toxoplasma gondii‐derived heat shock protein 70. Infect Immun 2005734634–4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mun H S, Aosai F, Norose K.et al TLR2 as an essential molecule for protective immunity against Toxoplasma gondii infection. Int Immunol 2003151081–1087. [DOI] [PubMed] [Google Scholar]
- 28.Tsuzuki H, Tani T, Ueyama H.et al Lipopolysaccharide: neutralization by polymyxin B shuts down the signaling pathway of nuclear factor kappaB in peripheral blood mononuclear cells, even during activation. J Surg Res 2001100127–134. [DOI] [PubMed] [Google Scholar]
- 29.Poltorak A, Merlin T, Nielsen P J.et al A point mutation in the IL‐12R beta 2 gene underlies the IL‐12 unresponsiveness of Lps‐defective C57BL/10ScCr mice. J Immunol 20011672106–2111. [DOI] [PubMed] [Google Scholar]
- 30.Werts C, Tapping R I, Mathison J C.et al Leptospiral lipopolysaccharide activates cells through a TLR2‐dependent mechanism. Nat Immunol 20012346–352. [DOI] [PubMed] [Google Scholar]
- 31.Poltorak A, He X, Smirnova I.et al Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 19982822085–2088. [DOI] [PubMed] [Google Scholar]
- 32.Lembo A, Kalis C, Kirschning C J.et al Differential contribution of Toll‐like receptors 4 and 2 to the cytokine response to Salmonella enterica serovar Typhimurium and Staphylococcus aureus in mice. Infect Immun 2003716058–6062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Opitz B, Schröder N W, Spreitzer I.et al Toll‐like receptor‐2 mediates Treponema glycolipid and lipoteichoic acid‐induced NF‐kappaB translocation. J Biol Chem 200127622041–22047. [DOI] [PubMed] [Google Scholar]
- 34.Lange S, Delbro D S, Jennische E.et al The role of the Lps gene in experimental ulcerative colitis in mice. APMIS 1996104823–833. [DOI] [PubMed] [Google Scholar]
- 35.Fort M M, Mozaffarian A, Stover A G.et al A synthetic TLR4 antagonist has anti‐inflammatory effects in two murine models of inflammatory bowel disease. J Immunol 20051746416–6423. [DOI] [PubMed] [Google Scholar]
- 36.Obermeier F, Dunger N, Strauch U G.et al CpG motifs of bacterial DNA essentially contribute to the perpetuation of chronic intestinal inflammation. Gastroenterology 2005129913–927. [DOI] [PubMed] [Google Scholar]
- 37.Ohkawara T, Takeda H, Nishihira J.et al Macrophage migration inhibitory factor contributes to the development of acute dextran sulphate sodium‐induced colitis in Toll‐like receptor 4 knockout mice. Clin Exp Immunol 2005141412–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oostenbrug L E, Drenth J P, de Jong D J.et al Association between Toll‐like receptor 4 and inflammatory bowel disease. Inflamm Bowel Dis 200511567–575. [DOI] [PubMed] [Google Scholar]
- 39.Franchimont D, Vermeire S, El Housni H.et al Deficient host–bacteria interactions in inflammatory bowel disease? The Toll‐like receptor (TLR)‐4 Asp299gly polymorphism is associated with Crohn's disease and ulcerative colitis. Gut 200453987–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gazouli M, Mantzaris G, Kotsinas A.et al Association between polymorphisms in the Toll‐like receptor 4, CD14, and CARD15/NOD2 and inflammatory bowel disease in the Greek population. World J Gastroenterol 200511681–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Török H P, Glas J, Tonenchi L.et al Polymorphisms of the lipopolysaccharide‐signaling complex in inflammatory bowel disease: association of a mutation in the Toll‐like receptor 4 gene with ulcerative colitis. Clin Immunol 200411285–91. [DOI] [PubMed] [Google Scholar]