Abstract
Infectious mononucleosis (IM) is an immunopathological disease caused by EBV that occurs in young adults and is a risk factor for Hodgkin lymphoma (HL). An association between EBV-positive HL and genetic markers in the HLA class I locus has been identified, indicating that genetic differences in the HLA class I locus may alter disease phenotypes associated with EBV infection. To further determine whether HLA class I alleles may affect development of EBV-associated diseases, we analyzed 2 microsatellite markers and 2 SNPs located near the HLA class I locus in patients with acute IM and in asymptomatic EBV-seropositive and -seronegative individuals. Alleles of both microsatellite markers were significantly associated with development of IM. Specific alleles of the 2 SNPs were also significantly more frequent in patients with IM than in EBV-seronegative individuals. IM patients possessing the associated microsatellite allele had fewer lymphocytes and increased neutrophils relative to IM patients lacking the allele. These patients also displayed higher EBV titers and milder IM symptoms. The results of this study indicate that HLA class I polymorphisms may predispose patients to development of IM upon primary EBV infection, suggesting that genetic variation in T cell responses can influence the nature of primary EBV infection and the level of viral persistence.
Introduction
Infectious mononucleosis (IM) is a benign lymphoproliferative disease that commonly occurs in adolescence or early adulthood and is characterized by fever, lymphadenopathy, and pharyngitis. The symptoms are thought to be immunopathological in nature, resulting from cytokines such as IFN-γ and IL-2 being released from the large numbers of circulating, virus-specific, CD8+ CTLs typically seen in the acute disease (1). In support of this, a correlation between the level of activated CTL and the severity of IM symptoms has recently been reported (2). The symptoms of IM usually resolve within 6 weeks; however, rare chronic and fatal outcomes occur (3), and a recent survey suggests that the number of hospital admissions with severe IM is increasing in Western countries (4).
IM is caused by EBV, a ubiquitous γ herpesvirus that infects over 90% of the world’s population (5). Primary infection generally occurs in early childhood and is usually subclinical; however, in older patients, it manifests as IM in 25%–70% of cases (6, 7). After primary infection, EBV establishes latency in B lymphocytes with virus production in the oropharynx and is spread via saliva. This persistent infection is etiologically linked to a number of lymphoid and epithelial tumors, including Burkitt lymphoma, nasopharyngeal carcinoma (NPC), and Hodgkin lymphoma (HL).
HL is one of the most common tumors in young adults in the West, where its incidence is increasing (8). Approximately 1,500 new cases occur each year in the United Kingdom, and HL now accounts for 1 in 8 of all lymphomas diagnosed. In approximately 25%–50% of Western HL cases, malignant Reed-Sternberg cells carry the EBV genome (9) and express viral antigens. The etiological link between EBV and HL is further substantiated by the finding that a previous history of IM is a significant risk factor for EBV-associated HL with around 1 in 1,000 patients with IM later developing HL (10, 11).
The factors that determine the development of IM as opposed to silent primary EBV infection are unknown. One theory suggests that the size of the initial viral inoculum is a contributory factor in that a large dose, possibly acquired through sexual contact between young adults, would result in a high level of T cell stimulation and hence the immunopathological symptoms of IM (6, 7). This theory is supported by the significant association reported between severe symptoms, high viral load, and increased T cell activation (2). However, in another study, comparable virus loads were found in acute IM patients and subjects with asymptomatic primary EBV infection (12).
Another possibility is that IM development has a genetic basis. Polymorphisms in cytokine genes and their receptors can result in high or low cytokine production, and recently, low IL-10 production has been associated with susceptibility to EBV infection (13, 14) whereas polymorphisms in the IL-1 complex have been related to EBV seronegativity (15).
Genetic differences in the HLA locus are of interest since HLA class 1 alleles may affect the efficiency of viral peptide presentation to T cells, with resultant differences in the effectiveness of the immune response. Clearance of hepatitis C virus, for example, has been associated with HLA-A*03 and B*27 alleles while CTLs expressing different but closely related HLA molecules have shown significant functional differences when targeting identical HIV epitopes (16–18). Recent studies have highlighted HLA class I associations with both EBV-positive HL and NPC. Diepstra et al. identified alleles of 2 microsatellite markers (D6S265: 126-bp allele; and D6S510: 284-bp allele) that are significantly associated with EBV-positive HL and a class III microsatellite, D6S273, that correlated with EBV-negative HL (19). Further work by the same group found SNPs within an 80-kb region, located near the HLA-A and HcG9 genes, which are also associated with EBV-positive HL (20). In another study, a region between the D6S510 and D6S211 markers of the HLA-A locus was associated with the development of NPC (21). Microsatellite markers show the highest degree of linkage disequilibrium with the HLA locus that is located nearest in the genome. In haplotype prediction studies, both the D6S510 and D6S265 microsatellite markers have been shown to have strong linkage disequilibrium with the HLA-A locus, with D6S510 associated with HLA-A1 subtype and D6S265 with HLA-A3 subtype (22).
Due to the well-substantiated association between IM and EBV-positive HL, we speculated that the development of IM during primary EBV infection may also be associated with HLA class I polymorphisms. We therefore analyzed 2 microsatellite markers from the HLA class I region (D6S510 and D6S265) previously associated with EBV-positive HL and 2 SNPs (rs2530388 and rs6457110) situated at either end of the 80-kb region of interest to identify links between IM and allele frequency. One further marker from the class III region (D6S273) associated with EBV-negative HL was analyzed as a control.
Results
HLA class I microsatellite polymorphisms predispose to IM.
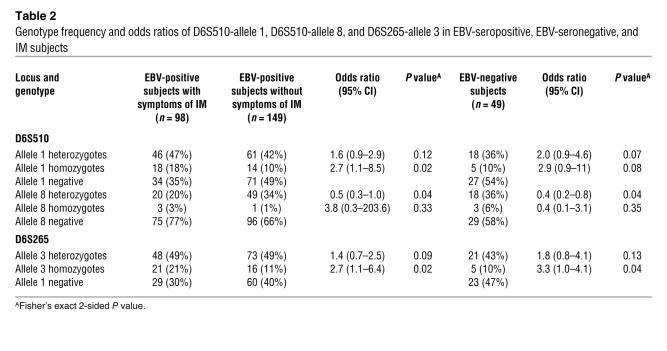
HLA genotyping for the 3 microsatellite markers (D6S265, D6S510, and D6S273) was performed on EBV-seropositive, EBV-seronegative, and IM subject groups. We identified 10 alleles (range 284-306 bp) for the D5S510 marker, 13 alleles (range 122-142 bp) for the D6S265 marker, and 9 alleles (range 128-144 bp) for the D6S273 marker (Table 1). All 3 markers were in Hardy-Weinberg equilibrium for each group. Among EBV-positive subjects, a significant difference between those with symptoms of IM and those without symptoms was observed for allele 1 of marker D6S510 (41.8% and 30.5% respectively, P = 0.01) and allele 3 of D6S265 (45.9% and 35.2%, P = 0.02) (Table 1). Significant differences were also found between EBV-seronegative subjects and IM patients for the same 2 alleles (allele 1: 28.0% versus 41.8%, P = 0.02; allele 3: 31.6% versus 45.9%, P = 0.02) as well as for allele 8 of D6S510 (24.0% versus 13.3%, P = 0.02) (Table 1). Comparison of IM and EBV-seropositive groups revealed that individuals homozygous for allele 1 of the D6S510 marker and allele 3 of the D6S265 marker had odds ratios for development of IM of 2.7 (95% confidence interval [CI]: 1.1-8.5) and 2.7 (95% CI: 1.1-6.4), respectively. The corresponding odds ratios for heterozygotes were 1.6 (95% CI: 0.9-2.9) and 1.4 (95% CI: 0.7-2.5) (Table 2). Odds ratios were also different upon comparison of IM and EBV-seronegative groups for allele 3 (D6S265 marker) with odds ratios of 3.3 (95% CI: 1.0-4.1) for homozygotes and 1.8 (95% CI: 0.8-4.1) for heterozygotes (Table 2). The increased odds ratio in IM subjects homozygous for these alleles indicates a codominant effect. No significant frequency differences were observed between IM patients and either of the other 2 groups (EBV seropositive without IM symptoms or EBV seronegative) for alleles of the control marker D6S273.
Table 1 .
Allele frequency of microsatellite markers D6S510, D6S265, and D6S273 in EBV-seropositive, EBV-seronegative, and IM subjects
Table 2 .
Genotype frequency and odds ratios of D6S510-allele 1, D6S510-allele 8, and D6S265-allele 3 in EBV-seropositive, EBV-seronegative, and IM subjects
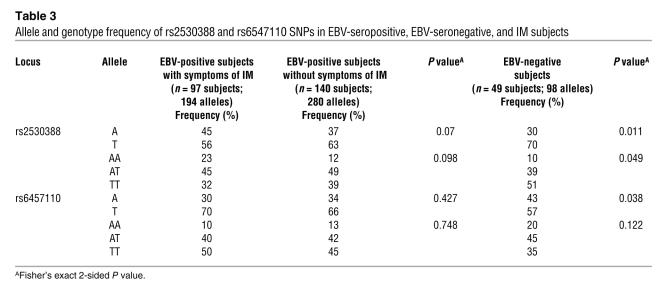
Allele A of SNP rs253088 and allele T of rs6457110 are associated with IM.
Analysis of SNP rs253088 (T/A) and rs6457110 (T/A) was performed on 97 EBV IM, 49 EBV-seronegative, and 140 EBV-seropositive subjects. Both SNPs were in Hardy-Weinberg equilibrium for all 3 subject groups. A significant difference in frequency was found between EBV-seronegative subjects and IM patients for allele A of SNP rs253088 (30% versus 45%; P = 0.011) and for allele T of SNP rs6457110 (57% versus 70%; P = 0.038) (Table 3). Genotype analysis between EBV-seronegative subjects and IM patients was also performed. For the SNP rs253088, the frequency of the A/A genotype was higher in the IM group compared with seronegative subjects (23% versus 10%) while the T/T genotype was lower in the IM group (51% versus 32%; P = 0.049) (Table 3). Comparison between EBV-seropositive subjects and IM patients was of borderline significance for allele A of rs253088 (37% versus 45%; P = 0.07) with no significant difference observed for allele T of rs6457110 (P = 0.42). Further comparison between these groups for the genotype A/A of SNP rs253088 revealed an increase in IM subjects carrying the A/A genotype (23%) compared with EBV-seropositive subjects without IM (12%) although this did not reach statistical significance (P = 0.098) (Table 3). No significant differences were observed between EBV-seronegative and EBV-seropositive individuals for any of the alleles or genotypes.
Table 3 .
Allele and genotype frequency of rs2530388 and rs6547110 SNPs in EBV-seropositive, EBV-seronegative, and IM subjects
Decreased total lymphocyte counts in IM patients positive for allele 1 (D6S510 marker) and allele 3 (D6S265 marker).
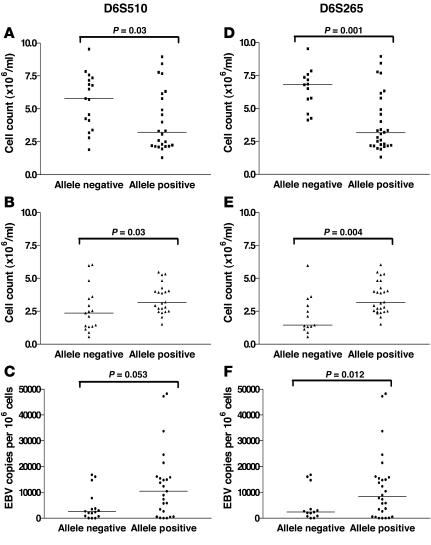
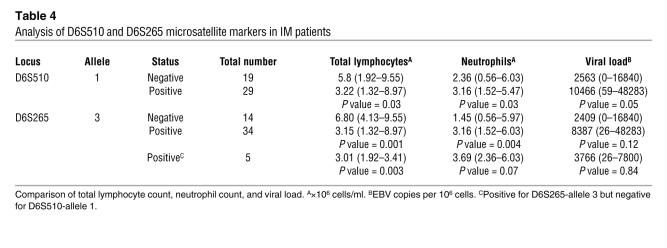
Clinical data and viral load estimations were available for 48 IM patients. For each allele studied, IM patients were grouped according to the presence or absence of each allele and counts of total lymphocytes, neutrophils, and monocytes; EB viral loads were assessed in relation to these groupings. The median total lymphocyte count was significantly lower (P = 0.03) among patients positive for allele 1 of marker D6S510 than among those who were negative for the allele (Figure 1A; Table 4). The association was even stronger (P = 0.001) with respect to allele 3 of marker D6S265. Those positive for the allele had a median total lymphocyte count of 3.15 compared with a count of 6.80 among those who were negative for the allele (Figure 1D; Table 4). The reduction in total lymphocyte counts was accounted for by a reduction in individual median CD3+, CD8+, and CD4+ cell counts in IM cases that were positive for allele 1 of D6S510 and allele 3 of D6S265 compared with those that were negative and were not due to variations in the time of bleeding (allele 1 positive and negative: median 9 days and 10 days, respectively, after onset of symptoms, P = 0.87; allele 3 positive and negative: median 8 days and 12 days, respectively, P = 0.25). All subjects who were positive for allele 1 of marker D6S510 were also positive for allele 3 of marker D6S265, so the apparent association observed for allele 1 of marker D6S510 may have been due to the linkage with the other allele. In contrast, the small number of subjects who were positive for allele 3 of marker D6S265 but negative for allele 1 of D6S510 still had significantly lower (P = 0.003) lymphocyte counts, indicating the importance of the latter allele even in the absence of the former (Table 4). Similar analyses were performed for the SNP alleles and genotypes. Allele A–positive (rs2530388) and allele T–positive (rs6457110) subjects had lower median total lymphocyte counts when compared with allele-negative subjects; however, this did not reach statistical significance (rs2530388, P = 0.31; rs6457110, P = 0.67). Individuals homozygous for each of these alleles also displayed lower median total lymphocyte counts when genotypes were compared.
Figure 1. Analysis of microsatellite markers with clinical features in IM subjects.
Panels A–C represent subjects positive and negative for the microsatellite marker D6S510-allele 1; panels D–F represent subjects positive and negative for microsatellite marker D6S265-allele 3. Total lymphocyte counts (× 106/ml) (A and D); neutrophil counts (× 106/ml) (B and E); viral load (EBV genome copies per 106 cells) (C and F).
Table 4 .
Analysis of D6S510 and D6S265 microsatellite markers in IM patients
Altered neutrophil counts in IM patients positive for allele 1 (D6S510 marker) and allele 3 (D6S265 marker).
Although within the normal range for the Coulter counter, significantly higher neutrophil counts were observed among subjects positive for allele 1 of D6S510 compared with those who were negative for the allele (P = 0.03). However, as with total lymphocyte counts, a much stronger association (P = 0.004) was observed for allele 3 of D6S265, in which the median count among those who were negative for the allele was below the normal range for the Coulter counter (Figure 1, B and E; Table 4). Again, the much stronger association for the latter allele suggests its relative importance. Similar analyses for the 2 SNPs revealed no significant differences.
No differences were observed in monocyte counts between any of the groups for the alleles examined. Allele 8 of the D6S510 marker was similarly analyzed with no significant differences observed.
Increased EB viral load in IM patients positive for allele 1 (D6S510 marker) and allele 3 (D6S265 marker).
There was a suggestion that viral loads were raised among subjects positive for allele 1 of D6S510 compared with those negative for the allele, but the association was of borderline significance (P = 0.053). Similarly, the viral load was raised among those who were positive for allele 3 of marker D6S265, but the association was not significant (P = 0.12) (Figure 1, C and F; Table 4). This trend was also observed with allele A (rs2530388) and allele T (rs6457110) and for the homozygous genotype of both alleles.
IM patients positive for allele 1 (D6S510 marker) and allele 3 (D6S265) marker present with milder symptoms.
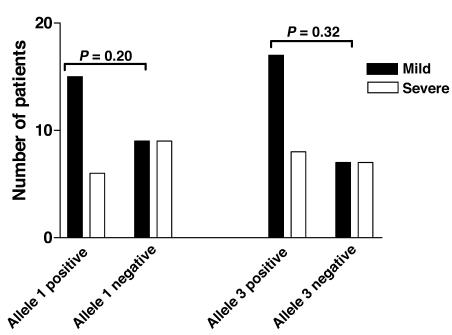
We analyzed the IM groupings in relation to the severity of clinical symptoms. Data on sore throats were available for 39 of the patients and were assessed as mild (able to swallow a normal diet) (24 subjects) or severe (unable to swallow a normal diet) (15 subjects). Among the 21 patients positive for allele 1 of the D6S510 marker, 6 (71%) reported that their sore throats were severe compared with 9 (50%) of the 18 cases negative for the allele (P = 0.20). Of the 25 patients positive for allele 3 of the D6S265 marker, 8 (32%) reported severe sore throats compared with 7 (50%) of the 14 cases negative for the allele (P = 0.32) (Figure 2).
Figure 2. Comparison of severe and mild sore throats in IM subjects.
IM subjects positive and negative for marker D6S510-allele 1 and marker D6S265-allele 3 were compared for differences in presentation of sore throats. Mild, black bars; severe, white bars. P values for comparison of mild symptoms between allele-positive and -negative subjects are indicated.
Discussion
In this study, we have assessed 2 HLA class I and 1 HLA class III microsatellite markers for evidence of an association with acute IM. Both HLA class I markers were found to be significantly associated with the development of IM, with allele 1 (284 bp) of marker D6S510 and allele 3 (126 bp) of marker D6S265 showing the strongest effects. Furthermore, a codominant effect was observed in IM subjects homozygous for these alleles. An unexpected observation was that the frequency of marker D6S510-allele 8 (302 bp) was significantly reduced in the IM group when compared with the seronegative group, perhaps indicating a role for this allele in protection from IM. However, the lack of any significant difference in allele frequency between the EBV-seropositive subjects without symptoms of IM and the seronegative subjects for all 3 alleles is an indication that these alleles do not confer protection from subclinical EBV infection. No association was observed between the subject groups and the control HLA class III marker D6S273. These findings are similar to recent reports associating the HLA class I region, in particular the D6S510 (284-bp allele) and D6S265 (126-bp allele) microsatellite markers, with EBV-positive HL (19, 20) and provide further evidence for an etiological link between IM and EBV-positive HL.
Also in line with recent reports (20) showing an association between several SNPs from the HLA class I region and EBV-positive HL is our finding that allele A of SNP rs2530388 and allele T of rs6457110 are more frequent in the IM group compared with EBV-seronegative individuals. The A/A genotype for SNP rs2530388 is also more frequent in IM compared with our EBV-seronegative group. The results suggest a genetic link between IM- and EBV-associated HL although the association between IM and the markers analyzed in this study is slightly weaker than the reported association with EBV-positive HL.
The markers we have identified as conferring a predisposition to IM are known to be in linkage disequilibrium with the HLA-A locus (22). A number of studies have shown that specific EBV peptides are presented in the context of specific HLA class I alleles (23, 24), and this association of HLA class I markers with IM suggests that the genetic makeup of an individual’s HLA class I locus dictates the efficiency of viral peptide presentation and the recruitment of T cells during the immune response to primary EBV infection. Thus, as IM is assumed to be immunopathological in nature, caused by T cell–derived cytokines, possession of these markers might be expected to result in an exuberant T cell response and severe IM. However, our results show that in individuals with these alleles, lymphocyte counts are significantly lower than in those without. Allele-positive patients also had higher viral loads than the allele-negative group. This increase was not accounted for by a higher proportion of B cells within the lymphocyte population in allele-positive subjects (allele 1 and 3 positive: 5%; allele 1 and 3 negative: 7%), suggesting that perhaps the virus-specific T cell response itself was insufficient to control the virus infection effectively. In addition, there is a suggestion that IM patients with the alleles had milder IM symptoms than those without, and this could result from low cytokine production by their virus-specific T cells. Thus, possession of either of the alleles we have identified appears to result in a weakened or aberrant immune response to EBV that predisposes to a mild form of IM.
The significantly higher neutrophil counts observed in IM patients positive for the alleles are more difficult to explain since there is no known direct link between the HLA locus and neutrophil function. As part of the innate immune response, neutrophils, together with natural killer cells and monocytes, form the main effector cells early in primary EBV infection. By releasing cytokines and chemokines, they attract other immune cells to the site of infection and are thereafter depleted, mainly by apoptotic cell death. Recent reports suggest that EBV can directly infect neutrophils and alter their cytokine production (25). However, it seems more likely that the low neutrophil counts observed in allele-negative subjects are a direct result of neutrophil apoptosis following recruitment of T cells to sites of infection. In a recent case control study on IM, the median neutrophil count in IM subjects at time of diagnosis was significantly lower than that of age-matched controls, with 26% of IM cases having neutrophil counts below the lower limit of the normal range (P = 0.015, K. Macsween, unpublished observations). Thus, finding a significantly higher median neutrophil count (although still within the normal range) in allele-positive subjects in the present study may just reflect the milder IM symptoms in these subjects.
The classic Reed-Sternberg cells of HL represent postgerminal center B cells, which contain nonfunctional immunoglobulin genes (26, 27). In EBV-positive HL, these abnormal cells are thought to have been rescued from apoptosis in germinal centers by the expression of EBV latent membrane proteins 1 and 2, which provide crucial survival signals (28). We postulate that the HLA class I markers D6S510 and D6S265 predispose to EBV-associated HL by inducing a suboptimal T cell immune response to the virus during IM, which in turn would result in poor virus clearance, allowing a high level of virus persistence in B cells. Elevated levels of viral persistence may increase the chance of EBV infection and survival of abnormal B cells with malignant potential, providing an explanation for the link between HL and IM.
In summary, we have demonstrated an association between polymorphisms in the HLA class I region and IM and show evidence suggestive of reduced T cell control of EBV during primary infection in allele-positive cases.
Methods
Study population.
EBV-seropositive and -seronegative individuals were recruited as part of an epidemiological study carried out at Edinburgh University. The study was approved by the Lothian Research Ethics Committee, and all participants provided written signed consent. Details for enrollment and serotyping have been published elsewhere (6, 7). In brief, all students enrolling at the university during 1999 and 2000 were approached to take part in the study, and upon recruitment, provided a blood sample for EBV serology. EBV-seronegative individuals were monitored for development of IM and followed clinically until symptoms resolved (2, 29). Upon diagnosis (median 10 days after onset of symptoms: range 3–42 days), IM patients were examined and asked to provide a blood sample for full blood count, which was carried out on a Coulter counter (Beckman Coulter). Those students seronegative at enrollment who did not report symptoms of IM were asked to return for further testing upon exit from university. Students who remained EBV negative formed the seronegative group while students who tested as EBV positive were regarded as asymptomatic seroconverters and joined the EBV-seropositive group. A total of 150 EBV-seropositive asymptomatic seroconverters, 50 EBV-seronegative subjects, and 98 IM patients were available for subsequent analysis. There was no variation in ethnicity between groups: the majority of subjects were of white European descent (EBV seropositive: 96.5%; EBV seronegative: 94%; IM: 92%); therefore, infection with different EBV types was assumed to be minimal.
HLA microsatellite genotyping.
PBMCs were separated from whole blood by density gradient centrifugation, then washed and stored at –70°C. DNA was extracted from PBMCs using the Easy DNA Extraction Kit (Invitrogen) per the manufacturer’s instructions and stored at –20°C until required. Three microsatellite markers, 2 from the HLA class I (D6S265 and D6S510) and 1 from the HLA class III regions (D6S273), were amplified using PCR performed in a total volume of 10 μl containing 25 ng of DNA (19). The reaction mix contained 1× GoTaq Flexi buffer (pH 8.5), 2.5 mM magnesium chloride, 200 μM dNTP (GE Healthcare), 25 μM of forward and reverse primer, and 0.5 units of GoTaq Flexi polymerase (Promega). Each primer pair had 1 primer 5′ labeled with fluorochrome 6-FAM (Table 5). The cycling conditions consisted of an initial denaturation at 94°C for 5 minutes followed by 35 cycles of 94°C for 30 seconds, 55°C or 62°C for 30 seconds, 72°C for 45 seconds, and a final extension at 72°C for 5 minutes. One microliter of PCR product was diluted in 50 μl of nuclease-free water and then further diluted 1 in 10 with Hi-Di formamide (containing GeneScan 500LIZ size standard, ABI; Applied Biosystems) before electrophoresis in an ABI 3730 (dye set 5) automated sequencer. ABI GeneMapper software (version 3.7; Applied Biosystems) was used to analyze the data.
Table 5 .
Primer and PCR information for microsatellite markers
SNP analysis.
SNP analysis was carried out using TaqMan SNP Genotyping Assays from Applied Biosystems. Reactions were performed in a final volume of 25 μl containing 1× TaqMan Universal Master Mix with no AmpEraseUNG, 900 nM of each primer (Applied Biosystems), 200 nM of probes labeled with either FAM or VIC, and 20 ng of extracted DNA. Thermal cycling (2 minutes at 50°C, 10 minutes at 95°C followed by 40 cycles of 15 seconds at 95°C and 1 minute at 60°C) and allelic discrimination were performed on an ABI 7500 Real Time PCR System (Applied Biosystems).
Virus load quantification.
The EBV-DNA load was determined by quantitative competitive PCR as described by Stevens et al (30). The amplification reaction contained 50 mM KCL, 1.5 mM MgCl, 10 mM Tris-HCl (pH 8.5), 200 μM dNTPs, 25 pmol of each primer (5′ primer labeled with biotin), and 1 U of Taq polymerase. Cycling conditions were 4 minutes at 95°C; 40 cycles at 95°C, 55°C, and 72°C for 1 minute each; and 3 minutes at 72°C. Products were captured on a streptavidin-coated plate and probed with digoxigenin-labeled wild-type and internal standard probes. Optical density was measured and used to calculate the number of copies.
Statistics.
Microsatellite markers and SNPs were tested for Hardy-Weinberg disequilibrium by comparing the observed allele frequency with the expected frequency if equilibrium applied. Classical association analysis was conducted to compare allele frequencies between the groups (EBV-positive with symptoms of IM, EBV-positive without symptoms of IM, EBV-negative) of subjects and tested using the Fisher’s exact test (this was not adjusted for multiple testing). Clinical characteristics (total lymphocyte count, neutrophils, and viral load) were compared across groups using the Mann-Whitney U test. The Fisher’s exact test was used to compare the severity of symptoms (as measured by sore throat) among IM cases who were positive compared with those who were negative for particular alleles.
Acknowledgments
We thank the staff from the Edinburgh University Health Service, Edinburgh, for recruitment of study subjects and the students of Edinburgh University for participating in the study. This study was supported by the UK Medical Research Council (MRC grant G 9826804).
Footnotes
Nonstandard abbreviations used: CI, confidence interval; HL, Hodgkin lymphoma; IM, infectious mononucleosis; NPC, nasopharyngeal carcinoma.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 117:3042–3048 (2007). doi:10.1172/JCI32377
See the related Commentary beginning on page 2756.
References
- 1.Callan M.F.C., Steven N., Krausa P. Large clonal expansions of CD8+ T cells in acute infectious mononucleosis. Nat. Med. 1996;2:906–911. doi: 10.1038/nm0896-906. [DOI] [PubMed] [Google Scholar]
- 2.Williams H., et al. Analysis of immune activation and clinical events in acute infectious mononucleosis. J. Infect. Dis. 2004;190:63–71. doi: 10.1086/421276. [DOI] [PubMed] [Google Scholar]
- 3.Cohen J.I. Epstein-Barr virus infection. N. Engl. J. Med. 2006;343:481–492. doi: 10.1056/NEJM200008173430707. [DOI] [PubMed] [Google Scholar]
- 4.Tattevin P., et al. Increasing incidence of severe Epstein-Barr virus-related infectious mononucleosis: surveillance study. J. Clin. Microbiol. 2006;44:1873–1874. doi: 10.1128/JCM.44.5.1873-1874.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henle G., Henle W. Immunofluorescence in cells derived from Burkitt’s lymphoma. J. Bacteriol. 1966;91:1248–1256. doi: 10.1128/jb.91.3.1248-1256.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crawford D.H., et al. A cohort study among university students: identification of risk factors for Epstein-Barr virus seroconversion and infectious mononucleosis. Clin. Infect. Dis. 2006;43:276–282. doi: 10.1086/505400. [DOI] [PubMed] [Google Scholar]
- 7.Crawford D.H., et al. Sexual history and Epstein-Barr virus infection. J. Infect. Dis. 2002;186:731–736. doi: 10.1086/342596. [DOI] [PubMed] [Google Scholar]
- 8.Swerdlow A.J. Epidemiology of Hodgkin’s disease and non-Hodgkin’s lymphoma. Eur. Nucl. Med. Mol. Imaging. 2003;30(Suppl. 1):S3–S12. doi: 10.1007/s00259-003-1154-9. [DOI] [PubMed] [Google Scholar]
- 9.Andersson J. Epstein-Barr virus and Hodgkin’s lymphoma. Herpes. 2006;13:12–16. [PubMed] [Google Scholar]
- 10.Jarrett R.F., et al. Impact of tumor Epstein-Barr virus status on presenting features and outcome in age-defined subgroups of patients with classic Hodgkin lymphoma: a population-based study. Blood. 2005;106:2444–2451. doi: 10.1182/blood-2004-09-3759. [DOI] [PubMed] [Google Scholar]
- 11.Hjalgrim H., et al. Characteristics of Hodgkin’s lymphoma after infectious mononucleosis. N. Engl. J. Med. 2003;349:1324–1332. doi: 10.1056/NEJMoa023141. [DOI] [PubMed] [Google Scholar]
- 12.Silins S.L., et al. Asymptomatic primary Epstein-Barr virus infection occurs in the absence of blood T-cell repertoire perturbations despite high levels of systemic viral load. Blood. 2001;98:3739–3744. doi: 10.1182/blood.v98.13.3739. [DOI] [PubMed] [Google Scholar]
- 13.Helminen M., Lahdenpohja N., Hurme M. Polymorphism of the Interleukin-10 gene is associated with susceptibility to Epstein-Barr virus infection. J. Infect. Dis. 1999;180:496–499. doi: 10.1086/314883. [DOI] [PubMed] [Google Scholar]
- 14.Wu M.S., et al. Tumour necrosis factor-alpha and interleukin-10 promoter polymorphisms in Epstein-Barr virus associated gastric carcinoma. J. Infect. Dis. 2002;185:106–109. doi: 10.1086/324771. [DOI] [PubMed] [Google Scholar]
- 15.Hurme M., Helminen M. Polymorphism of the IL-1 gene complex in Epstein-Barr virus seronegative and seropositive adult blood donors. Scand. J. Immunol. 1998;48:219–222. doi: 10.1046/j.1365-3083.1998.00406.x. [DOI] [PubMed] [Google Scholar]
- 16.McKiernan S.M., et al. Distinct MHC Class I and II alleles are associated with hepatitis C viral clearance, originating from a single source. Hepatology. 2004;40:108–114. doi: 10.1002/hep.20261. [DOI] [PubMed] [Google Scholar]
- 17.Frahm N., et al. HLA-B63 presents HLA-B57/B58-restricted cytotxic T-lymphocyte epitopes and is associated with low human immunodeficiency virus load. J. Virol. 2005;79:10218–10225. doi: 10.1128/JVI.79.16.10218-10225.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leslie A., et al. Differential selection pressure exerted on HIV by CTL targeting identical epitopes but restricted by distinct HLA alleles from the same HLA supertype. J. Immunol. 2006;177:4699–4708. doi: 10.4049/jimmunol.177.7.4699. [DOI] [PubMed] [Google Scholar]
- 19.Diepstra A., et al. Association with HLA class I in Epstein-Barr-virus-positive and with HLA class III in Epstein-Barr-virus-negative Hodgkin’s lymphoma. Lancet. 2005;365:2216–2224. doi: 10.1016/S0140-6736(05)66780-3. [DOI] [PubMed] [Google Scholar]
- 20.Niens M., et al. The human leukocyte antigen class I region is associated with EBV-positive Hodgkin’s lymphoma: HLA-A and HLA complex group 9 are putative candidate genes. Cancer Epidemiol. Biomarkers Prev. 2006;15:2280–2284. doi: 10.1158/1055-9965.EPI-06-0476. [DOI] [PubMed] [Google Scholar]
- 21.Lu C.C., et al. Nasopharyngeal carcinoma-susceptibility locus is localised to a 132 kb segment containing HLA-A using high-resolution microsatellite mapping. Int. J. Cancer. 2005;115:742–746. doi: 10.1002/ijc.20946. [DOI] [PubMed] [Google Scholar]
- 22.Malkki M., Single R., Carrington M., Thomson G., Petersdorf E. MHC microsatellite diversity and linkage disequilibrium among common HLA-A, HLA-B, DRB1 haplotypes: implications for unrelated donor hematopoietic transplantation and disease association studies. Tissue Antigens. 2005;66:114–124. doi: 10.1111/j.1399-0039.2005.00453.x. [DOI] [PubMed] [Google Scholar]
- 23.Rickinson A.B., Moss D.J. Human cytotoxic T lymphocyte responses to Epstein-Barr virus infection. Annu. Rev. Immunol. 1997;15:405–431. doi: 10.1146/annurev.immunol.15.1.405. [DOI] [PubMed] [Google Scholar]
- 24.Moss D.J., Burrows S.R., Silins S.L., Misko I., Khanna R. The immunology of Epstein-Barr virus infection. Philos. Trans. R. Soc. Lond. B Biol. Sci. . 2001;356:475–488. doi: 10.1098/rstb.2000.0784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Savard M., Gosselin J. Epstein-Barr virus immunossuppression of innate immunity mediated by phagocytes. Virus Res. 2006;119:134–145. doi: 10.1016/j.virusres.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 26.Kuppers R., Rajewsky K. The origin of Hodgkin and Reed/Sternberg cells in Hodgkin’s disease. Annu. Rev. Immunol. 1998;16:471–493. doi: 10.1146/annurev.immunol.16.1.471. [DOI] [PubMed] [Google Scholar]
- 27.Tamaru J., Hummel M., Zemlin M., Kalvelage B., Stein H. Hodgkin’s disease with a B-cell phenotype often shows a VDJ rearrangement and somatic mutations in the VH genes. Blood. 1994;84:708–715. [PubMed] [Google Scholar]
- 28.Brauninger A., et al. Molecular biology of Hodgkin‘s and Reed/Sternberg cells in Hodgkin’s lymphoma. Int. J. Cancer. 2006;118:1853–1861. doi: 10.1002/ijc.21716. [DOI] [PubMed] [Google Scholar]
- 29.Williams H., et al. The immune response to primary EBV infection: a role for natural killer cells. Br. J. Haematol. 2005;129:266–274. doi: 10.1111/j.1365-2141.2005.05452.x. [DOI] [PubMed] [Google Scholar]
- 30.Stevens S.J.C., et al. Monitoring of Epstein-Barr virus DNA load in peripheral blood by quantitative competitive PCR. J. Clin. Microbiol. 1999;37:2852–2857. doi: 10.1128/jcm.37.9.2852-2857.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]