Abstract
Human/rodent CYP1A1 and CYP1A2 orthologs are well known to exhibit species-specific differences in substrate preferences and rates of metabolism. This lab previously characterized a BAC-transgenic mouse carrying the human CYP1A1_CYP1A2 locus; in this line, human dioxin-inducible CYP1A1 and basal vs dioxin-inducible CYP1A2 have been shown to be expressed normally (with regard to mRNAs, proteins and three enzyme activities) in every one of nine mouse tissues studied. The mouse Cyp1a1 and Cyp1a2 genes are oriented head-to-head and share a bidirectional promoter region of 13,954 bp. Using Cre recombinase and loxP sites inserted 3′ of the stop codons of both genes, we show here a successful interchromosomal excision of 26,173 bp that ablated both genes on the same allele. The Cyp1a1/1a2(−) double-knockout allele was bred with the “humanized” line; the final product is the hCYP1A1_1A2_Cyp1a1/1a2(−/−) line on a theoretically >99.8% C57BL/6J genetic background—having both human genes replacing the mouse orthologs. This line will be valuable for human risk assessment studies involving any environmental toxicant or drug that is a substrate for CYP1A1 or CYP1A2.
Keywords: Humanized mouse line, Human CYP1A1_1A2 locus, Mouse Cyp1a1_1a2 locus, loxP, Cre recombinase, Interchromosomal excision, Benzo[a]pyrene, Dioxin
The human and mouse cytochrome P450 (CYP) gene superfamilies contain 57 and 102 protein-coding genes, respectively; one of the 18 mammalian CYP families is CYP1, having three members in both human and mouse—CYP1A1, CYP1A2, and CYP1B1 [1–3]. The CYP1A and CYP1B subfamily ancestors diverged from one another probably more than 500 million years ago; CYP1A2 arose as a gene duplication event from CYP1A1 about 450 million years ago. Thus, land animals (including fowl) carry both CYP1A1 and CYP1A2; sea animals do not have the CYP1A2 gene [4]. Accordingly, the CYP1A1 and CYP1A2 genes are located at human chromosome 15q24.1, in head-to-head orientation, 23,306 bases from one transcription initiation start-site to the other [5]. Of three mammalian genomes studied, estimates are that about 10% of gene duplication pairs share bidirectional promoters [6]. The latest data from the UCSC browser assembled by NCBI and the Mouse Genome Sequencing Consortium puts the Cyp1a1 and Cyp1a2 genes on mouse chromosome 9 at cM 31.0, also in a head-to-head orientation, 13,954 bases from one transcription start-site to the other. In contrast, CYP1B1 is located on human chromosome 2p22.2, and Cyp1b1 (syntenic with human CYP1B1) is located on mouse chromosome 17.
Cyp1a1(−/−) [7], Cyp1a2(−/−) [8], and Cyp1b1(−/−) [9] knockout mouse lines have previously been generated. Because of location on different chromosomes for the Cyp1a1_Cyp1a2 locus and the Cyp1b1 gene, the Cyp1a1/1b1(−/−) and Cyp1a2/1b1(−/−) double-knockout lines were easy to produce, and have been used in toxicity studies [10]. This lab discovered a recombination “hot spot” in the human CYP1A1_CYP1A2 spacer region [5]. Therefore, we felt it likely that a “hot spot” would also exist in the mouse Cyp1a1_Cyp1a2 bidirectional promoter, which should aid us in creating the Cyp1a1/1a2(−) allele. Despite breeding the Cyp1a1(+/−) × Cyp1a2(+/−) cross and genotyping several hundred offspring, however, our laboratory was unable to identify any Cyp1a1/1a2(−) allele. The present study describes how we were successful in generating the Cyp1a1/1a2(−/−) double-knockout mouse.
Human/rodent CYP1A2 orthologs are well known to exhibit species-specific differences in the rates by which various substrates are metabolized [11]. For example, human and mouse CYP1A2 differ by 3- to 7-fold in catalyzing ethoxyresorufin O-deethylation [12] and uroporphyrinogen oxidation [13].
Using bacterial artificial chromosomes (BACs), we and others have been able to generate “humanized” hCYP1A1_1A2 BAC-transgenic lines [5,14]. Studies have been carried out with the hCYP1A1_1A2_Cyp1a1(−/−) line vs the hCYP1A1_1A2_Cyp1a2(−/−) line; for example, these lines were used both for theophylline [15] and for the food mutagen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) [14]. In both cases, human CYP1A2 was shown to be responsible for the “human metabolite profile” in the absence of mouse CYP1A2.
Examining drug or carcinogen metabolism in the rodent, and then extrapolating results to human populations, is therefore prone to error. It would be very much preferred if human metabolism gene(s) could be inserted in place of the mouse orthologous gene(s). Studying the hCYP1A1_1A2_ Cyp1a1(−/−) and hCYP1A1_1A2_Cyp1a2(−/−) lines separately is cumbersome. It would be much better to have a mouse line carrying the human CYP1A1 and CYP1A2 genes in the absence of both mouse orthologs. The present study reports generation of such a mouse line.
Materials and methods
Mice
C57BL/6J (B6) and DBA/2J (D2) mice were purchased from The Jackson Laboratory (Bar Harbor, ME). Creation of conventional [7] and conditional [16] Cyp1a1(−/−) knockout lines has been described. Production of the Cyp1a1(−/−) [7], Cyp1a2(−/−) [8], and Cyp1b1(−/−) [9] knockout mouse lines have been reported. Development of two “humanized” hCYP1A1_1A2 BAC-transgenic lines have been detailed [5,14]. All experiments involving mice were conducted in accordance with the National Institutes of Health (NIH) standards for the care and use of experimental animals and the University of Cincinnati Institutional Animal Care and Use Committee.
Treatment of the mice
Benzo[a]pyrene (BaP), dissolved in corn oil, was given daily in normal rodent chow at a dose calculated to be 125 mg/kg/day [10,17,18]. Mice of five genotypes were sacrificed after 18 days on this diet. In some cases, mice were treated with 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) (15 μg/kg i.p. 48 h), vs corn oil for untreated. At least three groups (N = 3 each time) were studied to ensure reproducibility.
DNA preparations and polymerase-chain reaction (PCR) analysis
DNA for PCR analysis was isolated from embryonic stem (ES) cells, or from a 5-mm tail biopsy taken from mice on or before postnatal day 21, using previously described methodologies [7].
Cre recombinase/loxP technology
Cre recombinase is an enzyme and loxP is a DNA recognition site, present in bacteriophage P1 [19]. A loxP site represents 34 bases: inverted repeats of 13 bp, plus an 8-bp spacer sequence that imposes directionality to the recombination event: same orientation results in excision; opposite orientation, inversion [20]. Two loxP sites as far apart as 200 kb on the same chromosome have been recognized by Cre recombinase, which then excised all DNA between [21]. Successes at Cre-mediated interchromosomal recombination have also been reported—in which two genes in tandem spanning 40 kb were removed [22], 45 exons spanning 108 kb were excised [23], and nonhomologous chromosomes were joined to make novel chimeric chromosomes [24–28].
Generation of the Cyp1a1/1a2(−) allele
To eliminate both Cyp1a genes on a single chromosome, we relied on Cre recombinase-mediated interchromosomal recombination between loxP sites placed 3′ of the stop codon in both genes (Fig. 1A). Previously, we had deleted the “floxed” (flanked by loxP sites) Cyp1a1(f) coding sequence using Cre-mediated recombination (lines 2 and 4 of Fig. 1A); this included a floxed HPRT minigene in intron 1 and a loxP site 187 bp 3′ of the termination codon in exon 7 [16]. Cre-mediated excision gave us the resultant floxed null allele, Cyp1a1(−), containing only exon 1, a portion of intron 1, and one remaining loxP site 5′ of the distal 760 bp of the exon 7 untranslated region (UTR) (line 4 of Fig. 1A).
Fig. 1.
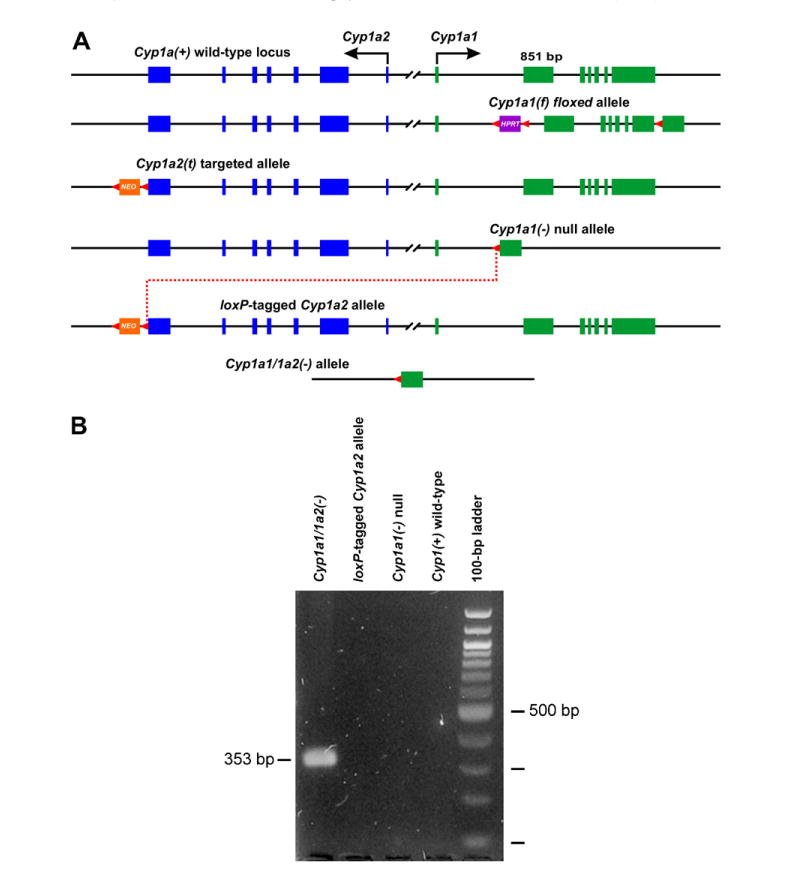
(A) Generation of the floxed Cyp1a1(f) allele, the targeted Cyp1a2(t) allele, and the Cre-mediated interchromosomal excision (dotted red line) between the Cyp1a1(−/−) null allele and the loxP-tagged Cyp1a2 allele. Cyp1a2 exons are shown in blue; Cyp1a1 exons are shown in green. All exons and introns are drawn to scale (Cyp1a1 exon 2 is 851 bp). All loxP sites (and their orientation) are shown as red arrowheads. HPRT, hypoxanthine-guanine phosphoribosyltransferase minigene used as selection marker. NEO, phosphoglycerate kinase-driven neomycin (PGK-NEO) minigene used as selection marker. For reason of clarity, most of the 13,954 bp 5′-flanking spacer region shared by both genes is omitted. (B) Confirmation of the Cyp1a1/1a2(−) allele: as expected, a 353-bp PCR fragment is generated (lane 1), compared with no such fragment for the loxP-tagged Cyp1a2 allele, the Cyp1a1(−) null allele, or the wild-type allele (lanes 2–4). The 100-bp ladder of markers is shown in far right lane. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this paper.)
Next, we generated the targeted Cyp1a2(t) allele containing a floxed PGK-NEO gene [29], placed 350 bp 3′ of the stop codon, using standard gene-targeting techniques; the loxP site is located 48 bp 3′ beyond the end of exon 7 (line 3 of Fig. 1A). We prepared a targeting construct with a long arm for recombination encompassing 4429 bp of Cyp1a2 genomic sequence plus 48 bp of 3′-flanking sequence. This was followed by a floxed PGK-NEO gene [22] and then by 1452 bp of 3′-flanking sequence for the short arm. Outside the 3′ Cyp1a2 homology arm, we placed the HSK-TK negative-selection cassette [30]. All targeting vector sequences were assembled in Bluescript SK(−). The targeting vector was linearized by cutting 4 bp downstream of the HSV-TK cassette. Following chloroform extraction and ethanol precipitation, this construct was used to electroporate ES cells derived from 129S6/SvEvTac mice [31]. ES cells were selected by using G418 and ganciclovir, and resistant colonies were harvested as described [31]. DNA was prepared from individual colonies and screened by PCR, using two primer sets. One set amplified a fragment of 1617 bp, using primer A in the PGK-NEO gene and primer B immediately downstream of the 3′-most Cyp1a2 sequence that was present in the targeting construct. These primer sequences are:
A: 5′-TCCTCTTGAAAACCACACTGCTCGAC-3′
B: 5′-GATTAGCTGATGGGTGTGTCTATGGG-3′
PCRs included 35 cycles, with initial denaturation at 94 °C for 3 min, followed by cycling at 94 °C for 30 s, 60 °C for 40 s, and then 72 °C for 3 min and 45 s. The second PCR amplified a fragment of 5050 bp, using primer C in the PGK-NEO gene and primer D just upstream of the 5′-most Cyp1a2 sequence that was present in the targeting construct. These primer sequences are:
C: 5′-TGGCTACCCGTGATATTGCTGAAGAG-3′
D: 5′-TAACCCACATCACCGCCAGAGAAATG-3′
PCR conditions were the same as above, except the annealing times were 60 s, and extension times were 6 min.
ES colonies positive for both PCRs were considered “targeted”. Several colonies were expanded and used for injecting into B6 blastocysts, as previously described [7,8,20,31]. Coat-color chimeric mice were bred with B6 mice, and agouti offspring were screened by PCR for the targeted Cyp1a2(t) allele.
Cyp1a2(+/t) heterozygotes were mated with Cyp1a1(+/−) heterozygotes that also carried the universal deleting CAGGS-CRE gene [32,33]. CAGGS-Cre is a Cre recombinase-expressing transgene driven by a globally expressed Actb (β-actin) promoter, which can function developmentally as early as the fertilized ovum [32]. Mice with the genotype Cyp1a1(+/−)/Cyp1a2(+/t)/CAGGS-CRE(+/−) were identified using PCR, and these mice were then bred with B6 mice. To detect Cre recombinase-mediated interchromosomal recombination between the loxP sites 3′ beyond the stop codons of the Cyp1a1 and Cyp1a2 genes (bottom line of Fig. 1A), we used PCR and a primer set in which each primer was 3′ of the loxP site of its respective Cyp1a gene (Fig. 1A). The sequence of these primers, which also have been used subsequently to genotype the Cyp1a1/1a2(−) double-knockout allele, are:
E: 5′-GACATAGGAGCTACCTACAC-3′ (3′ of loxP site of Cyp1a2)
F: 5′-GTCAAAGTAACCAGACACATCCTGC-3′ (3′ of loxP site of Cyp1a1)
The Cyp1a1/1a2(−) allele was backcrossed for 10 generations onto a B6 background, after which the Cyp1a1/1a2(+/−) mice (now theoretically on a >99.8% B6 background), were intercrossed to generate the homozygous Cyp1a1/1a2(−/−) mouse line. For Cyp1(+/+) wild-type mice, the B6 inbred strain can therefore be used [10,18].
Production of the hCYP1A1_1A2_Cyp1a1/1a2(−/−) mouse line
We previously had isolated a 180-kb BAC containing the human CYP1A1_CYP1A2 locus—including the 23.3-kb spacer region, ∼90 kb of CYP1A2 3′-flanking region and ∼53 kb of CYP1A1 3′-flanking region [5]. By zygotic pronuclear microinjection, one copy of the BAC was introduced into (B6D2)F1 oocytes, and the hCYP1A1_1A2 allele was then backcrossed for 10 generations onto a B6 background. This “humanized” hCYP1A1_1A2 line, theoretically on a >99.8% B6 background, had been bred separately with Cyp1a1(−/−) or Cyp1a2(−/−) mice. By Northern hybridization, Western immunoblot analysis, and three enzyme assays, human dioxin-inducible CYP1A1 and both human basal and dioxin-inducible CYP1A2 levels of expression (mRNA, protein, and enzyme activity)—in every one of nine tissues examined—paralleled very closely those seen with the mouse homologs [5]. The hCYP1A1_1A2(+/−) mouse, containing one copy of the human BAC, was bred with the Cyp1a1/1a2(−/−) mouse (described above), to produce the hCYP1A1_1A2_Cyp1a1/1a2(−/−) mouse line.
Real-time quantitative PCR (Q-PCR), Western immunoblot, and BaP toxicity studies
Daily treatment of mice with oral BaP for 18 days, TCDD or corn oil treatment for 48 h, total RNA preparation, reverse transcription, Q-PCR analysis, microsomal protein immunoblots, determinations of the alanine aminotransferase (ALT) and aspartate aminotransferase (AST) plasma enzymes, and histology of peripheral blood were performed—as described previously [10,18].
Results and discussion
Generation of the hCYP1A1_1A2_Cyp1a1/1a2(−/−) line
The Cyp1a1/1a2(−) double-knockout allele could perhaps have been generated by conventional gene-targeting techniques, using a construct in which both alleles lack function; however, the size of the Cyp1a1_1a2 locus is too bulky to be accommodated in a plasmid-based cloning vector—without deleting a large portion of the two-gene locus, which might negatively affect the targeting frequency. An alternative approach to ablating both genes separately was to use Cre recombinase to effect an interchromosomal recombination.
Fig. 1A illustrates the successful generation of the Cyp1a1/1a2(−) allele. Fortunately, the Cyp1a1(−) null allele [7] (including position and orientation of the one remaining loxP site) was tailor-made for using in our interchromosomal deletion strategy (Fig. 1A, line 4). From tail DNA of Cyp1a1(+/−)/Cyp1a2(+/t)/CAGGS-Cre(+/−) mice, we detected by PCR the interchromosomal recombination of Cyp1a1(−) with the loxP-tagged Cyp1a2 allele to produce the Cyp1a1/1a2(−) allele (Fig. 1B). We also detected the presence of the intact Cyp1a1(−) allele and the Cyp1a2(t) in which the PGK-NEO had been deleted; this suggests that interchromosomal recombination occurred late in development and perhaps was still occurring. We next backcrossed Cyp1a1(+/−)/Cyp1a2(+/t)/CAGGS-Cre(+/−) mice into B6 mice and identified the Cyp1a1/1a2(−) allele in offspring, using antisense primers (PCR product of 353 bp; Fig. 1B) in the 3′-UTR of Cyp1a1 and 3′-flanking region of Cyp1a2. The Cyp1a1/1a2(−) allele was found in offspring, independent of inheritance of the CAGGS-CRE transgene (not shown), suggesting germ line inheritance; this result was confirmed further by our observing normal Mendelian transmission of the Cyp1a1/1a2(−) allele in F1 offspring (not shown). Compared with expected frequencies, we found that the observed frequency of interchromosomal recombination was statistically not significantly different from that of intrachromosomal recombination (i.e., between 80% and 100%). To propagate the Cyp1a1/1a2(−) allele, we used offspring that did not possess the CAGGS-CRE transgene. Taken together, we conclude that Cre-mediated interchromosomal excision of 26,173 bp was successful and resulted in a Cyp1a1/1a2(−) double-knockout allele—lacking all of the Cyp1a2 gene plus 48 bp of its 3′-flanking region, the entire spacer region, and all but the last 760 bp of Cyp1a1 exon 7 (last line of Fig. 1A). It should be noted that, when the interchromosomal recombination occurs, there is a reciprocal Cyp1a2 gene duplication on the chromosome other than that shown (last line of Fig. 1A).
Levels of mRNA in liver and intestine
In all cases, basal vs TCDD-maximally induced levels of mRNA (or protein) were compared with that in mice receiving oral BaP for 18 days. In liver, comparing oral BaP with TCDD treatment (Fig. 2, upper left), mouse CYP1A1 mRNA was extremely low in Cyp1(+/+) and Cyp1a2(−/−) and not detected in Cyp1a1(−/−), Cyp1a1/1a2(−/−) or hCYP1A1_1A2_Cyp1a1/1a2(−/−) mice. Mouse CYP1A2 mRNA was also very low in oral BaP-treated Cyp1(+/+), compared with that after TCDD treatment. The same trend was seen for human CYP1A1 but not human CYP1A2 mRNA (Fig. 2, lower left) or mouse CYP1A2 mRNA in Cyp1a1(−/−) mice (Fig. 2, left, second panel).
Fig. 2.
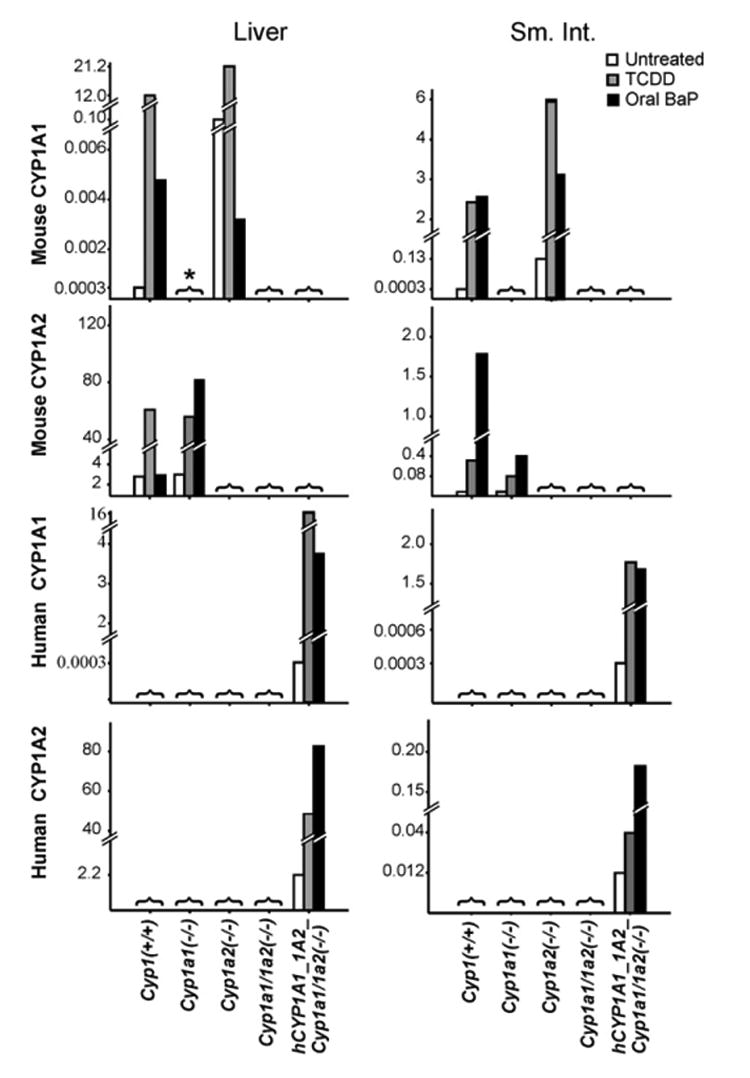
Q-PCR analysis of mouse CYP1A1 and CYP1A2 (top two panels) and human CYP1A1 and CYP1A2 (lower two panels) mRNA in liver (left) and proximal small intestine (right) from the five genotypes under study. Untreated, TCDD-treated, and oral BaP-treated mice are shown. The estimated mRNA levels are expressed as relative amounts after normalization to β-actin mRNA; note these levels on the ordinates vary dramatically––from 0.003 to 80. N = 3 per group. *[with bracket], absolutely not detected at all, which is to be expected due to the global genetic ablation of that particular gene.
In small intestine (Fig. 2, upper right), mouse CYP1A1 mRNA was induced to similar high levels in either oral BaP- or TCDD-treated Cyp1(+/+) and Cyp1a2(−/−) but not detected in Cyp1a1(−/−), Cyp1a1/1a2(−/−) or hCYP1A1_1A2_Cyp1a1/1a2(−/−) mice. Mouse CYP1A2 mRNA was not detected in Cyp1a2(−/−), Cyp1a1/1a2(−/−) or hCYP1A1_1A2_Cyp1a1/1a2(−/−) mice. Curiously, mouse CYP1A2 mRNA levels in Cyp1(+/+) and Cyp1a2(−/−) mice were about five times higher after oral BaP treatment than after TCDD treatment. The same magnitudes of change were also seen with the human CYP1A1 and CYP1A2 mRNAs, respectively (Fig. 2, lower right) in the hCYP1A1_1A2_Cyp1a1/1a2(−/−) line.
Any precise comparison of mRNA levels between liver and small intestine cannot be made, because the common denominator used was β-actin mRNA, whose levels might differ between liver and small intestine; moreover, the efficiency of a PCR is dependent on primers used and the mRNA being measured. Detailed kinetics of oral BaP-induced CYP1A1 and CYP1A2 levels will be addressed in a subsequent publication; briefly summarized, we have found that both intestinal and hepatic CYP1A1 and CYP1A2 of Cyp1(+/+) mice become highly induced within the first 12–48 h, after which hepatic CYP1A1 and CYP1A2 levels return towards that seen in untreated wild-type mice, whereas CYP1A1 and CYP1A2 in the small intestine remain highly induced—because of the daily intake of oral BaP. It seems clear that induced CYP1A1 (and CYP1B1; see below) levels in the small intestine are so efficient at BaP metabolism and clearance, that substantial amounts of BaP no longer reach liver and therefore the inducing effects are lost.
We found a small but significant increase of CYP1B1 mRNA levels in both the liver and small intestine of untreated Cyp1a1(−/−) and Cyp1a1/1a2(−/−) mice (not shown). On the other hand, the total level of CYP1B1 mRNA in liver and intestine was very low, i.e., never more than 5–10% of CYP1A1 mRNA levels. These results suggest that the genetic absence of CYP1A1 in mouse liver and intestine results in a compensatory increase in basal CYP1B1 expression.
Levels of protein in liver and intestine
In the liver and intestine of oral BaP- (Fig. 3A) vs TCDD-treated mice [5,15], mouse CYP1A1 and CYP1A2 protein levels in Cyp1(+/+), Cyp1a1(−/−), and Cyp1a2(−/−) mice––as well as human CYP1A1 and CYP1A2 protein levels in hCYP1A1_1A2_Cyp1a1/1a2(−/−) mice––were remarkably similar, in all instances, to the mRNA levels measured by Q-PCR (Fig. 2). These data indicate that posttranslational stabilization is not a major contributory factor in determining the ultimate level of enzyme activity, but rather that protein levels closely reflect mRNA levels for the two mouse genes and the two human genes in these mice.
Fig. 3.
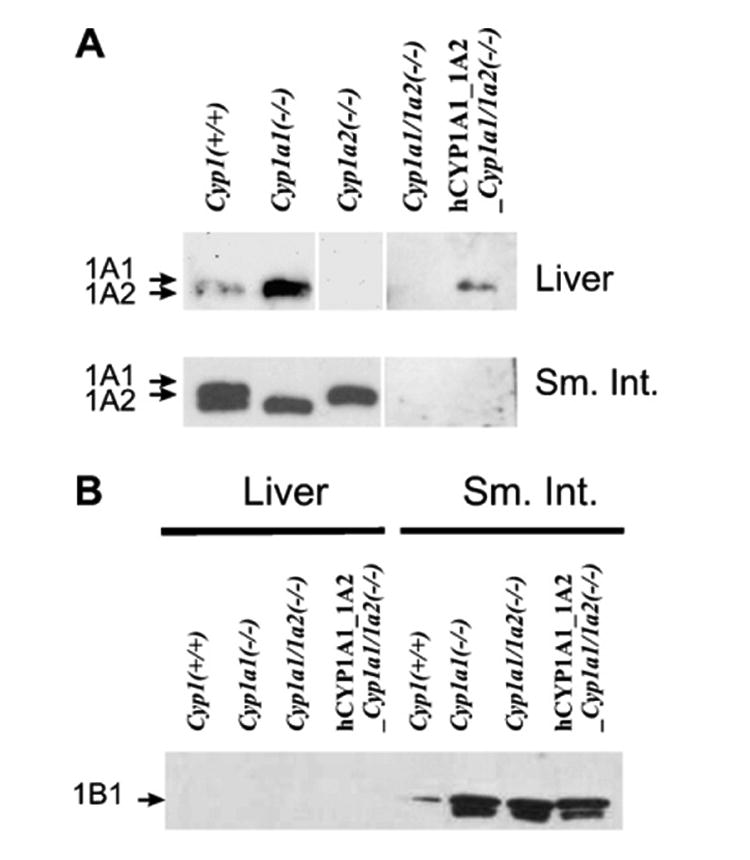
Western immunoblot analysis. (A) Mouse or human CYP1A1 and CYP1A2 proteins in the liver (top row) and proximal small intestine (lower row) from the five genotypes under study. The human CYP1A1 and CYP1A2 proteins migrate between that of the mouse CYP1A1 and CYP1A2 proteins [5]. The same regimen (oral BaP-treated) was employed. The rabbit polyclonal anti-rat CYP1A1/1A2 antibody (Daiichi Pharmaceuticals Co. Ltd., Tokyo) was used; this antibody recognizes both mouse and human CYP1A proteins. (B) Mouse CYP1B1 protein in liver (left) and intestine (right) of the oral BaP-treated mice. Liver and intestine CYP1B1 levels in Cyp1a2(−/−) mice (not shown) exhibited levels similar to that in Cyp1(+/+) mice. A rabbit polyclonal anti-mouse CYP1B1 antibody was developed in our laboratory, in collaboration with Alpha Diagnostic International (San Antonio, TX); this antibody recognizes both mouse and human CYP1B1 protein. Each lane was loaded with 0.25 and 10 μg microsomal proteins for liver and small intestine, respectively. The reason for the faster-moving band in this doublet is not known, but might represent mitochondrial CYP1B1 protein contamination [34] of the small intestine microsomes.
As we had previously found [5], it is difficult to resolve the human CYP1A1 and CYP1A2 proteins [5]; in fact, both of these bands run in the gap between mouse CYP1A1 and CYP1A2. The antibody used here was raised against rat CYP1A1/1A2, and therefore is expected to react better with mouse than with human CYP1A1/1A2 proteins. Consequently, no direct quantifiable comparisons can be made between the mouse CYP1A and human CYP1A levels in Fig. 3A.
CYP1B1 levels in liver and intestine
In the liver of all genotypes after 18 days of oral BaP (Fig. 3B, left), CYP1B1 protein was virtually undetectable. In the proximal small intestine (Fig. 3B, right), however, CYP1B1 protein was detectable in Cyp1(+/+) and Cyp1a2(−/−) mice and highly induced in Cyp1a1(−/−), Cyp1a1/1a2(−/−), and hCYP1A1_1A2_Cyp1a1/1a2(−/−) mice. These data suggest that the absence of mouse CYP1A1 causes a striking BaP-mediated induction of CYP1B1 protein in the small intestine. The presence of human CYP1A1 (Fig. 3B, last lane) does not completely return CYP1B1 protein levels in the intestine to that seen in the presence of mouse CYP1A1.
Oral BaP toxicity studies
Cyp1(+/+) mice are able to survive more than a year on a diet of oral BaP at 125 mg/kg/day; in fact, they live even slightly longer than Cyp1(+/+) mice receiving no BaP in their diet [17]. In contrast, Cyp1a1(−/−) mice become sickly and die between 28 and 34 days––due to severe immunosuppression as well as toxic chemical depression of the bone marrow [10,18]. These findings strongly suggest that CYP1A1 (in intestine and/or liver) is more important in detoxication than in metabolic activation of daily oral BaP. A large BaP body burden was found in Cyp1a1(−/−) and Cyp1a1/1a2(−/−) animals (Table 1), but not in Cyp1(+/+), Cyp1a2(−/−) or hCYP1A1_1A2_Cyp1a1/1a2(−/−) mice. Increased liver size (sign of chronic AHR activation) was seen in Cyp1a1(−/−) but not in Cyp1(+/+), Cyp1a2(−/−) or Cyp1a1/1a2(−/−) mice. Does the absence of CYP1A2 override the BaP effect when CYP1A1 is ablated? Surprisingly, an increased liver size was also exhibited by hCYP1A1_1A2_Cyp1a1/1a2(−/−) mice. Decreased spleen and thymus size, elevated ALT and AST levels, and leukocytopenia (signs of immunosuppression) were found in Cyp1a1(−/−) and Cyp1a1/1a2(−/−) mice (Table 1), compared with that in Cyp1(+/+), Cyp1a2(−/−), and hCYP1A1_1A2_Cyp1a1/1a2(−/−) mice. These findings indicate that substitution of the human CYP1A1 and CYP1A2 enzymes in place of the mouse orthologs is able to return the phenotype of the Cyp1a1/1a2(−/−) double-knockout line back to approximately that of the wild-type.
Table 1.
Various parameters of control vs oral BaP-treated mice of the five genotypes under study
| Genotype: | Cyp1(+/+) wild-type | Cyp1a1(−/−) | Cyp1a2(−/−) | Cyp1a1/1a2(−/−) | hCYP1A1_1A2_Cyp1a1/1a2(−/−) |
|---|---|---|---|---|---|
| Clinical appearance | Healthy | Sickly | Healthy | Sickly | Healthy |
| Blood [BaP] (ng/ml) after 5 days | 1.3 ± 0.7 | *29 ± 19 | †0.51 ± 0.20 | *62 ± 5.2 | †0.80 ± 0.10 |
| Liver size (g)/body weight (g) | 4.3 ± 0.28 | *5.9 ± 0.18 | †4.2 ± 0.65 | 4.5 ± 0.31 | *6.0 ± 0.31 |
| Spleen size (g)/body weight (g) | 0.26 ± 0.01 | *0.11 ± 0.01 | †0.27 ± 0.02 | *0.11 ± 0.01 | †0.21 ± 0.03 |
| Thymus size (g)/body weight (g) | 0.14 ± 0.01 | *0.03 ± 0.002 | †0.13 ± 0.2 | *0.03 ± 0.02 | *,†0.07 ± 0.006 |
| Plasma ALT (U/Liter) | 75 ± 25 | *390 ± 130 | †60 ± 10 | *240 ± 23 | †53 ± 9.3 |
| Plasma AST (U/Liter) | 140 ± 12 | *290 ± 91 | †130 ± 20 | *360 ± 77 | *,†180 ± 19 |
| % lymphocytes in peripheral blood | 93 ± 3.0 | *62 ± 1.1 | †94 ± 3.6 | *54 ± 18 | †81 ± 7.3 |
Except for whole blood BaP levels after 5 days, all other parameters were determined after 18 days of oral BaP (125 mg/kg/day). Groups of three or four male mice of the same age (5 weeks) for each genotype were studied. Percent wet weight (g) of liver, spleen, and thymus was measured per gram of total body weight (g). Values are given as means ± SEM.
P value <0.05, relative to the Cyp1(+/+) wild-type.
P value <0.05, relative to the Cyp1a1(−/−) knockout mouse; all parameters of Cyp1a1(−/−) mice were significantly different from those of Cyp1(+/+) mice.
Conclusions
There are two major breakthroughs in this study. First, although the Cyp1a1 and Cyp1a2 genes are in close proximity to one another (13,954 bp), we have succeeded in generating the Cyp1a1/1a2(−) allele via a Cre recombinase-mediated interchromosomal excision; this activity removed all of the Cyp1a2 gene and all but the last 760 bp in the UTR of exon 7 of the Cyp1a1 gene. Second, by way of further breeding, we have generated the humanized hCYP1A1_1A2_Cyp1a1/1a2(−/−) line, in which both mouse Cyp1a genes have been replaced by both human CYP1A genes. Both the Cyp1a1/1a2(−/−) double-knockout and this humanized line are viable, fertile, and show normal longevity. Herein we have demonstrated the presence of human CYP1A1 and CYP1A2 mRNAs and proteins, and absence of mouse CYP1A1 and CYP1A2 mRNAs and proteins; presence of human CYP1A1 and CYP1A2 mRNAs, proteins and enzyme activities in the hCYP1A1_1A2 line were also shown previously [5,15].
Although the human CYP1A1 in place of mouse CYP1A1 only partially “rescues” the animal from oral BaP-induced immunosuppression, it appears that human CYP1A1 does as well as its mouse counterpart in eliminating BaP (Table 1). In liver oral BaP-induced human CYP1A1 is as high as mouse CYP1A1; this conclusion is counter to the toxicological data, showing systemic effects of BaP in the humanized mice. The thymus is smaller, and the liver larger, in the hCYP1A1_1A2_Cyp1a1/1a2(−/−) line, compared with the Cyp1(+/+) wild-type. Also, plasma AST (perhaps a measure of peripheral toxicity) is significantly higher. The aforementioned toxicity and induction of CYP1A1 are seemingly at odds with the low BaP found in the bloodstream of the humanized mice. Is the human CYP1A1 as good as the mouse CYP1A1 in detoxication? Perhaps the human relies on liver for detoxication more so than the mouse, whereas the mouse might rely more on intestinal CYP1A1 than the human. Future studies will address these questions.
The hCYP1A1_1A2(+) transgene, combined with the Cyp1a1/1a2(−) double-knockout allele, has been backcrossed 10 times into the B6 genetic background, resulting in mice that theoretically contain >99.8% B6 genes. We believe that this newly developed hCYP1A1_1A2_Cyp1a1/1a2(−/−) line will be convenient to use, and invaluable to researchers carrying out human risk assessment studies—involving any drug or environmental toxicant that is a substrate for CYP1A1 or CYP1A2. Upon publication of this report, this line will be made commercially available by The Jackson Laboratories as soon as reasonably possible.
We are also backcrossing the hCYP1A1_1A2_Cyp1a1/1a2(−/−) genotype 10 times into the D2 genetic background. Given the fact that the human poor-affinity AHR resembles more closely the poor-affinity AHR of D2 mice (rather than the high-affinity AHR of B6 mice) [2,3], this future line (expected to be completed within two more years) could provide the investigator with answers to questions that studies in the humanized line, characterized herein, might not be able to answer. The line having the hCYP1A1_1A2_Cyp1a1/1a2(−/−) genotype in the presence of theoretically >99.8% D2 genes will also be made commercially available by The Jackson Laboratories as soon as possible.
Acknowledgments
We thank our colleagues for many fruitful discussions and careful readings of this manuscript. We especially appreciate Marian Miller for help with graphics, Tom Doetschman and Phil Sanford in the Transgenic Mouse Core for designing constructs for the interchromosomal excision, and David R. Nelson for genomics help with the mouse Cyp1a1_Cyp1a2 locus. Supported, in part, by NIH Grants R01 ES08147 (D.W.N.), R01 ES014403 (D.W.N.), P30 ES06096 (T.P.D.; D.W.N.), and Ministry of Education, Science, Sports & Culture, Japan; contact 16790089 (S.U.).
References
- 1.Nelson DR, Zeldin DC, Hoffman SM, Maltais LJ, Wain HM, Nebert DW. Comparison of cytochrome P450 (CYP) genes from the mouse and human genomes, including nomenclature recommendations for genes, pseudogenes and alternative-splice variants. Pharmacogenetics. 2004;14:1–18. doi: 10.1097/00008571-200401000-00001. [DOI] [PubMed] [Google Scholar]
- 2.Nebert DW, Dalton TP, Okey AB, Gonzalez FJ. Role of aryl hydrocarbon receptor-mediated induction of the CYP1 enzymes in environmental toxicity and cancer. J Biol Chem. 2004;279:23847–23850. doi: 10.1074/jbc.R400004200. [DOI] [PubMed] [Google Scholar]
- 3.Nebert DW, Dalton TP. The role of cytochrome P450 enzymes in endogenous signalling pathways and environmental carcinogenesis. Nat Rev Cancer. 2006;6:947–960. doi: 10.1038/nrc2015. [DOI] [PubMed] [Google Scholar]
- 4.Nelson DR, Koymans L, Kamataki T, Stegeman JJ, Feyereisen R, Waxman DJ, Waterman MR, Gotoh O, Coon MJ, Estabrook RW, Gunsalus IC, Nebert DW. P450 superfamily: update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics. 1996;6:1–42. doi: 10.1097/00008571-199602000-00002. [DOI] [PubMed] [Google Scholar]
- 5.Jiang Z, Dalton TP, Jin L, Wang B, Tsuneoka Y, Shertzer HG, Deka R, Nebert DW. Toward the evaluation of function in genetic variability: characterizing human SNP frequencies and establishing BAC-transgenic mice carrying the human CYP1A1_CYP1A2 locus. Hum Mutat. 2005;25:196–206. doi: 10.1002/humu.20134. [DOI] [PubMed] [Google Scholar]
- 6.Li YY, Yu H, Guo ZM, Guo TQ, Tu K, Li YX. Systematic analysis of head-to-head gene organization: evolutionary conservation and potential biological relevance. PLoS Comput Biol. 2006;2:e74. doi: 10.1371/journal.pcbi.0020074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dalton TP, Dieter MZ, Matlib RS, Childs NL, Shertzer HG, Genter MB, Nebert DW. Targeted knockout of Cyp1a1 gene does not alter hepatic constitutive expression of other genes in the mouse [Ah] battery. Biochem Biophys Res Commun. 2000;267:184–189. doi: 10.1006/bbrc.1999.1913. [DOI] [PubMed] [Google Scholar]
- 8.Liang HC, Li H, McKinnon RA, Duffy JJ, Potter SS, Puga A, Nebert DW. Cyp1a2(−/−) null mutant mice develop normally but show deficient drug metabolism. Proc Natl Acad Sci USA. 1996;93:1671–1676. doi: 10.1073/pnas.93.4.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buters JT, Sakai S, Richter T, Pineau T, Alexander DL, Savas U, Doehmer J, Ward JM, Jefcoate CR, Gonzalez FJ. Cytochrome P450 CYP1B1 determines susceptibility to 7,12-dimethylbenz[a]anthracene-induced lymphomas. Proc Natl Acad Sci USA. 1999;96:1977–1982. doi: 10.1073/pnas.96.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uno S, Dalton TP, Dragin N, Curran CP, Derkenne S, Miller ML, Shertzer HG, Gonzalez FJ, Nebert DW. Oral benzo[a]-pyrene in Cyp1 knockout mouse lines: CYP1A1 important in detoxication, CYP1B1 metabolism required for immune damage independent of total-body burden and clearance rate. Mol Pharmacol. 2006;69:1103–1114. doi: 10.1124/mol.105.021501. [DOI] [PubMed] [Google Scholar]
- 11.Turesky RJ. Interspecies metabolism of heterocyclic aromatic amines and the uncertainties in extrapolation of animal toxicity data for human risk assessment. Mol Nutr Food Res. 2005;49:101–117. doi: 10.1002/mnfr.200400076. [DOI] [PubMed] [Google Scholar]
- 12.Aoyama T, Gonzalez FJ, Gelboin HV. Human cDNA-expressed cytochrome P450 1A2: mutagen activation and substrate specificity. Mol Carcinog. 1989;2:192–198. doi: 10.1002/mc.2940020405. [DOI] [PubMed] [Google Scholar]
- 13.Nichols RC, Cooper S, Trask HW, Gorman N, Dalton TP, Nebert DW, Sinclair JF, Sinclair PR. Uroporphyrin accumulation in hepatoma cells expressing human or mouse CYP1A2: relation to the role of CYP1A2 in human porphyria cutanea tarda. Biochem Pharmacol. 2003;65:545–550. doi: 10.1016/s0006-2952(02)01550-2. [DOI] [PubMed] [Google Scholar]
- 14.Cheung C, Ma X, Krausz KW, Kimura S, Feigenbaum L, Dalton TP, Nebert DW, Idle JR, Gonzalez FJ. Differential metabolism of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) in mice humanized for CYP1A1 and CYP1A2. Chem Res Toxicol. 2005;18:1471–1478. doi: 10.1021/tx050136g. [DOI] [PubMed] [Google Scholar]
- 15.Derkenne S, Curran CP, Shertzer HG, Dalton TP, Dragin N, Nebert DW. Theophylline pharmacokinetics: comparison of Cyp1a1(−/−) and Cyp1a2(−/−) knockout mice, humanized hCYP1A1_1A2 knock-in mice lacking either the mouse Cyp1a1 or Cyp1a2 gene, and Cyp1(+/+) wild-type mice. Pharmacogenet Genomics. 2005;15:503–511. doi: 10.1097/01.fpc.0000167326.00411.50. [DOI] [PubMed] [Google Scholar]
- 16.Uno S, Wang B, Shertzer HG, Nebert DW, Dalton TP. Balancer-Cre transgenic mouse germ cells direct the incomplete resolution of a tri-loxP-targeted Cyp1a1 allele, producing a conditional knockout allele. Biochem Biophys Res Commun. 2003;312:494–499. doi: 10.1016/j.bbrc.2003.10.145. [DOI] [PubMed] [Google Scholar]
- 17.Robinson JR, Felton JS, Levitt RC, Thorgeirsson SS, Nebert DW. Relationship between “aromatic hydrocarbon responsiveness” and the survival times in mice treated with various drugs and environmental compounds. Mol Pharmacol. 1975;11:850–865. [PubMed] [Google Scholar]
- 18.Uno S, Dalton TP, Derkenne S, Curran CP, Miller ML, Shertzer HG, Nebert DW. Oral exposure to benzo[a]pyrene in the mouse: detoxication by inducible cytochrome P450 is more important than metabolic activation. Mol Pharmacol. 2004;65:1225–1237. doi: 10.1124/mol.65.5.1225. [DOI] [PubMed] [Google Scholar]
- 19.Gu H, Marth JD, Orban PC, Mossmann H, Rajewsky K. Deletion of a DNA polymerase beta gene segment in T cells using cell type-specific gene targeting. Science. 1994;265:103–106. doi: 10.1126/science.8016642. [DOI] [PubMed] [Google Scholar]
- 20.Nebert DW, Duffy JJ. How knockout mouse lines will be used to study the role of drug-metabolizing enzymes and their receptors during reproduction and development, and in environmental toxicity, cancer, and oxidative stress. Biochem Pharmacol. 1997;53:249–254. doi: 10.1016/s0006-2952(96)00740-x. [DOI] [PubMed] [Google Scholar]
- 21.Li ZW, Stark G, Gotz J, Rulicke T, Gschwind M, Huber G, Muller U, Weissmann C. Generation of mice with a 200-kb amyloid precursor protein gene deletion by Cre recombinase-mediated site-specific recombination in embryonic stem cells. Proc Natl Acad Sci USA. 1996;93:6158–6162. doi: 10.1073/pnas.93.12.6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsusaka T, Kon V, Takaya J, Katori H, Chen X, Miyazaki J, Homma T, Fogo A, Ichikawa I. Dual renin gene targeting by Cre-mediated interchromosomal recombination. Genomics. 2000;64:127–131. doi: 10.1006/geno.2000.6113. [DOI] [PubMed] [Google Scholar]
- 23.Khor B, Bredemeyer AL, Huang CY, Turnbull IR, Evans R, Maggi LB, Jr, White JM, Walker LM, Carnes K, Hess RA, Sleckman BP. Proteasome activator PA200 is required for normal spermatogenesis. Mol Cell Biol. 2006;26:2999–3007. doi: 10.1128/MCB.26.8.2999-3007.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Deursen J, Fornerod M, van Rees B, Grosveld G. Cre-mediated site-specific translocation between nonhomologous mouse chromosomes. Proc Natl Acad Sci USA. 1995;92:7376–7380. doi: 10.1073/pnas.92.16.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buchholz F, Refaeli Y, Trumpp A, Bishop JM. Inducible chromosomal translocation of AML1 and ETO genes through Cre/loxP-mediated recombination in the mouse. EMBO Rep. 2000;1:133–139. doi: 10.1093/embo-reports/kvd027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu Y, Bradley A. Engineering chromosomal rearrangements in mice. Nat Rev Genet. 2001;2:780–790. doi: 10.1038/35093564. [DOI] [PubMed] [Google Scholar]
- 27.Forster A, Pannell R, Drynan LF, McCormack M, Collins EC, Daser A, Rabbitts TH. Engineering de novo reciprocal chromosomal translocations associated with Mll to replicate primary events of human cancer. Cancer Cell. 2003;3:449–458. doi: 10.1016/s1535-6108(03)00106-5. [DOI] [PubMed] [Google Scholar]
- 28.Zong H, Espinosa JS, Su HH, Muzumdar MD, Luo L. Mosaic analysis with double markers in mice. Cell. 2005;121:479–492. doi: 10.1016/j.cell.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 29.Chen Y, Yang Y, Miller ML, Shen D, Shertzer HG, Stringer KF, Wang B, Schneider SN, Nebert DW, Dalton TP. Hepatocyte-specific deletion of the glutamate-cysteine ligase catalytic subunit leads to microvesicular steatosis with mitochondrial injury and liver failure. Hepatology. 2007;45:1118–1128. doi: 10.1002/hep.21635. [DOI] [PubMed] [Google Scholar]
- 30.Singhal S, Kaiser LR. Cancer chemotherapy using suicide genes. Surg Oncol Clin N Am. 1998;7:505–536. [PubMed] [Google Scholar]
- 31.Dalton TP, Dieter MZ, Yang Y, Shertzer HG, Nebert DW. Knockout of the mouse glutamate-cysteine ligase catalytic subunit (Gclc) gene: embryonic lethal when homozygous, and proposed model for moderate glutathione deficiency when heterozygous. Biochem Biophys Res Commun. 2000;279:324–329. doi: 10.1006/bbrc.2000.3930. [DOI] [PubMed] [Google Scholar]
- 32.Araki K, Araki M, Miyazaki J, Vassalli P. Site-specific recombination of a transgene in fertilized eggs by transient expression of Cre recombinase. Proc Natl Acad Sci USA. 1995;92:160–164. doi: 10.1073/pnas.92.1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sunaga S, Maki K, Komagata Y, Ikuta K, Miyazaki JI. Efficient removal of loxP-flanked DNA sequences in a gene-targeted locus by transient expression of Cre recombinase in fertilized eggs. Mol Reprod Dev. 1997;46:109–113. doi: 10.1002/(SICI)1098-2795(199702)46:2<109::AID-MRD1>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 34.Shayiq RM, Addya S, Avadhani NG. Constitutive and inducible forms of cytochrome P450 from hepatic mitochondria. Methods Enzymol. 1991;206:587–594. doi: 10.1016/0076-6879(91)06128-p. [DOI] [PubMed] [Google Scholar]


