Abstract
Chemokines and chemokine receptors have been posited to have important roles in several common malignancies, including breast and lung cancer. Here, we demonstrate that CXCR7 (RDC1, CCX-CKR2), recently deorphanized as a chemokine receptor that binds chemokines CXCL11 and CXCL12, can regulate these two common malignancies. Using a combination of overexpression and RNA interference, we establish that CXCR7 promotes growth of tumors formed from breast and lung cancer cells and enhances experimental lung metastases in immunodeficient as well as immunocompetent mouse models of cancer. These effects did not depend on expression of the related receptor CXCR4. Furthermore, immunohistochemistry of primary human tumor tissue demonstrates extensive CXCR7 expression in human breast and lung cancers, where it is highly expressed on a majority of tumor-associated blood vessels and malignant cells but not expressed on normal vasculature. In addition, a critical role for CXCR7 in vascular formation and angiogenesis during development is demonstrated by using morpholino-mediated knockdown of CXCR7 in zebrafish. Taken together, these data suggest that CXCR7 has key functions in promoting tumor development and progression.
Keywords: angiogenesis, cancer, chemokine
Certain chemokine receptors have been identified on tumor cells in many common malignancies, including breast and lung cancer, where these receptors have been implicated in multiple steps of tumorigenesis and progression to metastatic disease (reviewed in ref. 1). In particular, the chemokine CXCL12 (SDF-1) was thought to act through its “canonical” receptor, CXCR4, to promote growth of primary tumors and progression to metastatic disease in breast and lung cancer (reviewed in refs. 2 and 3). Myofibroblasts associated with breast cancer, but not those in normal breast tissue, produce CXCL12 and enhance growth of tumors through mechanisms that include proliferation and survival of malignant cells and angiogenesis (4, 5). Specific alleles of CXCL12 are associated with an increased risk of breast cancer (6), and CXCL12 has been shown to transactivate Her2/neu, an established oncogene in breast cancer (7). Furthermore, high levels of CXCL12 are produced in common sites of metastatic breast cancer, suggesting that gradients of this chemokine account for homing of breast cancer cells to specific organs (8).
Similar to the effects of CXCL12 and CXCR4 on breast cancer, chemokine signaling increases proliferation and pro-metastatic functions of other cancer cells. For example, CXCR4 is expressed in primary small-cell lung cancer cells and, on small-cell lung cancer cell lines, promotes migration, activation of integrins, and adhesion of malignant cells to bone marrow stromal cells (9). CXCR4 is also found in human non-small-cell lung cancer, and neutralizing antibodies to CXCL12 limit experimental metastases in mouse models (10). Finally, high levels of expression of CXCR4 correlate with increased metastases in patients with non-small-cell lung cancer (11).
Chemokine receptors other than CXCR4 may also regulate breast and lung cancer. Expression of CCR7 in breast cancer and non-small-cell lung cancer cells correlates with lymph node metastases and poor prognosis (1, 8, 12). Signaling mediated by the chemokine CCL5 and its receptor CCR5 in breast cancer cells activates the tumor suppressor p53, and patients with a nonfunctional allele of CCR5 and bearing wild-type p53 have larger primary breast tumors and reduced disease-free survival (13). CXCR3 also has been identified on some cultured breast cancer cell lines, but the significance of this receptor in breast cancer remains to be determined (14). Furthermore, expression of CXCR2 on cells in the tumor microenvironment appears important for angiogenesis and spontaneous metastases of lung cancer cells in a mouse model (15). Notwithstanding the collective reports, many questions remain regarding the direct mechanism of action of chemokines and their receptors in cancer progression, particularly surrounding the CXCL12/CXCR4 axis in common malignancies.
Recently, we demonstrated that the orphan receptor RDC1 (CCX-CKR2) functions as a chemokine receptor as demonstrated by its ability to bind both CXCL11 and CXCL12 and mediate enhanced growth and adhesion of cells in vitro (16, 17). We have designated this receptor “CXCR7.” We observed that the introduction of CXCR7 into cell lines correlated with an escape from apoptosis, that the receptor could be induced to be expressed on endothelial cells in culture models, and that systemic administration of a small molecule antagonist of CXCR7 correlated with a decrease in tumor size in both xenograft and syngenic in vivo tumor growth studies. More recently, ectopic expression of CXCR7 has been shown to increase cell proliferation of NIH 3T3 in vitro and enhance tumor formation in nude mice in vivo (18). We wished to understand whether CXCR7 could function directly to control tumor development in vivo; to assess whether such control was manifest in an array of tumor types, particularly breast and lung tumors; and to determine CXCR7's potential relevance to human disease by assessing its presence in a variety of primary human tumors. Using RNA interference, stable receptor expression studies, and developmental genetic experiments, we more precisely interrogated CXCR7's role in tumor growth. In animal models, a causal connection between CXCR7 expression and in vivo tumor progression was determined. Furthermore, to establish a link between animal models and human disease, primary malignant and normal biopsy tissues from human patients were surveyed to assess expression of CXCR7 on human tumor cells and tumor vasculature.
Results
Expression of CXCR7 on Breast and Lung Cancer Cell Lines.
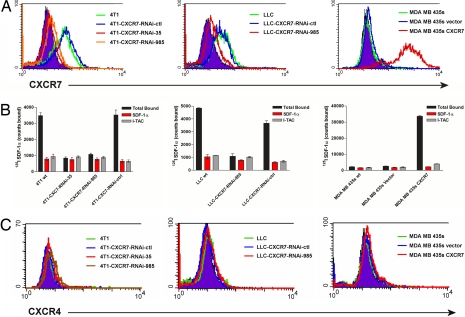
We first analyzed surface expression of CXCR7 in selected lung and breast cancer cell lines, using flow cytometry employing a CXCR7-specific mAb (11G8) and radioligand binding assays that exploited the unique pattern of chemokine binding we defined for this receptor (16, 17): Specifically, both CXCL11 and CXCL12 interact with CXCR7, and each chemokine effectively competes with the other for binding. Detectable levels of CXCR7 were present on the surface of murine breast tumor 4T1 and Lewis lung carcinoma (LLC) cell lines, whereas expression of the receptor on the surface of human breast tumor MDA MB 435s cells was undetectable, in agreement with our previous studies (16) (Fig. 1 A and B).
Fig. 1.
Surface expression of CXCR7 in breast and lung cancer cell lines. (A) Surface expression of CXCR7 on 4T1, LLC, and MDA MB 435s cell lines was quantified by flow cytometry, using CXCR7 antibody 11G8. (B) Binding of radiolabeled CXCL12 to 4T1, LLC, and MDA MB 435s cells was quantified in the presence or absence of nonradiolabeled chemokines and corresponds to levels of CXCR7 measured by flow cytometry. (C) Surface expression of CXCR4 on 4T1, LLC, and MDA MB 435s cell lines was quantified by flow cytometry using CXCR4 Ab 12G5. wt, wild type.
Both murine 4T1 and LLC cell lines are known to form primary and metastatic tumors in mice (19, 20). To quantify the direct contribution of CXCR7 expression in mouse models of primary and metastatic breast and lung cancer, we generated clonal populations of 4T1 and LLC cells harboring stable CXCR7 RNA interference (RNAi) expression vectors. RNAi expression vectors were developed against two independent sites beginning at nucleotide 35 or 985, respectively, in murine CXCR7 to inhibit expression of endogenous CXCR7. We isolated clonal populations of 4T1 cells with reduced expression of this receptor, which are referred to as 4T1-CXCR7-RNAi-35 and -985 cells, respectively, based on the RNAi target site. 4T1-CXCR7-RNAi-35 cells had essentially undetectable levels of CXCR7, whereas levels of the receptor were substantially reduced in 4T1-CXCR7-RNAi-985 cells by both flow cytometry and radioligand binding (Fig. 1 A and B Left). As quantified by branched DNA-based QuantiGene assay, expression of CXCR7 mRNA in each cell line was reduced to ≈10% of that expressed in parental 4T1 cells [supporting information (SI) Fig. 6]. RNAi of CXCR7 did not induce an IFN response, as measured by expression of IFN-induced protein with tetratricopeptide repeats-1 or oligoadenylate synthetase (SI Fig. 6). A similar LLC-based clone was generated with reduced expression of CXCR7 (LLC-CXCR7-RNAi-985) (Fig. 1 A and B Center). Importantly, none of these cell lines expressed mRNA or protein for CXCR4, allowing us to interrogate CXCR7 without potential confounding effects of CXCL12 interacting with CXCR4 (Fig. 1C and SI Fig. 6). In both 4T1 and the LLC cells, we also attempted to reduce expression of CXCR7 with an RNAi molecule targeting CXCR7 at nucleotide 200 (4T1-CXCR7-RNAi-control, LLC-CXCR7-RNAi-control). Neither CXCR7 mRNA nor protein was reduced by this RNA interference molecule, making these cells ideal controls for further studies (Fig. 1 A and B).
To test the effects of increased expression of CXCR7, we generated MDA MB 435s cells that stably overexpressed this receptor (MDA MB 435s CXCR7) and an empty expression vector control cell line (MDA MB 435s vector) (Fig. 1 A and B Right). Similar to parental MDA MB 435s cells, stable MDA MB 435s CXCR7 cells did not express CXCR4, once again allowing us to interrogate CXCR7 role in isolation (Fig. 1C).
CXCR7 Promotes Growth of Breast Tumors in Mice.
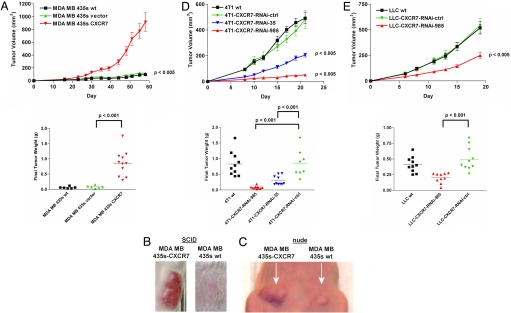
We implanted MDA MB 435s WT, MDA MB 435s vector, or MDA MB 435s CXCR7 cells s.c. into SCID mice to investigate the extent to which CXCR7 affects growth of cell-derived breast tumors (Fig. 2 A and B). MDA MB 435s CXCR7 cells formed larger tumors than WT or MDA MB 435s vector control cells, as shown by changes in tumor volumes over time and tumor weights measured at the end of the experiment (Fig. 2 A and B). Similar results were obtained by mammary fat pad implantation of MDA MB 435s clones into nude mice (Fig. 2C). To verify that breast cancer cells maintained relative differences in expression of CXCR7 in vivo, cells derived from MDA MB 435s WT, MDA MB 435s vector, or MDA MB 435s CXCR7 tumors were analyzed by flow cytometry and radioligand binding assay after resection and tumor dispersion. MDA MB 435s CXCR7 cells maintained high levels of CXCR7 expression in vivo as evidenced by surface staining with mAb 11G8 and by chemokine binding pattern, whereas cells from MDA MB 435s WT or MDA MB 435s vector tumors did not (data not shown). All in vivo-grown tumor cells were CXCR4-negative by flow cytometry with antibody 12G5, confirming that CXCR4 expression was not induced after growth in vivo (data not shown).
Fig. 2.
CXCR7 promotes growth of breast and lung cancer cell-derived tumors. (A–C) Tumor growth was monitored after WT and transfected, and control MDA MB 435s cells were implanted s.c. into the flank (A and B) of scid mice or mammary fat pad of nude mice (C). (D and E) 4T1 (D) and LLC (E) cells were implanted s.c. into the flank of BALB/c or C57Bl6 mice, respectively. Tumor progression was quantified by tumor volume over time and tumor weight at the end of the experiment. Studies were repeated twice with n ≥ 6 mice per group.
To establish that CXCR7 promoted growth of cell-derived tumors in immunocompetent mice, we performed similar experiments with 4T1 RNAi cell lines (Fig. 2D). Upon s.c. implantation of 4T1 cells into BALB/c mice, CXCR7 knockdown lines formed significantly smaller tumors than WT or 4T1-CXCR7-RNAi-control cells (Fig. 2D). Growth of 4T1-CXCR7-RNAi-control cells was identical to WT cells, indicating that the reduced size of the 4T1-CXCR7-RNAi tumors was not due to cell manipulation by RNAi methods. Ex vivo assays of tumor-derived cells demonstrated that the differences in levels of CXCR7 in various 4T1 cell lines were maintained in tumors, further supporting a direct correlation between breast cancer growth and CXCR7 levels in vivo (data not shown). Collectively, data from human MDA MB 435s and mouse 4T1 cells demonstrated that CXCR7 expression dramatically enhanced growth of cell-derived breast tumors.
CXCR7 Promotes Growth of Lung Cancer Cells in Immunocompetent Mice.
To interrogate the effects of CXCR7 on growth of lung cancer cells in vivo, we implanted LLC WT, LLC-CXCR7-RNAi-985, and LLC-control cells s.c. into C57BL/6 mice. Similar to results with breast cancer cells, LLC-CXCR7-RNAi-985 cells formed significantly smaller tumors than LLC WT or LLC-CXCR7-RNAi-control cells, with differences in tumor volumes evident as early as 6–8 days after implantation (Fig. 2E). Final weights of LLC-CXCR7-RNAi-985-derived tumors were significantly less than those of WT or control cells (Fig. 2E). Collectively, these data demonstrate that CXCR7 promotes tumor growth in a mouse model of lung and breast cancers.
CXCR7 Enhances Progression of Experimental Lung Metastases.
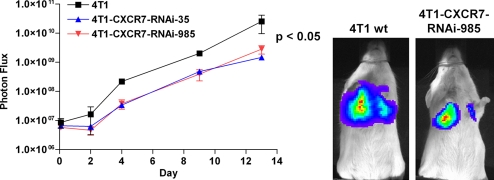
Having established that CXCR7 promotes growth of tumors derived from breast- and lung-transformed cells, we next analyzed the effect of the receptor on experimentally induced lung metastases. After tail vein injection of 4T1 WT or 4T1-CXCR7-RNAi cells, overall growth of 4T1 WT cells in the lung was greater than that of 4T1-CXCR7-RNAi-35 or -985 cells, as measured by bioluminescence imaging of luciferase activity (Fig. 3). By area-under-curve analysis of luciferase activity, total bioluminescence from 4T1 WT cells was significantly greater than that of 4T1-CXCR7-RNAi-35 and -985 cells. Necropsy indicated that the lungs from 4T1 WT cell-injected animals had more tumor mass (load) than 4T1-CXCR7-RNAi-35 or -985 cells (data not shown). In addition, mice injected with 4T1 WT cells had to be killed at earlier time points than RNAi lines because of morbidity (data not shown). We also investigated MDA MB 435s and MDA MB 435s CXCR7 cells in the experimental lung metastasis model. Overexpression of CXCR7 enhanced initial growth of MDA MB 435s cells relative to vector control, similar to effects of the receptor in increased proliferation of 4T1 cells in the lung (data not shown). These data demonstrate that expression of CXCR7 on breast cancer cells enhances the ability of these cells to seed and proliferate in the lung, a common site of metastatic breast cancer.
Fig. 3.
RNA interference of CXCR7 reduces growth of experimental lung metastases. WT and CXCR7 RNAi 4T1 lines were injected intravenously into BALB/c mice via tail vein. Tumor growth in lung was quantified by bioluminescence imaging. The study was repeated twice with n = 4 mice per group.
CXCR7 Expression Marks Various Human Malignancies.
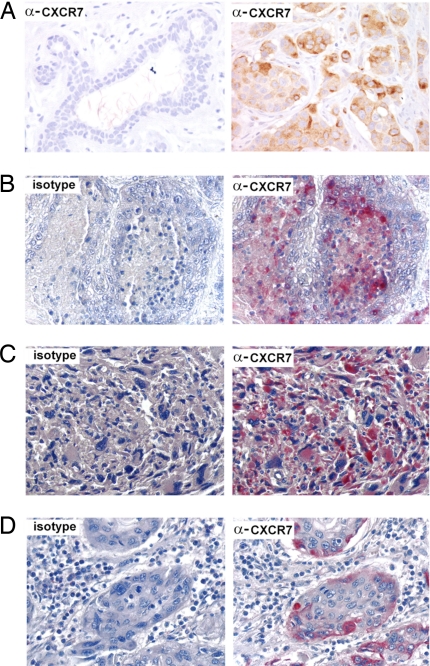
To correlate data from animal models with human malignancy, we analyzed expression of CXCR7 in breast and lung tissue samples from patients by immunohistochemistry, using the CXCR7 specific antibody 11G8 (16). In breast tissue obtained from reduction mammoplasties, CXCR7 was undetectable or present at very low levels in normal breast epithelium (Fig. 4A Left). In contrast, expression of CXCR7 was clearly detected on the transformed cells in >30% of breast tumor sections from different individuals, including those from both in situ and invasive ductal and lobular carcinomas (example Fig. 4A Right). Intensity of staining ranged from 2+ to 4+ on a 4-point semiquantitative scale and did not differ between in situ versus invasive malignancies (data not shown). Isotype control antibody did not react with any samples tested (Fig. 4 B–D Left). In lung cancer sections, CXCR7-specific reactivity was also readily apparent in multiple patients, primarily in squamous cell carcinomas but also occasionally in adenocarcinomas (Fig. 4B Right). Immunodetectable CXCR7 was expressed in soft-tissue tumors, such as rhabdomyosarcoma (Fig. 4C) as well as present in other malignancies, including cervical (Fig. 4D), renal, and esophageal tumors such as rhabdomyosarcoma. Overall, our data demonstrate that CXCR7 is expressed by tumor cells in a substantial number of patients with breast and lung cancer. Furthermore, our analysis of other common human cancers suggests that the receptor is expressed in a broad range of clinical malignancies.
Fig. 4.
CXCR7 expression in human breast, lung, and other cancers. (A) (Left) Undetectable expression of CXCR7 in normal breast tissue from a reduction mammoplasty. (Right) Primary invasive ductal carcinoma of the breast with increased amounts of CXCR7. Isotype control (Left) versus CXCR7 (Right) staining of sections from lung adenocarcinoma (B), Rhabdomyosarcoma (C), or Cervix squamous cell carcinoma (D). Nuclei were counterstained with hematoxylin (blue). (Magnification: ×400.)
CXCR7 Is Highly Expressed on Tumor Vasculature in Model Systems and Human Tumors.
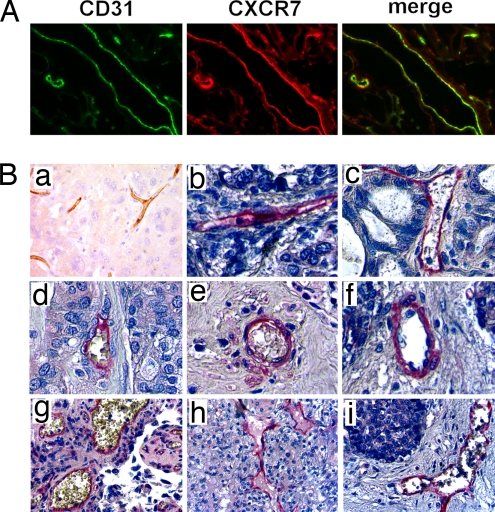
During the course of CXCR7 protein expression analysis by immunohistochemistry in mouse tumor models, we consistently observed staining of tumor vasculature. For example, in the MDA MB 435s xenograft model, extensive colocalization was observed between CXCR7 and the established endothelial marker CD31 (Fig. 5A). This result was observed regardless of CXCR7 expression levels on implanted tumor cells themselves (Fig. 5A). Similar colocalization was also detected in tumor endothelium associated with 4T1 cell-derived tumors in immunocompetent BALB/c mice (data not shown). Collectively, these data demonstrated that CXCR7 was expressed in endothelium of tumor blood vessels in addition to its expression on the transformed cells directly and irrespective of immune status of the animal.
Fig. 5.
CXCR7 is expressed in tumor vasculature. (A) Tumors formed from MDA MB 435s cells were stained with antibodies to the vascular endothelial marker CD31 (green) or CXCR7 (red). Merged image shows colocalization of CXCR7 and CD31 in tumor blood vessels (yellow). (B) Intense CXCR7 staining is observed on the tumor vascular of sections from breast carcinoma (a), lung adenocarcinoma (b), ovary mucinous adenocarcinoma (c), breast adenocarcinoma (d), lung squamous cell carcinoma (e), liver hepatocellular carcinoma (f), bladder transitional cell carcinoma (g), kidney renal cell carcinoma (h), and liver cholangiocarcinoma (i). No staining was observed with isotype control antibody.
Upon analysis of human sections, CXCR7 protein was expressed on blood vessels within human tumors (Fig. 5B). For example, human breast cancer specimens exhibited robust CXCR7 staining in 97% (106 of 109) of the samples, whereas it was undetectable or nearly undetectable in blood vessels associated with normal breast tissues derived from reduction mammoplasties and histologically normal breast tissues in patients with breast cancer (Fig. 4A Left). Similar to the data from primary breast cancers, we identified CXCR7 in vascular endothelium associated with other malignancies. These data suggest that CXCR7 is present on a variety of breast, lung, and other cancers and on a large percentage of tumor vasculature from human malignancies.
Discussion
Early studies examining the role of chemokines in cancer have focused on CXCL12 and its receptor CXCR4, and how they affect overall prognosis for patients. Our data establish that a second receptor for CXCL12, namely CXCR7, heretofore an orphan receptor that we recently characterized as a chemokine receptor (16, 17), promotes the growth of both breast and lung cancer. CXCR7 expression correlated with overall growth of cell-derived breast and lung cancers and experimentally induced lung metastases in mouse models. These results are supported by a recent publication detailing CXCR7's up-regulation in Kaposi's sarcoma-associated herpesvirus-infected endothelial cells and its ability to promote tumor growth of ectopically expressing cells in mice (18). CXCR7 is expressed on malignant cells in a substantial percentage of sections from primary human breast and lung cancers and other common malignancies. In addition, and perhaps most striking, CXCR7 is also detected on tumor-associated blood vessels in nearly all specimens of breast and lung cancer analyzed but not in blood vessels from nonmalignant tissue.
These studies indicate that CXCR7 plays a critical role in tumor growth in murine models of disease. Despite this, questions clearly remain about the mechanism by which CXCR7 mediates these effects. We have noted that CXCR7 expression levels correlate with levels of a number of secreted proteins, most notably matrix metalloproteinase 3, suggesting a role in regulating extra cellular matrix modifying proteins (our unpublished data). In addition, the observation that CXCR7 expressed is on both the tumor vasculature and malignant cells suggests a possible role in chemokine presentation or adhesion in the tumor microenvironment. This hypothesis is supported by adhesion studies (16) demonstrating, in vitro, that CXCR7 expression is regulated by inflammatory cytokines on endothelial cells and promotes maximal cell–cell interactions when expressed concomitantly on both tumor cells and activated endothelium. In addition, preliminary studies using morpholino oligo-mediated knockdown of CXCR7 in zebrafish have suggested a role for this receptor in angiogenesis leading to vascular organization during development (SI Fig. 7 and SI Movies 1–4). Indeed, CXCR7 morphant embryos strongly resemble VEGF-A morphants in the development of enlarged pericardium and major blood vessel deficiencies (21). Further studies are ongoing to determine the specific pathway(s) and mechanisms by which CXCR7 mediates its effect.
The hypothesis that CXCR7 plays a role in human disease is supported not only by animal models but also by the observation that CXCR7 is expressed on malignant but not normal human tissue biopsies. We observed CXCR7 expression on a wide variety of human malignancies, suggesting a wide role for this receptor in tumor promotion.
In summary, our data demonstrate that CXCR7 promotes breast and lung cancer growth through expression in malignant cells and may regulate tumorigenesis in a variety of other common malignancies. The significance of CXCR7 in cancer is also emphasized by the recent observation that a specific small molecule antagonist of this molecule limits growth of tumor in both syngenic and xenograft models (16). Collectively, these data show that therapeutic strategies to inhibit CXCR7 represent a unique opportunity to improve treatment of breast, lung, and possibly other cancers because of the potential to specifically target both malignant cells and tumor blood vessels.
Materials and Methods
Reagents.
Chemokines and anti-CXCR4 antibody 12G5 were purchased from R&D Systems and PeproTech. Anti-mCXCR4 antibody 2B11 was purchased from BD Biosciences. Anti-huCXCR7 antibody 11G8 was generated as reported in ref. 16. mRNA quantification QuantiGene assay kits were purchased from Genospectra. 125I-CXCL12-α and 125I-ITAC were purchased from PerkinElmer. All other reagents were from Sigma.
Cell Lines.
Mouse 4T1 and human MDA MB 435s breast cancer cell lines were purchased from American Type Culture Collection. Mouse LLC lung carcinoma cells were generously provided by C. Kuo (Stanford University, Stanford, CA). 4T1 cells were cultured in RPMI medium with 10% FBS, whereas MDA MB 435s and LLC cells were maintained in DMEM with 10% FBS. Generation of MDA MB 435s stable transfectants is detailed in ref. 16.
Cell Lines with Stable RNA Interference Against CXCR7.
We designed short hairpin RNA molecules targeted against sites beginning at nucleotides 35 and 985 in mouse CXCR7 (NM_007722). The following oligonucleotides were synthesized (Invitrogen): (i) position 35, 5′-caccGCAACTACTCTGACATCAACTcgaaAGTT G A TGTCAGAGTAGTTGC-3′ and 5′-aaaaGCAACTACTCTGACATCAACTttcgAGTTGATGTCAGAGTAGTTGC-3′ and (ii) position 985, 5′-caccGCCTTCATCTTCAAGTACTCGcgaaCGAGTACTTGAAGATGAAGGC-3′ and 5′-aaaaGCCTTCATCTTCAAGTACTCGttcgCGAGTACT T G A AGAT G A AGGC-3′ (lowercase letters represent linkers).
Oligonucleotides were annealed for subcloning into pBLOCK-iT U6 RNAi entry vector and inserted into pBLOCK-iT 3-DEST vectors (Invitrogen) by recombinational cloning. Efficiency of RNA interference against CXCR7 was validated by transient transfection shRNA pBLOCK-iT U6 RNAi entry vectors in 293 cells stably transfected with mouse CXCR7. 4T1 and LLC cells were transfected with shRNA constructs, using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Stable transfectants were selected and cultured in medium containing 1 mg/ml G418.
Lentiviruses.
To construct a lentiviral vector that expresses firefly luciferase and a monomeric orange fluorescent protein (mKO) (Stratech, Cary, NC), we removed the gene for firefly luciferase (FL) from pGL3 basic (Promega, Madison, WI) with NheI and XbaI and blunt end-ligated it into the BamHI site in the FUW lentiviral vector (22) (gift of D. Baltimore, California Institute of Technology, Pasadena, CA). mKO was removed with BamHI and HindIII and blunt end-ligated to the NotI site in pBUDCE4.1 (Invitrogen). The EF-1α promoter from pBUDCE4.1 and mKO were excised with NheI and BglII and blunt end-ligated into the PacI site of FUW. The resulting lentiviral transfer vector (FUW-FL-mKO) uses an EF-1α promoter to express mKO and a ubiquitin promoter to constitutively express FL, respectively.
Lentiviral stocks were prepared as described (20, 22) and used to transduce various cell lines. Stably transduced cell lines were identified by orange fluorescence and sorted by flow cytometry for experiments.
Flow Cytometry.
Cells were stained with mouse monoclonal antibodies against human CXCR7 (11G8, Chem; Centryxo), human CXCR4 (clone 12G5; R&D Systems), or mouse CXCR4 (clone 2B11; BD Biosciences), followed by a goat anti-mouse IgG antibody conjugated to PE (Beckman Coulter). Samples were analyzed on a BD Biosciences FACScan flow cytometer with Cell Quest software.
mRNA Quantification.
The branched-DNA-based QuantiGene assay (Genospectra) was used to quantify mRNAs in various samples after stable transfection of shRNA according to the manufacturer's instructions. Briefly, cells were harvested at the day for FACS and binding analysis. Cell lysates and specific probe sets were incubated in the capture plate and hybridized overnight at 53°C. Amplifier reagents and label probe sets were incubated in the capture plate for 60 min at 53°C after wash. Substrate reagents were added in the end for 30 min, and plates were read on a chemiluminescent plate reader.
Radioligand Binding Assays.
Assays to assess radioligand binding to CXCR7 expressed on various cells were performed as described in ref. 23. Cells were incubated for 3 h at 4°C with 125I-SDF1α (final concentration ≈0.05 nM) or 125I-CXCL11 (final concentration ≈0.01 nM) in buffer (25 mM Hepes/140 mM NaCl/1 mM CaCl2/5 mM MgCl2/0.2% BSA, adjusted to pH 7.1) in the presence of an excess (100 nM) unlabeled chemokine. Reactions were aspirated onto PEI-treated GF/B glass filters, using a cell harvester (Packard). Filters were washed twice (25 mM Hepes/500 mM NaCl/1 mM CaCl2/5 mM MgCl2, adjusted to pH 7.1). Scintillant (MicroScint-10; 35 μl) was added to the filters and counted in a Packard Topcount scintillation counter. Data were analyzed and plotted by using GraphPad software (GraphPad Software).
Animal Experiments.
All animal procedures were approved by ChemoCentryx Institutional Animal Care and Use Committee or the University of Michigan Committee on Use and Care of Animals. Implantation of breast tumor cells into inguinal mammary fat pads was performed as described in ref. 24. A total of 2.5 × 105 4T1 or 4T1-CXCR7-RNAi cells were implanted into 6- to 8-week-old female BALB/c mice (Taconic Farms), and 1 × 106 MDA MB 435s or MDA MB 435s CXCR7 cells were injected into 6- to 8-week-old female Ncr nude mice (Taconic) or SCID mice (Charles River Laboratories). For some experiments, volumes of cell-derived tumors were quantified as the product of caliper measurements in two dimensions and calculated by the equation of width (mm) × width (mm) × length (mm) × 0.52. Animals were killed when tumor volumes reached 1,000 mm3 or animals lost >20% of initial body weight. Tumor weights were taken at termination of each study. In other experiments, growth of viable breast cancer cells in viable tumors was quantified by bioluminescence imaging.
To produce experimental lung metastases, 1 × 106 4T1 breast cancer cells were injected intravenously via a tail vein in 100 μl of sterile 0.9% NaCl. Bioluminescence imaging was used to quantify overall proliferation of metastatic cells.
CXCR7 Knockdown Experiments in Zebrafish.
Morpholino phosphodiamidate oligonucleotides were designed against the 5′UTR region of zebrafish CXCR7: CXCR7mo1, 5′-TCACGTTCACACTCATCTTGGTCCG-3′; CXCR7mo2, 5′-TGTTATCGTCAACACTTCAGTGACC-3′. For the experiments shown here, a mixture of CXCR7mo1 (1 ng/embryo) and CXCR7mo2 (12 ng/embryo) was injected between the one- and eight-cell stages. Similar results were seen upon injection of higher concentrations of each morpholino phosphodiamidate oligonucleotide alone. For microangiography experiments, embryos were anesthetized in tricaine solution and injected with FITC-Dextran (20 mg/ml) into the sinus venosa. Data are representative of multiple experiments: microangiography (n = 67).
Bioluminescence Imaging.
Bioluminescence imaging and data analysis for photon flux produced by primary and metastatic tumors were performed as described in ref. 20. For experimental lung metastases, data for photon flux in the lung were normalized to values obtained 3 h after injection to normalize for variations in actual numbers of cells successfully injected (20).
Immunofluorescence and Immunohistochemistry.
For immunofluorescence microscopy, tumors were frozen in OCT compound and sectioned at 10-μm intervals. We processed specimens for immunofluorescence microscopy as described in ref. 24, using 1 μg/ml final concentrations of the mouse monoclonal antibody against CXCR7 or a rat polyclonal antibody against CD31 present on endothelium of blood vessels (eBioscience). Primary antibodies were detected with Cy3-conjugated donkey anti-mouse-IgG and Cy2-conjugated donkey anti-rat IgG secondary antibodies, respectively (Jackson ImmunoResearch). Images of each fluorophore were merged electronically using commercially available Spotfire software (Spotfire).
Immunohistochemistry was performed on paraffin-embedded breast tissue sections arrayed in a high-density tissue microarray as performed in ref. 25. Tissues were obtained from the surgical pathology files at the University of Michigan. The tissue microarray contained largely consecutive invasive carcinoma tissue samples characterized in ref. 25. In addition, normal breast tissues derived from five different reduction mammoplasties were used. Tumor microarrays were purchased from Imgenex, Zymed/Invitrogen, Cybrdi, US Biomax, Biochain, and Petagen/Telechem. Specimens were stained with 10 μg/ml CXCR7 antibody by using conventional methods; detection was performed with biotinylated rabbit anti-mouse IgG (Jackson ImmunoResearch) coupled with ABC-AP and fuchsin+ kits (Dako). Mayer's hematoxylin (Sigma) was used as a counterstain. An irrelevant mouse IgG1 antibody was used as an isotype control in all cases to demonstrate that staining was specific for CXCR7.
Statistical Analysis.
Area-under-the-curve analysis was done with Prism software (GraphPad). Data are reported as mean values ± SEM and compared with Student's t test. Values ≤ 0.05 were considered significant.
Supplementary Material
Acknowledgments
We thank Linda Ertl, Trageen Baumgart, Kevin Moore, Dan Dairaghi, Nu Lai, Niky Zhao, Ton Dang, Anita Melikian, and J. J. Kim Wright for contributions to this manuscript. This work was supported by National Institutes of Health National Institute of Allergy and Infectious Diseases Grant 1 U19 AI056690 (to ChemoCentryx); the Susan B. Komen Foundation and the Sidney Kimmel Foundation (G.D.L.); and National Institutes of Health Grants P50 CA93990 (to G.D.L. and A.R.), R01CA107469 (to C.G.K.), and R24CA083099 (for the University of Michigan Small Animal Imaging Resource).
Abbreviation
- LLC
Lewis lung carcinoma.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610444104/DC1.
References
- 1.Balkwill F. Nat Rev Cancer. 2004;4:540–550. doi: 10.1038/nrc1388. [DOI] [PubMed] [Google Scholar]
- 2.Hartmann T, Burger M, Burger J. J Biol Regul Homeost Agents. 2004;18:126–130. [PubMed] [Google Scholar]
- 3.Luker K, Luker G. Cancer Lett. 2006:23830–41. doi: 10.1016/j.canlet.2005.06.021. [DOI] [PubMed] [Google Scholar]
- 4.Allinen M, Beroukhim R, Cai L, Brennan C, Lahti-Domenici J, Huang H, Porter D, Hu M, Chin L, Richardson A, et al. Cancer Cell. 2004;6:17–32. doi: 10.1016/j.ccr.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 5.Orimo A, Gupta P, Sgroi D, Arenzana-Seisdedos F, Delaunay T, Naeem R, Carey V, Richardson A, Weinberg R. Cell. 2005;121:335–348. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 6.Razmkhah M, Talei A, Doroudchi M, Khalili-Azad T, Gharderi A. Cancer Lett. 2005;225:261–266. doi: 10.1016/j.canlet.2004.10.039. [DOI] [PubMed] [Google Scholar]
- 7.Cabioglu N, Summy J, Miller C, Parikh N, Sahin A, Tuzlali S, Pumiglia K, Gallick G, Price J. Cancer Res. 2005;65:6493–6497. doi: 10.1158/0008-5472.CAN-04-1303. [DOI] [PubMed] [Google Scholar]
- 8.Muller A, Homey B, Soto H, Ge N, Catron D, Buchanon M, McClanahan T, Murphy E, Yuan W, Wagner S, et al. Nature. 2001;410:50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 9.Burger M, Glodek A, Hartmann T, Schmitt-Graff A, Silberstein L, Fujii N, Kipps T, Burger J. Oncogene. 2003;22:8093–8101. doi: 10.1038/sj.onc.1207097. [DOI] [PubMed] [Google Scholar]
- 10.Phillips R, Burdick M, Lutz M, Belperio J, Keane M, Strieter R. Am J Respir Crit Care Med. 2003;167:1676–1686. doi: 10.1164/rccm.200301-071OC. [DOI] [PubMed] [Google Scholar]
- 11.Su L, Zhang J, Xu H, Wang Y, Chu Y, Liu R, Xiong S. Clin Cancer Res. 2005;11:8273–8280. doi: 10.1158/1078-0432.CCR-05-0537. [DOI] [PubMed] [Google Scholar]
- 12.Takanami I. Int J Cancer. 2003;105:186–189. doi: 10.1002/ijc.11063. [DOI] [PubMed] [Google Scholar]
- 13.Mañes S, Mira E, Colomer R, Montero S, Real LM, Gómez-Moutón C, Jiménez-Baranda S, Garzón A, Lacalle RA, Harshman K, et al. J Exp Med. 2003;198:1381–1389. doi: 10.1084/jem.20030580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldberg-Bittman L, Neumark E, Sagi-Assif O, Azenshtein E, Meshel T, Witz I, Ben-Baruch A. Immunol Lett. 2004;92:171–178. doi: 10.1016/j.imlet.2003.10.020. [DOI] [PubMed] [Google Scholar]
- 15.Keane M, Belperio J, Xue Y, Burdick M, Strieter R. J Immunol. 2004;172:2853–2860. doi: 10.4049/jimmunol.172.5.2853. [DOI] [PubMed] [Google Scholar]
- 16.Burns J, Summers B, Wang Y, Melikian A, Berahovich R, Miao Z, Penfold M, Sunshine M, Littman D, Kuo C, et al. J Exp Med. 2006;203:2201–2213. doi: 10.1084/jem.20052144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Melikian A, Burns J, McMaster B, Schall T, Wright J. WO04058705. Patent Cooperation Treaty Appl. 2004 (7/15/2004) and USA patent publication US 20040170634 (9/2/2004)
- 18.Raggo C, Ruhl R, McAllister S, Koon H, Dezube B, Fruh K, Moses A. Cancer Res. 2005;65:5084–5095. doi: 10.1158/0008-5472.CAN-04-2822. [DOI] [PubMed] [Google Scholar]
- 19.Young M, Duffie G, Lozano Y, Young M, Wright M. Cancer Res. 1990;50:2973–2978. [PubMed] [Google Scholar]
- 20.Smith M, Luker K, Garbow J, Prior J, Jackson E, Piwnica-Worms D, Luker G. Cancer Res. 2004;64:8604–8612. doi: 10.1158/0008-5472.CAN-04-1844. [DOI] [PubMed] [Google Scholar]
- 21.Nasevicius A, Larson J, Ekker S. Yeast. 2000;17:294–301. doi: 10.1002/1097-0061(200012)17:4<294::AID-YEA54>3.0.CO;2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lois C, Hong E, Pease S, Brown E, Baltimore D. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- 23.Dairaghi D, Fan R, McMaster B, Hanley M, Schall T. J Biol Chem. 1999;274:21569–21574. doi: 10.1074/jbc.274.31.21569. [DOI] [PubMed] [Google Scholar]
- 24.Luker G, Pica C, Kumar A, Covey D, Piwnica-Worms D. Biochemistry. 2000;39:7651–7661. doi: 10.1021/bi9928593. [DOI] [PubMed] [Google Scholar]
- 25.Kleer C, Cao Q, Varambally S, Shen R, Ota I, Tomlins S, Ghosh D, Sewalt R, Otte A, Hayes D, et al. Proc Natl Acad Sci USA. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.