Summary
Ubiquitin receptor proteins play an important role in delivering ubiquitylated protein substrates to the proteasome for degradation. HHR23a and hPLIC2 are two such ubiquitin receptors that contain ubiquitin-like (UBL) domains, which interact with the proteasome, and ubiquitin-associated (UBA) domains, which interact with ubiquitin. Depending on their abundance UBL/UBA family members can either promote or inhibit the degradation of other proteins, which suggests their participation in the delivery of substrates to the proteasome is highly regulated. In previous work, we determined UBL/UBA domain interactions to promote intramolecular interactions in hHR23a that are abrogated with the addition of either ubiquitin or the proteasome component S5a. In yeast, we determined the hHR23a ortholog (Rad23) to interact with another UBL/UBA family member (Ddi1) and to bind a common tetraubiquitin chain. Here, we use NMR spectroscopy to reveal that hHR23a interacts with hPLIC2 via UBL/UBA domain interactions and to map their binding surfaces. In addition, we demonstrate that these two proteins associate in mammalian cells. Intriguingly, inhibition of the proteasome mitigates hHR23a/hPLIC2 interaction.
Keywords: hHR23a, hPLIC2, ubiquitin receptor proteins, proteasome-mediated protein degradation, ubiquitin-associated domains
Introduction
The ubiquitin-proteasome pathway plays a key regulatory role in an astounding number of cellular events, including the removal of misfolded proteins,1 production of immunocompetent peptides,2 activation or repression of transcription,3, 4 and regulation of cell cycle progression.5 Through it, proteins are ubiquitylated and consequently delivered to the 26S proteasome for degradation.6 The moieties of a polyubiquitin chain are linked by isopeptide bonds between the carbonyl carbon of the C-terminal glycine and a lysine side chain NH group. Ubiquitin contains seven lysines, and among the factors that determine whether ubiquitylation leads to proteasomal degradation is the chain length and lysine used to link the ubiquitin subunits.7 Another key factor is the recognition of the ubiquitylated protein by a ubiquitin receptor family member. In particular, proteins with attached K48-linked chains are degraded by the proteasome; however, distinct receptor pathways are required to funnel them there.8, 9
Within the ubiquitin receptor family of proteins is a group that can directly connect ubiquitylated proteins to the proteasome as their UBL domains bind the proteasome 10,11,12,13 and UBA domains bind ubiquitin 14,15,16 simultaneously.16 Depending on their protein levels UBL/UBA containing proteins can promote or inhibit the degradation of ubiquitylated substrates.9 The inhibition is caused by the UBA domains, which sequester the polyubiquitin chain to in turn prevent deubiquitylation.9,17,18
Whether ubiquitin receptor proteins act independently or collaboratively has yet to be determined. However, in yeast, UBL/UBA family members interact with each other 19,20 via UBL/UBA domain interactions.21 No such intermolecular interactions have been reported yet in humans; however, the N-terminal UBL domain of hHR23a interacts dynamically with its internal and C-terminal UBA domains.22 Evidence exists for such intramolecular interactions in the yeast Dsk2 protein. In particular, a single domain protein construct of its UBA domain interacts with itself and its UBL domain when crystallized.23
In this manuscript, we used NMR spectroscopy to demonstrate that hHR23a binds in a bidentate manner to hPLIC2, a human ortholog of Dsk2. Dsk2 functions in spindle body duplication 24 and, like hHR23a’s ortholog Rad23, has been implicated in ER-associated degradation of certain substrates.25 Our studies indicate that the UBA domain of hPLIC2 binds to hHR23a’s UBL domain, and that its UBL domain binds hHR23a’s C-terminal UBA domain. In contrast to these interactions, hPLIC2’s UBL domain does not demonstrate binding to the internal UBA domain of hHR23a, even when at 2-fold molar excess and mM protein concentrations. Our data provide an explanation for hPLIC2’s ability to bind hHR23a’s C-terminal, but not internal UBA domain. Finally, we use immunoprecipitation experiments on endogenous hHR23a and hPLIC2 proteins to demonstrate that they associate in mammalian cells. Intriguingly, proteasome inhibition substantially reduces the interaction of these two proteins. This finding provides insights into the functional significance of their interaction.
Results
HHR23a and hPLIC2 interact via UBL/UBA domain interactions
Since hHR23a’s UBL domain interacts with its own UBA domains,22 we tested whether it binds that of hPLIC2 (Figure 1(a)). We acquired [1H,15N] HSQC experiments on 15N-labeled hHR23a with or without unlabeled hPLIC2 UBA domain (Figure 1(b)). [1H,15N] HSQC experiments detect all hydrogen atoms that are attached to nitrogen, and their chemical shift values are sensitive to their chemical environment.26 We determined that addition of equimolar quantities of hPLIC2 UBA domain causes several hHR23a resonances to shift in such spectra (Figure 1(b)). The assignment of the amide resonances in the [1H,15N] HSQC spectrum of hHR23a to specific atoms was achieved in a previous study,22 and this information was used to identify the resonances that shift. Consistent with our hypothesis, all of the resonances that shift dramatically are derived from the hHR23a UBL domain. This finding offers strong evidence that the UBA domain of hPLIC2 binds hHR23a’s UBL domain.
Figure 1.
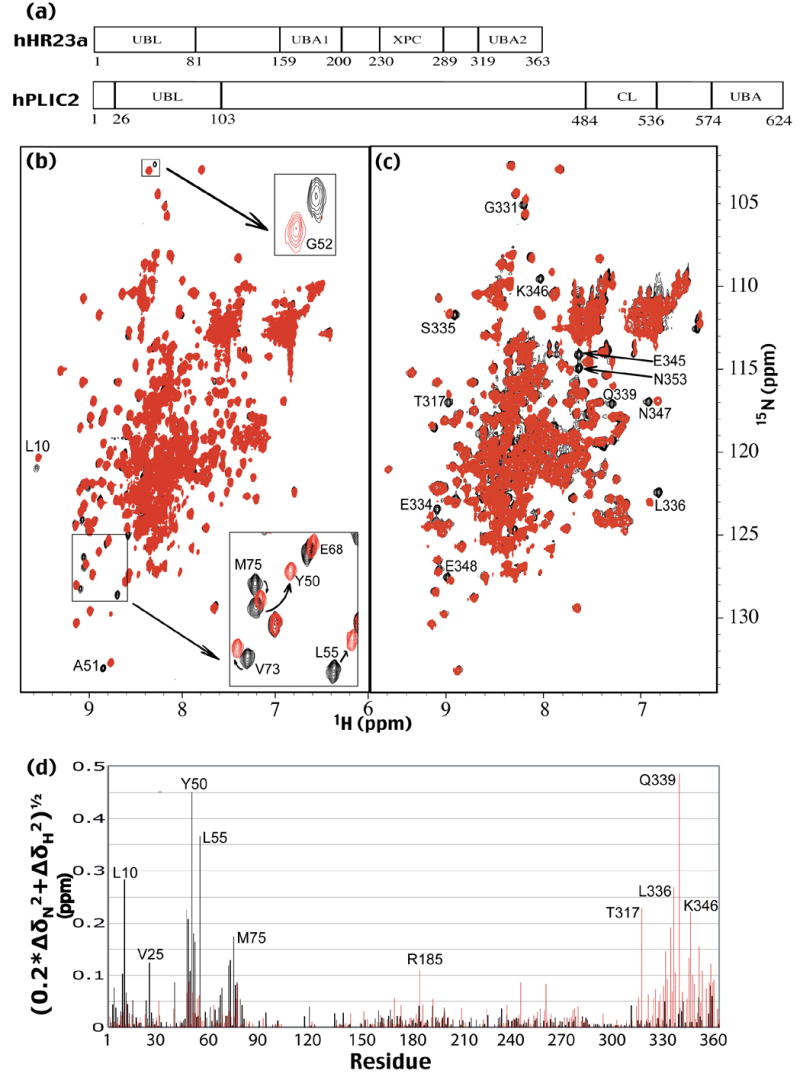
Ubiquitin recognition proteins hHR23a and hPLIC2 interact via UBL/UBA domain interactions. (a) The sequence location of hHR23a’s and hPLIC2’s UBL and UBA domains as well as hHR23a’s XPC binding domain and hPLIC2’s collagen-like domain is illustrated. (b) Comparison of the [1H,15N] HSQC spectrum of 15N-labeled hHR23a alone (black) to that acquired in the presence of equimolar amount of hPLIC2 UBA domain. The zoomed region highlights the chemical shift perturbations of hHR23a caused by hPLIC2 UBA domain addition. (c) [1H,15N] HSQC spectra are displayed of hHR23a alone (black) and with equimolar amount of hPLIC2’s UBL domain (red). Selected shifted resonances of hHR23a’s C-terminal UBA domain are labeled. Together with Figure 1(b) and (c), these data provide strong evidence for the UBL/UBA domain interactions of hHR23a and hPLIC2. (d) Chemical shift perturbation data of hHR23a caused by hPLIC2’s UBA domain (black) or hPLIC2’s UBL domain (red). The data were treated according to equation 1.
To test whether hPLIC2’s UBL domain likewise binds hHR23a’s UBA domains, we performed an analogous experiment to that described above. In particular, unlabeled hPLIC2 UBL domain was titrated into 15N-labeled hHR23a, which was monitored by [1H,15N] HSQC experiments. Interestingly, we observed significant perturbation of resonances derived from the C-terminal, but not the internal UBA domain of hHR23a when these two proteins are at equimolar concentration (Figure 1(c)). Surprisingly, only minor chemical shift perturbations are observed in the internal UBA domain even when hPLIC2 UBL domain is at 2-fold molar excess (Supplementary Figure 1). We quantified the data from Figures 1(b) and (c) according to Equation 1, where ΔδN and ΔδH represent the chemical shift perturbation value of the amide nitrogen and proton, respectively.
| (1) |
This analysis demonstrates the large chemical shift perturbations for hHR23a UBL or C-terminal UBA domain resonances upon hPLIC2 UBA (black) or UBL (red) domain addition, respectively (Figure 1(d)). It is noteworthy that the protein complex containing full-length hHR23a and hPLIC2 UBL domain is saturated at equimolar concentration, as only one residue (G331) experiences additional chemical shift perturbations when the hPLIC2 UBL domain concentration is increased from equimolar to 2-fold molar excess (Supplementary Figure 1). This finding demonstrates that the hPLIC2 UBA domain competes effectively with those of hHR23a for hHR23a’s UBL domain. Furthermore, minor chemical shift perturbations are observed for resonances derived from the UBA domains or UBL domain of hHR23a upon addition of hPLIC2’s UBA or UBL domain, respectively (Figure 1(d)). In a previous study, we demonstrated that monoubiquitin causes chemical shift perturbations for amide resonances of hHR23a’s UBL domain, as it binds hHR23a’s UBA domains and thereby abrogates the intramolecular UBL/UBA domain interactions.16 It is noteworthy that the hPLIC2 UBL domain exhibits a similar effect, and the profile of the chemical shift perturbations of hHR23a’s UBL domain mimics that which results from monoubiquitin addition. This phenomenon suggests that binding to hPLIC2, like ubiquitin binding, abrogates the intramolecular UBL/UBA domain interactions present in hHR23a.22 This hypothesis is confirmed below by using NMR relaxation experiments.
The C-terminal UBA domains of hPLIC2 and hHR23a bind to an analogous surface on each other’s UBL domain
The chemical shift perturbation data of Figure 1(d) was mapped onto a surface representation of hHR23a’s UBL domain (Figure 2(a)). In particular, the residues were colored according to a gradient derived from their chemical shift perturbation value, such that a darker blue represents a larger perturbation. This analysis reveals that the residues of the hHR23a UBL domain affected by the hPLIC2 UBA domain localize to a hydrophobic surface formed by β-strands. This surface is well known for its interactions with S5a 10, 22, 27 and its own UBA domains,22 which suggests that hPLIC2 UBA domain binding to hHR23a is mutually exclusive with these interactions.
Figure 2.
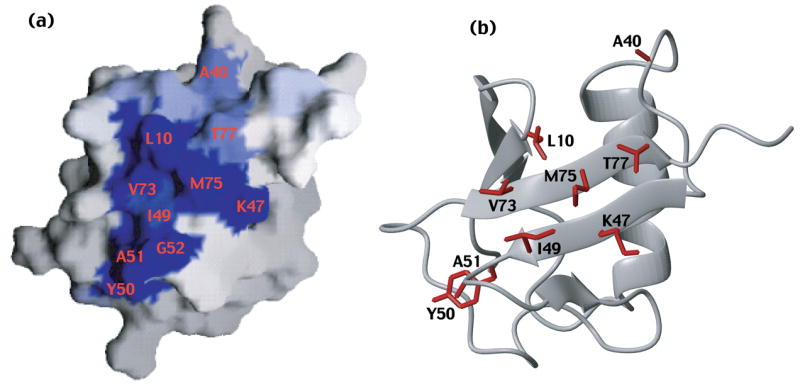
The surface of hHR23a’s UBL domain that binds hPLIC2’s UBA domain is provided. GRASP 33 and MOLMOL 34 were used to generate panels (a) and (b), respectively. The reported UBL domain coordinates for hHR23a 22 were used to generate this figure. The surface not shown, namely that rotated by 180° relative to that shown in (a) contains no perturbed residues, and is provided as Supplementary Figure 2(a).
HPLIC2 binds the proteasome and S5a with an analogous UBL domain surface compared to hHR23a.11,22,27 We tested whether this surface is also used to bind hHR23a’s C-terminal UBA domain. Unlabeled hHR23a was titrated into 15N-labeled hPLIC2 UBL domain, which was monitored by [1H,15N] HSQC experiments (Figure 3(a)). We quantified the chemical shift perturbation data according to Equation 1 (Figure 3(b)), and mapped the results onto a surface representation of hPLIC2’s UBL domain (Figure 3(c)). The assignment of the amide resonances in the [1H,15N] HSQC spectrum of hPLIC2’s UBL domain to specific atoms was achieved in a previous study,11 and prolines, which lack amide protons, were excluded from this analysis. This analysis revealed that the UBL domains of hPLIC2 and hHR23a use the same surface to bind each other’s C-terminal UBA domains (Figure 2(a) and 2(b), Figure 3(c) and 3(d)).
Figure 3.
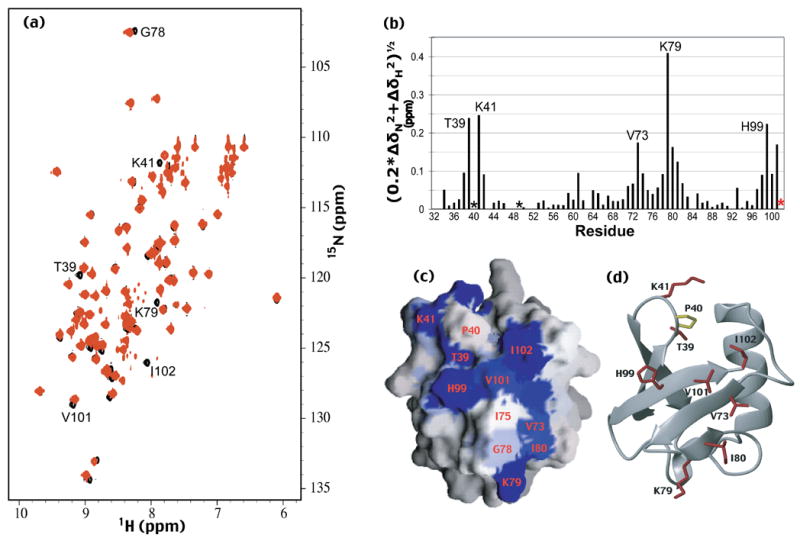
The surface of hPLIC2’s UBL domain that interacts with hHR23a is revealed. (a) [1H,15N] HSQC spectra of hPLIC2 (26-103) alone (black) and with equimolar quantities of hHR23a (red). Selected shifted or broadened resonances of hPLIC2’s UBL domain are labeled. (b) Chemical shift perturbation data for hPLIC2’s UBL domain caused by hHR23a addition. The data were treated according to equation 1. I102, whose resonance disappeared upon hHR23a addition, is indicated by a red star, whereas P40 and P49, which are excluded from this analysis, are indicated with black stars. (c and d) The surface of hPLIC2 UBL domain that binds hHR23a has been identified. GRASP 33 and MOLMOL 34 were used to generate panels (c) and (d) respectively. P40, which lacks an amide proton, is labeled in panel (c) and colored yellow in panel (d). The reported UBL domain coordinates for hPLIC2 11 were used to generate this figure. The surface not shown, namely that rotated by 180° relative to that shown in (a) contains no perturbed residues, and is provided as Supplementary Figure 2(b).
HHR23a adopts an opened conformation when bound to hPLIC2
The surface of hHR23a that binds the hPLIC2 UBA domain (Figure 2(a)) also binds S5a.22 Upon binding S5a, hHR23a adopts an opened conformation, in which its UBA domains no longer interact with the N-terminal UBL domain.22 We hypothesized hPLIC2 UBA domain binding to have the same effect. To test this, we performed 15N NMR relaxation experiments on the hHR23a/hPLIC2 UBA domain complex (Supplementary Figure 3(a) and 3(b)). These experiments probe the internal dynamics of each residue and can be used to identify flexible regions within a protein and to assess how rapidly each domain tumbles. In the unbound protein, the relaxation data reflect the UBL/UBA domain interaction by fast RN(NX) relaxation values.22 When complexed with the hPLIC2 UBA domain, the linker regions connecting each of the four structured domains of hHR23a have fast RN(NZ) and slow RN(NX), as in the free protein. Such trends indicate flexibility and, in fact, the values in these regions change little upon adding hPLIC2 UBA domain. In contrast, the RN(NX) relaxation values for residues in the UBA domains decreased substantially compared to their values in the free protein (Table 1) indicating that these domains are tumbling more labile when hHR23a is bound to hPLIC2 UBA domain. It is worth noting that the values for the UBL domain residues are also reduced, even though this domain is complexed with UBA from hPLIC2. RN(NX) relaxation values are sensitive to chemical exchange, which occurs in the free protein as each of the UBA domains competes for an overlapping surface of the UBL domain.22 Such exchange is not apparent in the hHR23a/hPLIC2 UBA domain complex, suggesting the presence of one stable protein complex in which hPLIC2’s UBA domain binds hHR23a’s UBL domain. Altogether, our chemical shift perturbation data and relaxation studies support our hypothesis that hPLIC2 UBA domain binding to hHR23a precludes intramolecular UBL/UBA domain interactions. Our data support a model in which the hHR23a/hPLIC2 complex is stabilized by intermolecular interactions between N-terminal UBL and C-terminal UBA domains (Figure 4).
Table 1.
Average Relaxation Values for Each Domain of hHR23a Alone 22 and in Complex with hPLIC2 UBA domain
| RN (NZ) (s−1)
|
RN (NX) (s−1)
|
|||
|---|---|---|---|---|
| Domain | Free | Complex | Free | Complex |
| UBL | 1.2 | 1.0 | 41.9 | 25.1 |
| UBA1 | 1.4 | 1.4 | 27.9 | 16.0 |
| XPC binding | 1.3 | 1.3 | 35.0 | 25.5 |
| UBA2 | 1.4 | 1.4 | 30.0 | 13.7 |
Figure 4.
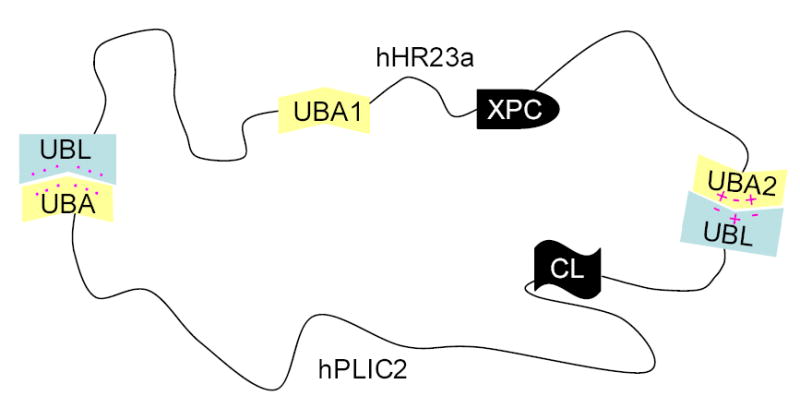
A schematic model of how hHR23a interacts with hPLIC2. A hydrophobic surface of hHR23a’s UBL domain interacts with hPLIC2’s UBA domain, whereas charged residues appear to play a role in its C-terminal UBA domain binding to hPLIC2’s UBL domain. Pink dots represent hydrophobic surfaces, whereas the “+” and “−“ symbols reflect electrostatic surfaces.
Endogenously expressed hHR23a and hPLIC2 interact in human cells
After establishing that hHR23a and hPLIC2 interact in vitro, we tested whether they associate in vivo. Towards this aim, HeLa cells were lysed and immunoprecipitated with anti-hPLIC2 antibody and protein G beads. After extensive washing, the remaining proteins were separated by SDS-PAGE and visualized with either anti-hPLIC2 or anti-hHR23a antibody. This experiment demonstrated endogenous hPLIC2 and hHR23a to interact (Figure 5 (a), lane 2). To exclude the possibility that the apparent interaction stems from nonspecific interactions with the protein G resin, we also performed a mock immunoprecipitated experiment with pre-bleed serum (Figure 5 (a), lane 1). No hHR23a protein was detected in this experiment and therefore we conclude that hPLIC2 and hHR23a interact in vivo.
Figure 5.
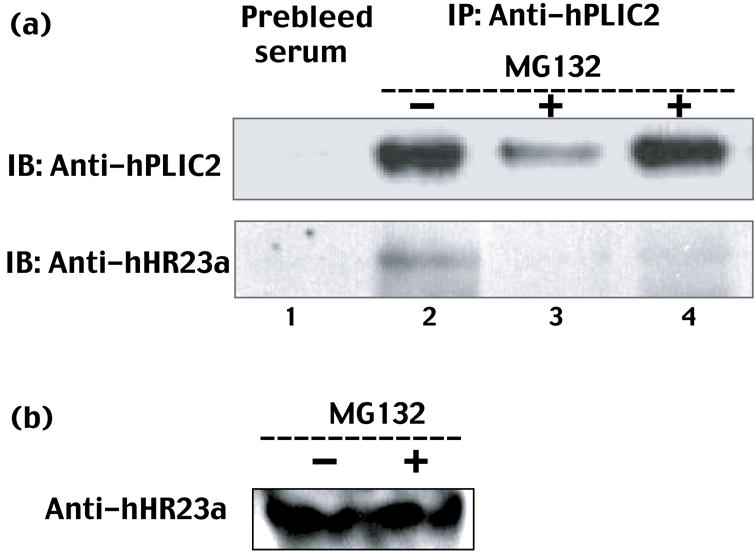
(a) Endogenously expressed hHR23a and hPLIC2 interact in human cells. Endogenous hPLIC2 from HeLa cells cultured in DMEM +10% FBS medium for 8 hours in the absence or presence of the proteasome inhibitor MG132 was immunoprecipitated with anti-hPLIC2 antibody (from Dr. Peter Howley). The immunoprecipitates were subjected to Western blot analysis using anti-hPLIC2 or anti-hHR23a (Abcam) antibody. The first lane represents a negative control in which the immunoprecipitation was performed using prebleed serum. 1000 μg of protein from the cell lysate was used for Lanes 2 and 3 whereas 5000 μg was used for Lane 4. (b) The amount of detectable endogenous hHR23a protein in HeLa cells is unperturbed by MG132 treatment. HeLa cell lysates containing 100 μg of total protein from (a) were TCA precipitated, disrupted with glass beads, and the resulting proteins resolved by SDS-PAGE and probed with anti-hHR23a (Abcam) antibody.
We tested whether hPLIC2 and hHR23a interact when the proteasome is inhibited by MG132. Under these conditions, polyubiquitylated substrates, to which these proteins bind,15,28 accumulate. Intriguingly, hHR23a association with hPLIC2 is dramatically reduced in cells treated with MG132 (Figure 5(a), lane 3 and 4). To demonstrate that the reduction is not due to either a decrease in hHR23a protein levels or to a change in its localization, we used anti-hHR23a antibody (Abcam) and Western blot analysis to detect the hHR23a protein levels in untreated and MG132 treated HeLa whole cell lysates. No obvious change to the level of detected hHR23a protein was observed after MG132 treatment (Figure 5(b)). This finding suggests that these two proteins dissociate to bind different ubiquitylated substrates.
Discussion
In this manuscript, we demonstrate the two human UBL/UBA family members hHR23a and hPLIC2 to interact in vitro as well as in vivo. Our study complements others, which demonstrate that the ubiquitin-proteasome pathway is plagued with packing problems, as the same β-sheet surface of ubiquitin and ubiquitin-like domains is required for intra- and inter-molecular interactions.10,11,22,27,29,30 Indeed, critical to understanding the contribution of these interactions to ubiquitin-mediated protein degradation is an understanding of how it is determined which interactions will dominate and when.
We attempted to use chemical shift perturbation analysis as was done for the UBL domains to determine the hHR23a UBA2 domain surface responsible for binding hPLIC2 UBL domain. This analysis was unable to reveal a single surface, most likely because residues undergo chemical shift perturbations from two distinct processes, namely the new interaction with the hPLIC2 UBL domain and lost one with the hHR23a UBL domain. However, T317 is among the hHR23a UBA2 domain residues that exhibit significant chemical shift perturbations upon hPLIC2 UBL domain addition. One of the key structural differences between hHR23a’s UBA domains is that the C-terminal one contains an ordered strand formed by V316-Q319 that extends towards this domain’s second α-helix. Unique interactions involving this region likely contribute to hPLIC2’s preference for the C-terminal UBA domain of hHR23a. In addition, our chemical shift perturbation data implicate L336, Q339 and K346 (Figure 1(d), red) as participating in binding the hPLIC2 UBL domain. These residues are not conserved in hHR23a’s internal UBA domain and only Q339 is conserved in hPLIC2’s UBA domain (Figure 6(a)). These changes likely contribute to hHR23a’s C-terminal UBA domain preference for hPLIC2’s UBL domain over its own (Figure 6(b)). In addition, hPLIC2’s UBA domain can displace those of hHR23a at equimolar concentration, suggesting that hHR23a’s UBL domain similarly prefers intermolecular interaction with hPLIC2. To date, full-length hPLIC2 has not been expressed and purified from E. coli, but we expect the full-length protein to exhibit an even higher affinity for hHR23a, as the two UBL/UBA domain binding modes can occur simultaneously. In addition, it is possible that higher order oligomers form as the UBL and UBA domains bind different molecules. This possibility is more likely however at high concentrations, above that expected in the cellular environment.
Figure 6.
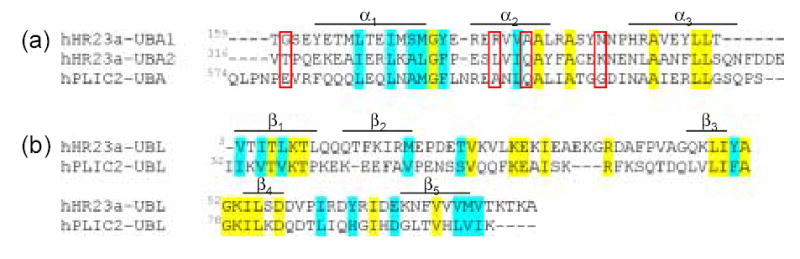
Sequence alignment of the UBA (a) and UBL (b) domains from hHR23a and hPLIC2. Identical and conserved residues are highlighted in yellow and blue, respectively. T317, L336, Q339 and K346 from hHR23a are implicated by NMR titration experiments (Figure 1(d)) as interacting with hPLIC2’s UBL domain and are boxed. Secondary structural elements for hHR23a’s C-terminal UBA (a) and UBL (b) domains are included.
Despite their interaction, UBL/UBA family members do not appear to interfere with each other’s ability to bind ubiquitin or ubiquitylated substrates. To the contrary, in yeast Rad23 and Ddi1 bind a common tetraubiquitin chain 21 and therefore may collaborate in delivering ubiquitylated substrates to the proteasome. We demonstrate here that endogenous hPLIC2 and hHR23a interaction is severely reduced in cells treated with MG132 (Figure 5(a)). This finding offers strong evidence that their protein complex dissociates to allow each protein to bind their ubiquitylated substrates. It is possible, however, that hHR23a and hPLIC2 can interfere with each other’s ability to bind the proteasome, which would impact the delivery of ubiquitylated substrates to the proteasome. At high protein levels, UBL/UBA family members do inhibit the degradation of ubiquitylated substrates.9 This effect, however, may be dominated by the UBA domains sequestering the polyubiquitin chains, and thereby preventing their deubiquitylation.9,17,18
Materials and Methods
Sample preparation
Dr. Peter Howley generously provided us with the plasmids for the expression of GST tagged hPLIC2 UBL domain [pGEX-2T-hPLIC2 (26–103)] and GST tagged hPLIC2 UBA domain [pGEX-2T-hPLIC2 (574–624)]. The plasmids containing these genes were each transformed into BL21 (DE3) cells and grown at 37°C in M9 minimal medium or in Luria broth containing ampicillin (100 μg/ml). The cells were harvested three hours after protein expression was induced with 0.4 mM isopropyl β-D-thiogalactoside (IPTG). The proteins were purified by using affinity purification on glutathione S-sepharose resin as described previously.22 Further purification was achieved on an FPLC system (Pharmacia), by either Superdex 200 (for hHR23a) or 75 (for UBL and UBA domains of hPLIC2) preparative columns. 15N labeled samples for NMR spectroscopy were produced by growth and expression in M9 minimal media with 15N NH4Cl as the only source of nitrogen.
NMR spectroscopy
All NMR samples were dissolved in 20 mM NaPO4 (pH 6.5), 30 mM NaCl, 0.1% NaN3, and 10% D2O. Spectra were acquired at 25°C on Varian NMR spectrometers operating at either 800 or 600 MHz. Processing was performed in NMRPipe 31 and the resulting spectra were visualized in XEASY.32 Protein concentrations were calculated by using extinction coefficients based on amino acid composition and absorbance at 280 nm for protein dissolved in 6M guanidium hydrochloride. Rates for 15N longitudinal (RN(NZ)) and transverse (RN(NX)) relaxation were recorded on hHR23a with 2-fold molar excess hPLIC2 UBA domain. The data were collected at 800 MHz with a cryogenically cooled probe and at 25°C and pH 6.5. In this complex, hHR23a was 0.4 mM and 15N-labeled whereas hPLIC2 UBA domain was unlabeled. RN(NZ) and RN(NX) were derived by fitting data acquired with different relaxation delays to a single exponential decay function and error values were determined by repeating one data point.
In vivo experiments
HeLa cells were cultured in DMEM (Fisher) +10% FBS (Invitrogen) medium for 8 hours, in the absence or presence of 20 μM MG132. Cells were harvested and then lysed with TBS buffer (50mM Tris-HCl pH7.4, 150mM NaCl) containing 1 mM EDTA, 1% Triton X-100, and protease inhibitor cocktail (Roche). Nuclei and insoluble debris were removed by centrifugation. In figure 5(a), immunoprecipitation of hPLIC2 was performed by using a previously published method.28 Briefly, 2 μl of anti-hPLIC2 or pre-bleed (as a control) rabbit serum was added to 1 ml of lysate containing 1000 μg total protein prepared from a 10 cm dish of adherent cells. After rotating for an hour at 4°C, 20 μl of protein G beads was added and the solution rotated for another 30 minutes at 4°C. The beads were subsequently spun down, washed several times in the lysis buffer, and the proteins separated by SDS-PAGE. HPLIC2 or hHR23a were visualized with either anti-hPLIC2 (from Dr. Peter Howley) or anti-hHR23a (Abcam) antibody. In figure 5(b), 200 μl of HeLa cell lysate containing 100 μg of total protein was precipitated by 10% TCA, disrupted with 80 μl of glass beads. The proteins were resolved by SDS-PAGE and probed with anti-hHR23a (Abcam) antibody.
Supplementary Material
Supplementary Figure 1: Comparison of the [1H,15N] HSQC spectrum of 15N-labeled hHR23a with equimolar quantities (black) or 2-fold molar excess of hPLIC2 UBL domain (red). G331 is labeled, as it is the only resonance that shifts from the presence of additional hPLIC2 UBL domain.
Supplementary Figure 2: The structure of (a) and (b) is rotated 180° relative to that of Figure 2a and 3a respectively. The program of GRASP 33 is used to generate this figure.
Supplementary Figure 3: Binding to the hPLIC2 UBA domain alters the hHR23a protein structure. Rates for 15N longitudinal [RN(NZ)] (a) and transverse [RN(NX)] (b) relaxation for the hHR23a-hPLIC2 (574-624) complex are provided. These data were collected at 800 MHz with a cryogenically cooled probe, 25°C and pH 6.5 on a sample containing 2-fold molar excess hPLIC2 UBA. Randomly coiled regions and structured domains are represented in black and red, respectively.
Acknowledgments
We are grateful to Dr. Peter M. Howley and Dr. Maurits Kleijnen for useful discussions and for providing us with anti-hPLIC2 antibody and hPLIC2 DNA constructs. NMR data were acquired in the NMR facility of the UMN and we thank Dr. David Live and Dr. Beverly Ostrowsky for their technical assistance. Data processing and visualization occurred in the Minnesota Supercomputing Institute Basic Sciences Computing Lab. This work was funded by grants from the National Institutes of Health CA097004 (KJW), Minnesota Medical Foundation (NMR facility), NSF BIR-961477 (NMR facility).
Abbreviations
- hHR23
human homologue of Rad23
- hPLIC2
human homolog 2 of protein linking integrin-associated protein and cytoskeleton
- XPC
xeroderma pigmentosum C
- CL
collagen like
- HSQC
heteronuclear single quantum coherence
- NMR
nuclear magnetic resonance
- UBA
ubiquitin-associated
- UBL
ubiquitin-like
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Schubert U, Anton LC, Gibbs J, Norbury CC, Yewdell JW, Bennink JR. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770–774. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- 2.Rock KL, Goldberg AL. Degradation of cell proteins and the generation of MHC class I-presented peptides. Annu Rev Immunol. 1999;17:739–779. doi: 10.1146/annurev.immunol.17.1.739. [DOI] [PubMed] [Google Scholar]
- 3.Conaway RC, Brower CS, Conaway JW. Emerging roles of ubiquitin in transcription regulation. Science. 2002;296:1254–1258. doi: 10.1126/science.1067466. [DOI] [PubMed] [Google Scholar]
- 4.Muratani M, Tansey WP. How the ubiquitin-proteasome system controls transcription. Nat Rev Mol Cell Biol. 2003;4:192–201. doi: 10.1038/nrm1049. [DOI] [PubMed] [Google Scholar]
- 5.Yamaguchi R, Dutta A. Proteasome inhibitors alter the orderly progression of DNA synthesis during S-phase in HeLa cells and lead to replication of DNA. Exp Cell Res. 2000;261:271–283. doi: 10.1006/excr.2000.5053. [DOI] [PubMed] [Google Scholar]
- 6.Ciechanover A. The ubiquitin-proteasome proteolytic pathway. Cell. 1994;79:13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- 7.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. Embo J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elsasser S, Chandler-Militello D, Muller B, Hanna J, Finley D. Rad23 and Rpn10 serve as alternative ubiquitin receptors for the proteasome. J Biol Chem. 2004;279:26817–26822. doi: 10.1074/jbc.M404020200. [DOI] [PubMed] [Google Scholar]
- 9.Verma R, Oania R, Graumann J, Deshaies RJ. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell. 2004;118:99–110. doi: 10.1016/j.cell.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 10.Hiyama H, Yokoi M, Masutani C, Sugasawa K, Maekawa T, Tanaka K, Hoeijmakers JH, Hanaoka F. Interaction of hHR23 with S5a. The ubiquitin-like domain of hHR23 mediates interaction with S5a subunit of 26 S proteasome. J Biol Chem. 1999;274:28019–28025. doi: 10.1074/jbc.274.39.28019. [DOI] [PubMed] [Google Scholar]
- 11.Walters KJ, Kleijnen MF, Goh AM, Wagner G, Howley PM. Structural studies of the interaction between ubiquitin family proteins and proteasome subunit S5a. Biochemistry. 2002;41:1767–1777. doi: 10.1021/bi011892y. [DOI] [PubMed] [Google Scholar]
- 12.Elsasser S, Gali RR, Schwickart M, Larsen CN, Leggett DS, Muller B, Feng MT, Tubing F, Dittmar GA, Finley D. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat Cell Biol. 2002;4:725–730. doi: 10.1038/ncb845. [DOI] [PubMed] [Google Scholar]
- 13.Saeki Y, Sone T, Toh-e A, Yokosawa H. Identification of ubiquitin-like protein-binding subunits of the 26S proteasome. Biochem Biophys Res Commun. 2002;296:813–819. doi: 10.1016/s0006-291x(02)02002-8. [DOI] [PubMed] [Google Scholar]
- 14.Wilkinson CR, Seeger M, Hartmann-Petersen R, Stone M, Wallace M, Semple C, Gordon C. Proteins containing the UBA domain are able to bind to multi-ubiquitin chains. Nat Cell Biol. 2001;3:939–943. doi: 10.1038/ncb1001-939. [DOI] [PubMed] [Google Scholar]
- 15.Bertolaet BL, Clarke DJ, Wolff M, Watson MH, Henze M, Divita G, Reed SI. UBA domains of DNA damage-inducible proteins interact with ubiquitin. Nat Struct Biol. 2001;8:417–422. doi: 10.1038/87575. [DOI] [PubMed] [Google Scholar]
- 16.Wang Q, Goh AM, Howley PM, Walters KJ. Ubiquitin recognition by the DNA repair protein hHR23a. Biochemistry. 2003;42:13529–13535. doi: 10.1021/bi035391j. [DOI] [PubMed] [Google Scholar]
- 17.Ortolan TG, Tongaonkar P, Lambertson D, Chen L, Schauber C, Madura K. The DNA repair protein Rad23 is a negative regulator of multi-ubiquitin chain assembly. Nat Cell Biol. 2000;2:601–608. doi: 10.1038/35023547. [DOI] [PubMed] [Google Scholar]
- 18.Raasi S, Pickart CM. Rad23 UBA domains inhibit 26S proteasome-catalyzed proteolysis by sequestering lysine 48-linked polyubiquitin chains. J Biol Chem. 2003;278:8951–8959. doi: 10.1074/jbc.m212841200. [DOI] [PubMed] [Google Scholar]
- 19.Bertolaet BL, Clarke DJ, Wolff M, Watson MH, Henze M, Divita G, Reed SI. UBA domains mediate protein-protein interactions between two DNA damage- inducible proteins. J Mol Biol. 2001;313:955–963. doi: 10.1006/jmbi.2001.5105. [DOI] [PubMed] [Google Scholar]
- 20.Rao H, Sastry A. Recognition of specific ubiquitin conjugates is important for the proteolytic functions of the ubiquitin-associated domain proteins Dsk2 and Rad23. J Biol Chem. 2002;277:11691–11695. doi: 10.1074/jbc.M200245200. [DOI] [PubMed] [Google Scholar]
- 21.Kang Y, Vossler RA, Diaz-Martinez LA, Winter NS, Clarke DJ, Walters KJ. UBL/UBA ubiquitin receptor proteins bind a common tetraubiquitin chain. J Mol Biol. 2006;356:1027–1035. doi: 10.1016/j.jmb.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 22.Walters KJ, Lech PJ, Goh AM, Wang Q, Howley PM. DNA-repair protein hHR23a alters its protein structure upon binding proteasomal subunit S5a. Proc Natl Acad Sci USA. 2003;100:12694–12699. doi: 10.1073/pnas.1634989100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lowe ED, Hasan N, Trempe JF, Fonso L, Noble ME, Endicott JA, Johnson LN, Brown NR. Structures of the Dsk2 UBL and UBA domains and their complex. Acta Crystallogr D Biol Crystallogr. 2006;62:177–188. doi: 10.1107/S0907444905037777. [DOI] [PubMed] [Google Scholar]
- 24.Biggins S, Ivanovska I, Rose MD. Yeast ubiquitin-like genes are involved in duplication of the microtubule organizing center. J Cell Biol. 1996;133:1331–1346. doi: 10.1083/jcb.133.6.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Medicherla B, Kostova Z, Schaefer A, Wolf DH. A genomic screen identifies Dsk2p and Rad23p as essential components of ER-associated degradation. EMBO Rep. 2004;5:692–697. doi: 10.1038/sj.embor.7400164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wüthrich K. NMR of Proteins and Nucleic Acids. Wiley; New York: 1986. [Google Scholar]
- 27.Mueller TD, Feigon J. Structural determinants for the binding of ubiquitin-like domains to the proteasome. EMBO J. 2003;22:4634–4645. doi: 10.1093/emboj/cdg467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kleijnen MF, Shih AH, Zhou P, Kumar S, Soccio RE, Kedersha NL, Gill G, Howley PM. The hPLIC proteins may provide a link between the ubiquitination machinery and the proteasome. Mol Cell. 2000;6:409–419. doi: 10.1016/s1097-2765(00)00040-x. [DOI] [PubMed] [Google Scholar]
- 29.Varadan R, Walker O, Pickart C, Fushman D. Structural properties of polyubiquitin chains in solution. J Mol Biol. 2002;324:637–647. doi: 10.1016/s0022-2836(02)01198-1. [DOI] [PubMed] [Google Scholar]
- 30.Wang Q, Young P, Walters KJ. Structure of S5a bound to monoubiquitin provides a model for polyubiquitin recognition. J Mol Biol. 2005;348:727–739. doi: 10.1016/j.jmb.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 31.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 32.Bartels C, Xia TH, Billeter M, Güntert P, Wüthrich K. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J Biomol NMR. 1995;6:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- 33.Nicholls AJ. GRASP Manual. Columbia University; New York: 1993. [Google Scholar]
- 34.Koradi R, Billeter M, Wüthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Comparison of the [1H,15N] HSQC spectrum of 15N-labeled hHR23a with equimolar quantities (black) or 2-fold molar excess of hPLIC2 UBL domain (red). G331 is labeled, as it is the only resonance that shifts from the presence of additional hPLIC2 UBL domain.
Supplementary Figure 2: The structure of (a) and (b) is rotated 180° relative to that of Figure 2a and 3a respectively. The program of GRASP 33 is used to generate this figure.
Supplementary Figure 3: Binding to the hPLIC2 UBA domain alters the hHR23a protein structure. Rates for 15N longitudinal [RN(NZ)] (a) and transverse [RN(NX)] (b) relaxation for the hHR23a-hPLIC2 (574-624) complex are provided. These data were collected at 800 MHz with a cryogenically cooled probe, 25°C and pH 6.5 on a sample containing 2-fold molar excess hPLIC2 UBA. Randomly coiled regions and structured domains are represented in black and red, respectively.


