Abstract
A dissection of longevity in Caenorhabditis elegans reveals that animal life span is influenced by genes, environment, and stochastic factors. From molecules to physiology, a remarkable degree of evolutionary conservation is seen.
Introduction
Over the last 20 years, fundamental insights into the biology of aging have emerged through the study of model genetic organisms, where undoubtedly the tiny nematode C. elegans has led the way. Importantly, the discovery that the single gene mutants age-1 and daf-2 could extend the short three-week life span 1–2-fold revealed that longevity is under genetic control [1,2]. Long life was shown to depend on another locus, daf-16, defining an epistasis pathway for this process. Moreover, the molecular identification of all three as components of insulin/IGF-I signaling (IIS) [3–6] eventually led to the striking realization that a modest downregulation of IIS promotes stress resistance and longevity across taxa [7–10].
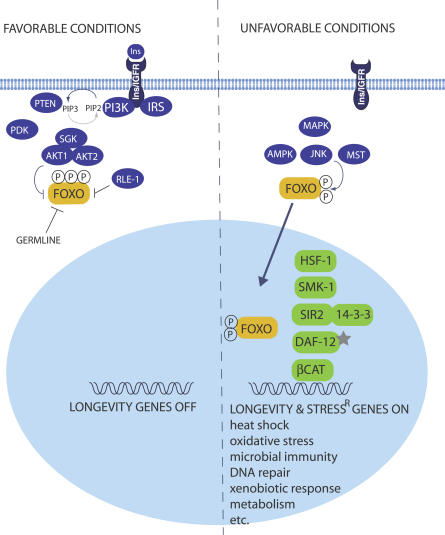
Molecular genetic studies suggest that in response to insulin-like peptides (ILPs) [11,12], activation of the DAF-2/insulin/IGF-1 receptor tyrosine kinase triggers a PI3/AKT/SGK kinase cascade that phosphorylates the DAF-16/FOXO transcription factor [13–16] (Figure 1). Consequently, DAF-16/FOXO is retained cytoplasmically and animals live normal life spans. In response to increased DAF-18/PTEN activity [14,17], stress (heat, oxidative, starvation) or reduced IIS, DAF-16/FOXO enters the nucleus [18–20], where it turns on survival genes, including those that manage oxidative stress, heat shock, innate immunity, metabolism, autophagy, and xenobiotic response, among others [21–25]. Indeed, over- or underexpression of such targets often impacts stress resistance and longevity [22,26]. Many of these points and more have been reviewed elsewhere [27,28].
Figure 1. A Model for Insulin/IGF-1 Signal Transduction, Showing Associated Kinase Inputs and Nuclear Factors (See Text).
Importantly, IIS is best understood as a signaling pathway selected in evolution to regulate organismal survival, not aging per se. Indeed, during larval development, DAF-16/FOXO specifies the dauer diapause, a long-lived stress-resistant larval stage specialized for survival, but can independently trigger longevity in adults [29]. Evidently, the primary role of IIS is to invest in somatic endurance to outlast hard times, and conversely specify growth and reproduction in good times, with secondary consequences on adult life span. Consistent with this, many longevity mutants have reduced Darwinian fitness under conditions favoring short-term rapid reproductive output [30].
Since the elucidation of this core pathway, scores of longevity genes have emerged from candidate approaches, unbiased genome-wide RNAi screens, and transcriptional profiles, some working through IIS, others not [22,31–35]. Because they cannot all be enumerated here, we have taken an admittedly “transduction-centric” view to highlight new developments. We start with upstream signaling events of sensory perception, walk through new signaling inputs, focus on transcriptional factors, and then discuss broad physiological processes (respiration, translation, checkpoint control, diet restriction), which may impact longevity through hormesis, the induction of enhanced robustness and survival by exposure to low levels of stress [36].
Sensory Input: The Senseless Liveth
Critical for making preemptive decisions that prepare the animal for times ahead, sensory perception can also dramatically influence life span. Mutants deficient in sensory neuron structure and function have extended longevity, which is largely but not entirely daf-16 dependent [37]. Moreover, such mutants cause DAF-16/FOXO to relocate to the nucleus [19]. Components of sensory cell signal transduction, including G-protein coupled receptors, G-proteins, cGMP channel subunits, ciliary proteins, Tubby, and proteins implicated in synaptic transmission [37–42] are thought to regulate release of ILPs from sensory cells, thereby influencing organismal physiology. Cell ablation studies show that both gustatory and olfactory neurons contribute, with specific neurons promoting longevity and others suppressing it [38]. The exact sensory cues governing ILP synthesis and release are likely to include nutrients, and various repellants or attractants, but this area remains largely unexplored. Related findings have recently emerged in Drosophila, in which or83b mutants, broadly deficient in olfaction, live long [43]. Moreover, odorants from yeast paste reduce longevity induced under conditions of dietary restriction. Thus, perception as well as ingestion may impact animal life span in diverse taxa.
In addition to sensory cues, serotonergic inputs may work upstream of IIS. Notably, mutants in the serotonergic receptor ser-1 are longer lived, in a daf-16/FOXO-dependent manner [44]. Surprisingly, mutants in ser-4, another serotonin receptor, are somewhat short lived, suggesting the receptors could act antagonistically, similar to the cells described above. Moreover, mutants deficient in serotonin production, such as tph-1, are stress resistant, but not long lived [44–46]; opposing effects of the receptors might explain this phenomenon. Because serotonergic signaling is phyletically ancient, it may have a conserved physiologic role in life span regulation, and be amenable to pharmacologic intervention.
Signal Transduction: Wiring Longevity
Aside from IIS, several other transduction pathways appear to impinge upon DAF-16/FOXO. Whereas the IIS kinase inputs described above inhibit DAF-16/FOXO, several kinases described below stimulate it (Figure 1). JUN kinase, a mediator of the stress response, results in life span extension, increased resistance to oxidative stress, and DAF-16/FOXO nuclear localization upon overexpression [47]. Longevity is daf-16/FOXO dependent, and FOXO is a JNK substrate. Because JNK overexpression further increases daf-2/IIR mutant longevity, it may work in parallel to PI3/AKT. Importantly, this role is evolutionarily conserved. In D. melanogaster, JNK activation specifically within insulin-producing neurons results in nuclear localization of dFOXO, suppression of insulin production, and extension of life span [48,49]. Intriguingly, this suggests that the organismal stress response can be coordinated centrally. MST is a ste20-like kinase implicated in growth control. Like JNK-1, overexpression of the C. elegans homolog slows aging, extends life span in a daf-16-dependent manner, and further extends longevity of daf-2/RNAi treated animals [50]. Moreover, loss of function reduces daf-2 longevity. Although JNK and MST phosphorylate different residues, it is unclear if they identify parallel pathways or the same pathway. The mammalian MST1 also activates FOXO, but instead stimulates apoptosis in primary neurons in response to oxidative stress, i.e., decreasing cellular survival [50]. Conceivably, the mammalian response may be tissue specific. The p38 MAP kinase pathway, implicated in the C. elegans stress response and innate immunity as well as RAS signaling, is required in part for daf-2 longevity [51,52]. Finally, mutants of AMP-activated protein kinase (AMPK), a fuel sensor sensitive to AMP/ATP ratios, suppress daf-2 longevity, while overexpression modestly extends life [53]. How these and other kinases interact to control FOXO activity is not well understood, and it will be fascinating to deduce the kinase inputs, decipher the signaling specificity, and elucidate the covalent code modulating FOXO activity.
Aside from phosphorylation, FOXO is also covalently modified by ubiquitination and acetylation (below). Recently, an evolutionarily conserved E3 ubiquitin ligase, called RLE-1, has been shown to ubiquitinate DAF-16/FOXO, leading to its destabilization [54]. Conversely, RLE-1 loss results in increased levels of FOXO protein, sustained nuclear localization, stress resistance, and longevity. As expected, the longevity of rle-1 mutants is daf-16 dependent. RLE-1 may be one of several ubiquitin ligases that controls FOXO stability.
Transcriptional Control: The Master Regulators of Survival
As the discussion above reveals, DAF-16/FOXO is a master regulator of survival that integrates multiple inputs. Likewise, several different transcription factors/cofactors work in concert with DAF-16/FOXO to modulate its output (Figure 1). The conserved nuclear factor, SMK-1, is responsible for mediating specific aspects of FOXO biology, namely IIS and germline longevity and pathogen, UV, and oxidative-stress resistance, but not heat resistance and dauer formation [55]. Conceivably, SMK-1 works as a coregulator or cofactor for DAF-16/FOXO, since it modulates its transcriptional output. Moreover, SMK-1 harbors LXXLL motifs typical of transcriptional coactivators, but its exact molecular activity remains to be determined. In response to thermal stress, HSF-1, a heat shock transcription factor, induces various heat shock chaperones, which clear misfolded proteins and protect cells. Loss of activity accelerates proteotoxicity and tissue aging, while overexpression increases heat resistance and extends life span in a daf-16(+)-dependent manner. Moreover, hsf-1 is required for daf-2 longevity [56,57] as well as the innate immune response, broadening the spectrum of its age-related functions [58].
Best known for its developmental role in wnt signal transduction, β-catenin also guards cells against oxidative stress. Mutants in the C. elegans homolog, bar-1, are more susceptible to oxidative stress and are short lived [59]. Interestingly, BAR-1 physically associates with DAF-16/FOXO, as do the mammalian counterparts. Moreover, β-catenin enhances superoxide dismutase expression in both systems. However, whether bar-1 or other components of wnt signaling mediate daf-2 longevity has not been directly tested.
Sirtuins are NAD+-dependent protein deacetylases whose increased dose augments longevity in yeast, worms, and flies [60–62]. In mammals, sirtuins deacetylate a number of targets, including p53, PGC-1alpha, and FOXO, to name a few [63–67], thereby modulating their transcriptional activity. Notably, extra copies of C. elegans sir-2.1 enhance longevity and stress resistance, in a manner dependent on daf-16/FOXO and 14-3-3 scaffold proteins. Evidence suggests SIR-2.1 may work in parallel to IIS [61,68,69]. Indeed, in response to stress (but not reduced IIS), SIR-2.1,14-3-3 protein and DAF-16/FOXO form a nuclear complex that activates FOXO transcriptional targets such as sod-3.
SIR-2.1 may also have FOXO-independent outputs. Sirtuins are stimulated by polyphenolics such as resveratrol [70], one of the salutary compounds found in red wine. Interestingly, resveratrol extends the life span of yeast, worms, flies, and perhaps mice, in a sirtuin-dependent manner [71,72]. In C. elegans, resveratrol induces abu-11 and other components of the endoplasmic reticulum (ER) stress response [73]. The ER stress response is stimulated as a result of misfolded or misglycosylated proteins in the secretory pathway. ABU-11 and related molecules are transmembrane proteins similar to scavenger receptors, and are proposed to target secretory components for lysosomal degradation [74]. Consistent with a rate-limiting role, abu-11 RNAi abrogates resveratrol-mediated longevity and overexpression extends life, while, surprisingly, daf-16/FOXO has little effect.
Nuclear hormone receptors (NHR) transcription factors regulate gene expression in response to lipophilic hormones, and are well poised to coordinate organismal physiology. C. elegans DAF-12/NHR, a relative of vertebrate vitamin D and LXR receptors, promotes longevity in several contexts. One is in the germline longevity pathway. When germline stem cells are removed, either by mutation or by laser microsurgery, animals live 50%–60% longer than wild type [75,76]. This is not due simply to sterility, since additional removal of somatic gonadal support cells produces infertile animals with normal life span. Conceivably, germline and somatic gonad produce opposing signals that coordinate organismal maturation, with secondary consequences on aging. Although distinct from IIS, germline longevity depends on functional DAF-16/FOXO as well as DAF-12/NHR [76]. Moreover, FOXO enters intestinal nuclei upon germline removal, indicating a signaling event [19]. Notably, mutants deficient in biosynthesis of DAF-12 ligands, as well as the conserved gene kri-1, also abolish germline longevity, and diminish FOXO nuclear accumulation [77–80]. Endogenous DAF-12 ligands include 3-keto bile acid-like steroids, called dafachronic acids [81], as well as the structurally related 25S-cholestenoic acid [82], which also serves as ligand for the mammalian homolog LXR [83]. As predicted from the genetics, ligand supplementation of hormone biosynthetic mutants restores DAF-16/FOXO nuclear localization and longevity of germlineless animals [80]. Recently, the steroid pregnenolone has been reportedly found in higher concentrations in germline-less animals compared to wild type [84]. Moreover, exogenous pregnenolone enhances longevity in a daf-12(+)-dependent fashion. However, pregnenolone is not a DAF-12/NHR ligand, suggesting an indirect effect. DAF-12 is also required for longevity in the dauer pathways. Biosynthetic mutants devoid of dafachronic acids, such as daf-9/cytochrome P450, manifest adult longevity and stress resistance that is daf-12(+) dependent [79,85,86]. Moreover, hormone supplementation restores normal life span and stress resistance [80]. Altogether, these findings provide crucial evidence for bile acid–like steroids modulating animal life span.
MicroRNAs: Timing Long Life?
The heterochronic loci control C. elegans larval developmental timing, specifying stage specific programs and the life plan, but curiously, a handful also impact adult life span. lin-4 encodes an evolutionarily conserved microRNA, which downregulates the nuclear factor LIN-14 via translational inhibition of the 3′ UTR [87,88]. These genes work in tandem in a timing switch that advances early larval stage programs. With respect to adult longevity, lin-4 mutants are short lived, while lin-14 mutants are long lived, suggesting that lin-4(+) retards aging through downregulation of lin-14(+) [89]. Interestingly, longevity is also daf-16/FOXO dependent. Conceivably, long life arises from perturbations of a timer in the adult. Alternatively, lin-4/lin-14 may regulate IIS directly, since the insulin-like gene, ins-33, is a bona fide LIN-14 transcriptional target [90]. Intriguingly, numerous microRNAs undergo distinct changes in expression pattern during adult life, including those predicted to target known regulators of survival [91]. Perhaps they too function in adult aging.
Checkpoint Control
A critical facet of cellular function is the response to DNA damage, genotoxic stress, and other insults. Aging in higher animals may be influenced by the balance of cell survival versus death, a decision often governed by checkpoint proteins in dividing cells. However, adult C. elegans animals lack dividing cells except for the germline. Surprisingly, then, deficiencies in checkpoint control—including cid-1 poly(A+) polymerase, chk-1 kinase, cdc-25 phosphatase, as well as clk-2/RAD5—result in longer adult life but reduced broods [92,93]. Moreover, checkpoint-deficient animals are typically thermotolerant and upregulate sod-3/MnSOD (in intestine) and the ER resident hsp-4/BiP (in vulva), showing molecular induction of a stress response. However, chk-1 RNAi longevity is daf-16/FOXO independent, and therefore likely works through another transcriptional regulator. A good candidate is cep-1, whose mammalian counterpart, p53, works downstream of chk1. In mammals, p53 loss increases tumorigenesis, while specific gain-of-function alleles reduce tumor incidence but accelerate aging, suggesting a trade-off between tumor surveillance and stem cell maintenance [94]. Intriguingly then, disabling checkpoint function in C. elegans mitotic germline cells or postmitotic somatic cells triggers states of enhanced organismal survival. Modulating checkpoints so that benefits are not outweighed by detriments remains a future challenge.
Mitochondrial-Induced Longevity
Mitochondrial function is central to cellular metabolism and apoptosis, while dysfunction causes numerous age-associated diseases, including diabetes, cardiomyopathy, and neurodegeneration. Surprisingly, then, a major class of C. elegans longevity mutants are deficient in mitochondrial function. The first described include clk-1, which is defective in ubiquinone biosynthesis, and isp-1, which lacks a Rieske FeS protein of complex III [93,95,96]. Genome-wide RNAi screens identified many other mitochondrial components, all of which extend life independent of daf-16 and daf-2 [33,34]. These mutants display increased developmental times, slowed behavior, and smaller broods. As expected, most diminish respiration, although it is controversial whether clk-1 does or not [97,98]. Evidently, levels of mitochondrial gene activity must be optimized, since severe loss of function results in lethality or shortevity [99]. How might disabling mitochondria provoke extended survival? Conceivably, it produces a lower rate of living, and consequently decreased production of reactive oxygen species (ROS) [96]. Alternately, perturbation of electron transport may actually increase ROS, provoking a hormetic adaptive response [99]. Or it may stimulate mitochondrial turnover and thrifty metabolism. In any case, longevity likely depends in part on signaling, since molecules such as aak-2/AMPK are required [100]. Can this example illuminate aging in higher organisms? Perhaps yes, since heterozygous knockouts of the mouse clk-1 are longer lived, suggesting that partial loss of function can confer benefits [101].
Dietary Restriction–Induced Longevity
Dietary restriction (DR), the reduction of dietary intake without malnutrition, extends life span from yeast to rodents, and likely involves evolutionarily conserved mechanisms. In C. elegans, DR is induced by various regimens, including limiting the amount of bacterial food in liquid culture [102], liquid growth in a defined synthetic medium [103], or limiting dietary intake with feeding mutants such as eat-2 [104]. Even shifting adults onto agar plates without bacterial food, but with residual nutrients from peptone and agar, robustly extends life [105,106]. Whether these regimens all pinpoint the same process is still unclear, and each one has its own merits and caveats.
Recently, some exciting progress has been made in identifying genes mediating DR. Namely, PHA-4/FOXA1 and SKN-1/NRF transcription factors have been shown to be required for DR induced longevity [107,108]. When these genes are inactivated in the adult, animals are no longer long lived under reduced dietary intake. The effect is specific, since these mutants have little effect on daf-2/IIR longevity; moreover, daf-16/FOXO mutants are still susceptible to DR. SKN-1 has an early role in pharynx and gut specification, and a later role in the response to oxidative stress in the gut [109,110]. Interestingly, however, SKN-1 activity specifically in two neurosensory cells, called the ASIs, mediates DR-induced longevity [108], implying sensory inputs into DR. Nonetheless, mutants deficient in sensory transduction still respond to DR. By inference, neuronal SKN-1 must regulate the organismal response to DR through a hormonal mechanism. Accordingly, DR globally stimulates respiration, presumably to maximize organismal energy efficiency. Similarly, PHA-4/FOXA1 has an early role in endo/mesoderm specification in worms and mammals, but also regulates mammalian glucose homeostasis later in life [111,112]. C. elegans PHA-4 is expressed in the adult gut, gonad, and nervous system, yet influences life span of the entire organism, also implying endocrine control. Consistent with this, the mammalian FOXA regulates pancreatic glucagon production [112]. Dissecting their respective signaling pathways promises to further open up DR to a molecular analysis.
Sirtuins and related molecules have been implicated as potential mediators of DR in yeast and flies, although this remains controversial [62,113,114]. Conflicting reports also leave this issue unresolved in worms [115,116]. Discrepancies might arise because of unknown differences in culture conditions. Evidence also suggests that TOR (target of rapamycin) kinase mediates DR in flies and yeast [117,118]. TOR kinase promotes growth and protein synthesis, while its reduction dampens translation and increases recycling of cellular components through autophagy. In worms, a reduction of TOR, the downstream S6 kinase, ribosomal initiation factors, as well as ribosomal protein subunits themselves, reduce translation and extend adult life span [115,119–123]. Consistent with a role in DR, TOR longevity is not further extended in eat-2 mutants. Surprisingly, however, downstream components do extend life span beyond eat-2, suggesting that TOR outputs independent of translation could be critical for DR. Interestingly, deletion of ribosomal subunits in S. cerevisiae also extends replicative life span [118]. It will be critical to understand whether longevity arises from benefits due to globally reduced translation itself or from the regulation of specific factors.
Aging and Age-Related Disease
With all these longevity genes, a critical question is how do worms age, and from what do they die? With age, there is a progressive decline in body movement and pharyngeal pumping [124]. Most striking, muscle integrity deteriorates dramatically [125], with derangement of muscle fibers and overt changes in nuclear morphology—phenotypes reminiscent of sarcopenia, a major contributor of age-related decline in people. Surprisingly, the worm nervous system appears resilient, with little obvious change in structure or reporter expression, although function has not been critically tested. Among other things, aging worms show an increase in the appearance of necrotic cells, crosslinked cuticle, lipid droplets [126], endomitotic DNA synthesis in the germline [127], AMP/ATP ratios [53], oxidized protein [128], and lipofuscin [129]. Worms are described to die of enteric bacterial infection, and antibiotics extend life [126]. Conceivably, infection may be secondary to a decline in enteric muscle function, which is necessary to expel bacteria.
Although genotype determines the mean life span of a population, individual longevity has a large stochastic component, with several-fold differences observed even with an isogenic population in a uniform environment. Nonetheless, specific markers serve as good predictors of individual life expectancy. For example, stochastic induction of a hsp-16::gfp reporter predicts survival, while premature appearance of lipofuscin predicts early death [129,130]. Significantly, long-lived genotypes also delay the onset of aging and age-related disease, e.g., daf-2 mutants curtail many of the age-dependent changes described above, and ameliorate models of polyQ repeat protein disease, Aβ-proteotoxicity, and germline tumorigenesis [56,131,132]. Conversely, daf-16 or hsf-1 mutants often accelerate pathology. Conceivably, multiple age-related diseases could be mitigated at once by targeting IIS or other longevity pathways.
Conclusions
Animal life span is unexpectedly plastic, reflecting regulatory pathways responsive to environmental signals such as nutrients and stress. The future challenge will be to determine how these different pathways map onto and interact with each other, and decipher their molecular mechanisms. For some, basic questions such as where and when are they required, and whether they work cell autonomously or nonautonomously need to be addressed. Another major challenge will be to clarify how global processes (e.g., DR, mitochondrial longevity, checkpoint control, translation, and others) impact aging. What tradeoffs do they entail? Do they invoke signaling events or do they passively divert resources to somatic maintenance? Finally, what are the fundamental causes of aging and how can they be offset? Is it altered protein metabolism, organellar turnover, immune function, metabolic efficiency, ROS or xenobiotic detoxification, genome stability, or all of the above? Answers to these and other questions will be key to understanding broadly conserved aspects of life span determination.
Acknowledgments
I thank Dan Magner for comments on the manuscript. My apologies to colleagues whose work I could not cover.
Abbreviations
- DR
dietary restriction
- ER
endoplasmic reticulum
- IIS
insulin/IGF-I signaling
- ILP
insulin-like peptide
- NHR
nuclear hormone receptor
- ROS
reactive oxygen species
- TOR
target of rapamycin
Footnotes
Funding. National Institutes of Health (NIH), National Institute on Aging (NIA), and Glenn/American Foundation for Aging Research/Breakthroughs in Gerontology Award.
Competing interests. The author has declared that no competing interests exist.
References
- Friedman DB, Johnson TE. A mutation in the age-1 gene in C. elegans lengthens life and reduces hermaphrodite fertility. Genetics. 1988;118:75–86. doi: 10.1093/genetics/118.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- Morris JZ, Tissenbaum HA, Ruvkun G. A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in C. elegans. Nature. 1996;382:536–539. doi: 10.1038/382536a0. [DOI] [PubMed] [Google Scholar]
- Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in C. elegans. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, et al. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- Lin K, Dorman JB, Rodan A, Kenyon C. daf-16: an HNF-3/forkhead family member that can function to double the life-span of C. elegans. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- Clancy DJ, Gems D, Harshman LG, Oldham S, Stocker H, et al. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science. 2001;292:104–106. doi: 10.1126/science.1057991. [DOI] [PubMed] [Google Scholar]
- Tatar M, Kopelman A, Epstein D, Tu MP, Yin CM, et al. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science. 2001;292:107–110. doi: 10.1126/science.1057987. [DOI] [PubMed] [Google Scholar]
- Bluher M, Kahn BB, Kahn CR. Extended longevity in mice lacking the insulin receptor in adipose tissue. Science. 2003;299:572–574. doi: 10.1126/science.1078223. [DOI] [PubMed] [Google Scholar]
- Holzenberger M, Dupont J, Ducos B, Leneuve P, Geloen A, et al. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature. 2003;421:182–187. doi: 10.1038/nature01298. [DOI] [PubMed] [Google Scholar]
- Pierce SB, Costa M, Wisotzkey R, Devadhar S, Homburger SA, et al. Regulation of DAF-2 receptor signaling by human insulin and ins-1, a member of the unusually large and diverse C. elegans insulin gene family. Genes Dev. 2001;15:672–686. doi: 10.1101/gad.867301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Kennedy SG, Ruvkun G. daf-28 encodes a C. elegans insulin superfamily member that is regulated by environmental cues and acts in the DAF-2 signaling pathway. Genes Dev. 2003;17:844–858. doi: 10.1101/gad.1066503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis S, Ruvkun G. C. elegans Akt/PKB transduces insulin receptor-like signals from AGE-1 PI3 kinase to the DAF-16 transcription factor. Genes Dev. 1998;12:2488–2498. doi: 10.1101/gad.12.16.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogg S, Ruvkun G. The C. elegans PTEN homolog, DAF-18, acts in the insulin receptor-like metabolic signaling pathway. Mol Cell. 1998;2:887–893. doi: 10.1016/s1097-2765(00)80303-2. [DOI] [PubMed] [Google Scholar]
- Paradis S, Ailion M, Toker A, Thomas JH, Ruvkun G. A PDK1 homolog is necessary and sufficient to transduce AGE-1 PI3 kinase signals that regulate diapause in C. elegans. Genes Dev. 1999;13:1438–1452. doi: 10.1101/gad.13.11.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertweck M, Gobel C, Baumeister R. C. elegans SGK-1 is the critical component in the Akt/PKB kinase complex to control stress response and life span. Dev Cell. 2004;6:577–588. doi: 10.1016/s1534-5807(04)00095-4. [DOI] [PubMed] [Google Scholar]
- Mihaylova VT, Borland CZ, Manjarrez L, Stern MJ, Sun H. The PTEN tumor suppressor homolog in C. elegans regulates longevity and dauer formation in an insulin receptor-like signaling pathway. Proc Natl Acad Sci U S A. 1999;96:7427–7432. doi: 10.1073/pnas.96.13.7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RY, Hench J, Ruvkun G. Regulation of C. elegans DAF-16 and its human ortholog FKHRL1 by the daf-2 insulin-like signaling pathway. Curr Biol. 2001;11:1950–1957. doi: 10.1016/s0960-9822(01)00595-4. [DOI] [PubMed] [Google Scholar]
- Lin K, Hsin H, Libina N, Kenyon C. Regulation of the C. elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat Genet. 2001;28:139–145. doi: 10.1038/88850. [DOI] [PubMed] [Google Scholar]
- Henderson ST, Johnson TE. daf-16 integrates developmental and environmental inputs to mediate aging in the nematode C. elegans. Curr Biol. 2001;11:1975–1980. doi: 10.1016/s0960-9822(01)00594-2. [DOI] [PubMed] [Google Scholar]
- Honda Y, Honda S. The daf-2 gene network for longevity regulates oxidative stress resistance and Mn-superoxide dismutase gene expression in C. elegans. Faseb J. 1999;13:1385–1393. [PubMed] [Google Scholar]
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, et al. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- Lee SS, Kennedy S, Tolonen AC, Ruvkun G. DAF-16 target genes that control C. elegans life-span and metabolism. Science. 2003;300:644–647. doi: 10.1126/science.1083614. [DOI] [PubMed] [Google Scholar]
- Melendez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, et al. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- McElwee JJ, Schuster E, Blanc E, Thomas JH, Gems D. Shared transcriptional signature in Caenorhabditis elegans Dauer larvae and long-lived daf-2 mutants implicates detoxification system in longevity assurance. J Biol Chem. 2004;279:44533–44543. doi: 10.1074/jbc.M406207200. [DOI] [PubMed] [Google Scholar]
- Walker GA, Lithgow GJ. Lifespan extension in C. elegans by a molecular chaperone dependent upon insulin-like signals. Aging Cell. 2003;2:131–139. doi: 10.1046/j.1474-9728.2003.00045.x. [DOI] [PubMed] [Google Scholar]
- Kenyon C. The plasticity of aging: insights from long-lived mutants. Cell. 2005;120:449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Tatar M, Bartke A, Antebi A. The endocrine regulation of aging by insulin-like signals. Science. 2003;299:1346–1351. doi: 10.1126/science.1081447. [DOI] [PubMed] [Google Scholar]
- Dillin A, Crawford DK, Kenyon C. Timing requirements for insulin/IGF-1 signaling in C. elegans. Science. 2002;298:830–834. doi: 10.1126/science.1074240. [DOI] [PubMed] [Google Scholar]
- Jenkins NL, McColl G, Lithgow GJ. Fitness cost of extended lifespan in Caenorhabditis elegans. Proc Biol Sci. 2004;271:2523–2526. doi: 10.1098/rspb.2004.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton B, Dong Y, Shindo M, Liu W, Odell I, et al. A systematic RNAi screen for longevity genes in C. elegans. Genes Dev. 2005;19:1544–1555. doi: 10.1101/gad.1308205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Hsu AL, Dillin A, Kenyon C. New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet. 2005;1:119–128. doi: 10.1371/journal.pgen.0010017. doi: 10.1371/journal.pgen.0010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillin A, Hsu AL, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, et al. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298:2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- Lee SS, Lee RY, Fraser AG, Kamath RS, Ahringer J, et al. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat Genet. 2003;33:40–48. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- Oh SW, Mukhopadhyay A, Dixit BL, Raha T, Green MR, et al. Identification of direct DAF-16 targets controlling longevity, metabolism and diapause by chromatin immunoprecipitation. Nat Genet. 2006;38:251–257. doi: 10.1038/ng1723. [DOI] [PubMed] [Google Scholar]
- Cypser JR, Tedesco P, Johnson TE. Hormesis and aging in Caenorhabditis elegans. Exp Gerontol. 2006;41:935–939. doi: 10.1016/j.exger.2006.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apfeld J, Kenyon C. Regulation of lifespan by sensory perception in Caenorhabditis elegans. Nature. 1999;402:804–809. doi: 10.1038/45544. [DOI] [PubMed] [Google Scholar]
- Alcedo J, Kenyon C. Regulation of C. elegans longevity by specific gustatory and olfactory neurons. Neuron. 2004;41:45–55. doi: 10.1016/s0896-6273(03)00816-x. [DOI] [PubMed] [Google Scholar]
- Ailion M, Inoue T, Weaver CI, Holdcraft RW, Thomas JH. Neurosecretory control of aging in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1999;96:7394–7397. doi: 10.1073/pnas.96.13.7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz MJ, Riddle DL. Positive selection of C. elegans mutants with increased stress resistance and longevity. Genetics. 2003;163:171–180. doi: 10.1093/genetics/163.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay A, Deplancke B, Walhout AJ, Tissenbaum HA. C. elegans tubby regulates life span and fat storage by two independent mechanisms. Cell Metab. 2005;2:35–42. doi: 10.1016/j.cmet.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Lans H, Jansen G. Multiple sensory G proteins in the olfactory, gustatory and nociceptive neurons modulate longevity in Caenorhabditis elegans. Dev Biol. 2007;303:474–482. doi: 10.1016/j.ydbio.2006.11.028. [DOI] [PubMed] [Google Scholar]
- Libert S, Zwiener J, Chu X, Vanvoorhies W, Roman G, et al. Regulation of Drosophila life span by olfaction and food-derived odors. Science. 2007;315:1133–1137. doi: 10.1126/science.1136610. [DOI] [PubMed] [Google Scholar]
- Murakami H, Murakami S. Serotonin receptors antagonistically modulate C. elegans longevity. Aging Cell. 2007;6:483–488. doi: 10.1111/j.1474-9726.2007.00303.x. [DOI] [PubMed] [Google Scholar]
- Sze JY, Victor M, Loer C, Shi Y, Ruvkun G. Food and metabolic signalling defects in a C. elegans serotonin-synthesis mutant. Nature. 2000;403:560–564. doi: 10.1038/35000609. [DOI] [PubMed] [Google Scholar]
- Liang B, Moussaif M, Kuan CJ, Gargus JJ, Sze JY. Serotonin targets the DAF-16/FOXO signaling pathway to modulate stress responses. Cell Metab. 2006;4:429–440. doi: 10.1016/j.cmet.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Oh SW, Mukhopadhyay A, Svrzikapa N, Jiang F, Davis RJ, et al. JNK regulates lifespan in Caenorhabditis elegans by modulating nuclear translocation of forkhead transcription factor/DAF-16. Proc Natl Acad Sci U S A. 2005;102:4494–4499. doi: 10.1073/pnas.0500749102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MC, Bohmann D, Jasper H. JNK extends life span and limits growth by antagonizing cellular and organism-wide responses to insulin signaling. Cell. 2005;121:115–125. doi: 10.1016/j.cell.2005.02.030. [DOI] [PubMed] [Google Scholar]
- Wang MC, Bohmann D, Jasper H. JNK signaling confers tolerance to oxidative stress and extends lifespan in Drosophila. Dev Cell. 2003;5:811–816. doi: 10.1016/s1534-5807(03)00323-x. [DOI] [PubMed] [Google Scholar]
- Lehtinen MK, Yuan Z, Boag PR, Yang Y, Villen J, et al. A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell. 2006;125:987–1001. doi: 10.1016/j.cell.2006.03.046. [DOI] [PubMed] [Google Scholar]
- Troemel ER, Chu SW, Reinke V, Lee SS, Ausubel FM, et al. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet. 2006;2:e183. doi: 10.1371/journal.pgen.0020183. doi: 10.1371/journal.pgen.0020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanji M, Hopper NA, Gems D. LET-60 RAS modulates effects of insulin/IGF-1 signaling on development and aging in Caenorhabditis elegans. Aging Cell. 2005;4:235–245. doi: 10.1111/j.1474-9726.2005.00166.x. [DOI] [PubMed] [Google Scholar]
- Apfeld J, O'Connor G, McDonagh T, DiStefano PS, Curtis R. The AMP-activated protein kinase AAK-2 links energy levels and insulin-like signals to lifespan in C. elegans. Genes Dev. 2004;18:3004–3009. doi: 10.1101/gad.1255404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Gao B, Lee SM, Bennett K, Fang D. RLE-1, an E3 ubiquitin ligase, regulates C. elegans aging by catalyzing DAF-16 polyubiquitination. Dev Cell. 2007;12:235–246. doi: 10.1016/j.devcel.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Wolff S, Ma H, Burch D, Maciel GA, Hunter T, et al. SMK-1, an essential regulator of DAF-16-mediated longevity. Cell. 2006;124:1039–1053. doi: 10.1016/j.cell.2005.12.042. [DOI] [PubMed] [Google Scholar]
- Hsu AL, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- Morley JF, Morimoto RI. Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol Biol Cell. 2004;15:657–664. doi: 10.1091/mbc.E03-07-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh V, Aballay A. Heat-shock transcription factor (HSF)-1 pathway required for Caenorhabditis elegans immunity. Proc Natl Acad Sci U S A. 2006;103:13092–13097. doi: 10.1073/pnas.0604050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers MA, de Vries-Smits LM, Barker N, Polderman PE, Burgering BM, et al. Functional interaction between beta-catenin and FOXO in oxidative stress signaling. Science. 2005;308:1181–1184. doi: 10.1126/science.1109083. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in C. elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci U S A. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Nikolaev AY, Imai S, Chen D, Su F, et al. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, et al. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- Nemoto S, Fergusson MM, Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1(alpha} J Biol Chem. 2005;280:16456–16460. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- Berdichevsky A, Viswanathan M, Horvitz HR, Guarente L. C. elegans SIR-2.1 interacts with 14-3-3 proteins to activate DAF-16 and extend life span. Cell. 2006;125:1165–1177. doi: 10.1016/j.cell.2006.04.036. [DOI] [PubMed] [Google Scholar]
- Wang Y, Oh SW, Deplancke B, Luo J, Walhout AJ, et al. C. elegans 14-3-3 proteins regulate life span and interact with SIR-2.1 and DAF-16/FOXO. Mech Ageing Dev. 2006;127:741–747. doi: 10.1016/j.mad.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- Wood JG, Rogina B, Lavu S, Howitz K, Helfand SL, et al. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430:686–689. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan M, Kim SK, Berdichevsky A, Guarente L. A role for SIR-2.1 regulation of ER stress response genes in determining C. elegans life span. Dev Cell. 2005;9:605–615. doi: 10.1016/j.devcel.2005.09.017. [DOI] [PubMed] [Google Scholar]
- Urano F, Calfon M, Yoneda T, Yun C, Kiraly M, et al. A survival pathway for Caenorhabditis elegans with a blocked unfolded protein response. J Cell Biol. 2002;158:639–646. doi: 10.1083/jcb.200203086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arantes-Oliveira N, Apfeld J, Dillin A, Kenyon C. Regulation of life-span by germ-line stem cells in Caenorhabditis elegans. Science. 2002;295:502–505. doi: 10.1126/science.1065768. [DOI] [PubMed] [Google Scholar]
- Hsin H, Kenyon C. Signals from the reproductive system regulate the lifespan of C. elegans. Nature. 1999;399:362–366. doi: 10.1038/20694. [DOI] [PubMed] [Google Scholar]
- Berman JR, Kenyon C. Germ-cell loss extends C. elegans life span through regulation of DAF-16 by kri-1 and lipophilic-hormone signaling. Cell. 2006;124:1055–1068. doi: 10.1016/j.cell.2006.01.039. [DOI] [PubMed] [Google Scholar]
- Rottiers V, Motola DL, Gerisch B, Cummins CL, Nishiwaki K, et al. Hormonal control of C. elegans dauer formation and life span by a Rieske-like oxygenase. Developmental Cell. 2006;10:473–482. doi: 10.1016/j.devcel.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Gerisch B, Weitzel C, Kober-Eisermann C, Rottiers V, Antebi A. A hormonal signaling pathway influencing C. elegans metabolism, reproductive development, and life span. Dev Cell. 2001;1:841–851. doi: 10.1016/s1534-5807(01)00085-5. [DOI] [PubMed] [Google Scholar]
- Gerisch B, Rottiers V, Li D, Motola DL, Cummins CL, et al. A bile acid-like steroid modulates C. elegans lifespan through nuclear receptor signaling. PNAS. 2007;104:5014–5019. doi: 10.1073/pnas.0700847104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motola DL, Cummins CL, Rottiers V, Sharma K, Sunino K, et al. Identification of DAF-12 ligands that govern dauer formation and reproduction in C. elegans. Cell. 2006;124:1209–1223. doi: 10.1016/j.cell.2006.01.037. [DOI] [PubMed] [Google Scholar]
- Held JM, White MP, Fisher AL, Gibson BW, Lithgow GJ, et al. DAF-12-dependent rescue of dauer formation in Caenorhabditis elegans by (25S)-cholestenoic acid. Aging Cell. 2006;5:283–291. doi: 10.1111/j.1474-9726.2006.00218.x. [DOI] [PubMed] [Google Scholar]
- Song C, Liao S. Cholestenoic acid is a naturally occurring ligand for liver X receptor alpha. Endocrinology. 2000;141:4180–4184. doi: 10.1210/endo.141.11.7772. [DOI] [PubMed] [Google Scholar]
- Broue F, Liere P, Kenyon C, Baulieu EE. A steroid hormone that extends the lifespan of Caenorhabditis elegans. Aging Cell. 2007;6:87–94. doi: 10.1111/j.1474-9726.2006.00268.x. [DOI] [PubMed] [Google Scholar]
- Jia K, Albert PS, Riddle DL. DAF-9, a cytochrome P450 regulating C. elegans larval development and adult longevity. Development. 2002;129:221–231. doi: 10.1242/dev.129.1.221. [DOI] [PubMed] [Google Scholar]
- Ludewig AH, Kober-Eisermann C, Weitzel C, Bethke A, Neubert K, et al. A novel nuclear receptor/coregulator complex controls C. elegans lipid metabolism, larval development, and aging. Genes Dev. 2004;18:2120–2133. doi: 10.1101/gad.312604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Boehm M, Slack F. A developmental timing microRNA and its target regulate life span in C. elegans. Science. 2005;310:1954–1957. doi: 10.1126/science.1115596. [DOI] [PubMed] [Google Scholar]
- Hristova M, Birse D, Hong Y, Ambros V. The Caenorhabditis elegans heterochronic regulator LIN-14 is a novel transcription factor that controls the developmental timing of transcription from the insulin/insulin-like growth factor gene ins-33 by direct DNA binding. Mol Cell Biol. 2005;25:11059–11072. doi: 10.1128/MCB.25.24.11059-11072.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibanez-Ventoso C, Yang M, Guo S, Robins H, Padgett RW, et al. Modulated microRNA expression during adult lifespan in Caenorhabditis elegans. Aging Cell. 2006;5:235–246. doi: 10.1111/j.1474-9726.2006.00210.x. [DOI] [PubMed] [Google Scholar]
- Olsen A, Vantipalli MC, Lithgow GJ. Checkpoint proteins control survival of the postmitotic cells in Caenorhabditis elegans. Science. 2006;312:1381–1385. doi: 10.1126/science.1124981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakowski B, Hekimi S. Determination of life-span in Caenorhabditis elegans by four clock genes. Science. 1996;272:1010–1013. doi: 10.1126/science.272.5264.1010. [DOI] [PubMed] [Google Scholar]
- Campisi J. Cancer and ageing: rival demons? Nat Rev Cancer. 2003;3:339–349. doi: 10.1038/nrc1073. [DOI] [PubMed] [Google Scholar]
- Jonassen T, Proft M, Randez-Gil F, Schultz JR, Marbois BN, et al. Yeast Clk-1 homologue (Coq7/Cat5) is a mitochondrial protein in coenzyme Q synthesis. J Biol Chem. 1998;273:3351–3357. doi: 10.1074/jbc.273.6.3351. [DOI] [PubMed] [Google Scholar]
- Feng J, Bussiere F, Hekimi S. Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Dev Cell. 2001;1:633–644. doi: 10.1016/s1534-5807(01)00071-5. [DOI] [PubMed] [Google Scholar]
- Braeckman BP, Houthoofd K, Brys K, Lenaerts I, De Vreese A, et al. No reduction of energy metabolism in Clk mutants. Mech Ageing Dev. 2002;123:1447–1456. doi: 10.1016/s0047-6374(02)00085-4. [DOI] [PubMed] [Google Scholar]
- Kayser EB, Sedensky MM, Morgan PG, Hoppel CL. Mitochondrial oxidative phosphorylation is defective in the long-lived mutant clk-1. J Biol Chem. 2004;279:54479–54486. doi: 10.1074/jbc.M403066200. [DOI] [PubMed] [Google Scholar]
- Ventura N, Rea SL, Testi R. Long-lived C. elegans mitochondrial mutants as a model for human mitochondrial-associated diseases. Exp Gerontol. 2006;41:974–991. doi: 10.1016/j.exger.2006.06.060. [DOI] [PubMed] [Google Scholar]
- Curtis R, O'Connor G, DiStefano PS. Aging networks in Caenorhabditis elegans: AMP-activated protein kinase (aak-2) links multiple aging and metabolism pathways. Aging Cell. 2006;5:119–126. doi: 10.1111/j.1474-9726.2006.00205.x. [DOI] [PubMed] [Google Scholar]
- Liu X, Jiang N, Hughes B, Bigras E, Shoubridge E, et al. Evolutionary conservation of the clk-1-dependent mechanism of longevity: loss of mclk1 increases cellular fitness and lifespan in mice. Genes Dev. 2005;19:2424–2434. doi: 10.1101/gad.1352905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klass MR. Aging in the nematode C. elegans: major biological and environmental factors influencing life span. Mech Ageing Dev. 1977;6:413–429. doi: 10.1016/0047-6374(77)90043-4. [DOI] [PubMed] [Google Scholar]
- Houthoofd K, Braeckman BP, Johnson TE, Vanfleteren JR. Life extension via dietary restriction is independent of the Ins/IGF-1 signalling pathway in Caenorhabditis elegans. Exp Gerontol. 2003;38:947–954. doi: 10.1016/s0531-5565(03)00161-x. [DOI] [PubMed] [Google Scholar]
- Lakowski B, Hekimi S. The genetics of caloric restriction in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1998;95:13091–13096. doi: 10.1073/pnas.95.22.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GD, Wilson MA, Zhu M, Wolkow CA, de Cabo R, et al. Dietary deprivation extends lifespan in Caenorhabditis elegans. Aging Cell. 2006;5:515–524. doi: 10.1111/j.1474-9726.2006.00241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein TL, Smith ED, Tsuchiya M, Welton KL, Thomas JH, et al. Lifespan extension in Caenorhabditis elegans by complete removal of food. Aging Cell. 2006;5:487–494. doi: 10.1111/j.1474-9726.2006.00238.x. [DOI] [PubMed] [Google Scholar]
- Panowski SH, Wolff S, Aguilaniu H, Durieux J, Dillin A. PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature. 2007;447:550–555. doi: 10.1038/nature05837. [DOI] [PubMed] [Google Scholar]
- Bishop NA, Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature. 2007;447:545–549. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- Bowerman B, Eaton BA, Priess JR. skn-1, a maternally expressed gene required to specify the fate of ventral blastomeres in the early C. elegans embryo. Cell. 1992;68:1061–1075. doi: 10.1016/0092-8674(92)90078-q. [DOI] [PubMed] [Google Scholar]
- An JH, Blackwell TK. SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev. 2003;17:1882–1893. doi: 10.1101/gad.1107803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mango SE, Lambie EJ, Kimble J. The pha-4 gene is required to generate the pharyngeal primordium of Caenorhabditis elegans. Development. 1994;120:3019–3031. doi: 10.1242/dev.120.10.3019. [DOI] [PubMed] [Google Scholar]
- Friedman JR, Kaestner KH. The Foxa family of transcription factors in development and metabolism. Cell Mol Life Sci. 2006;63:2317–2328. doi: 10.1007/s00018-006-6095-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Sir2-independent life span extension by calorie restriction in yeast. PLoS Biol. 2004;2:e296. doi: 10.1371/journal.pbio.0020296. doi: 10.1371/journal.pbio.0020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Taubert S, Crawford D, Libina N, Lee SJ, et al. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6:95–110. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Tissenbaum HA. Overlapping and distinct functions for a Caenorhabditis elegans SIR2 and DAF-16/FOXO. Mech Ageing Dev. 2006;127:48–56. doi: 10.1016/j.mad.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, et al. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr Biol. 2004;14:885–890. doi: 10.1016/j.cub.2004.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Powers RW, 3rd, Steffen KK, Westman EA, Hu D, et al. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, et al. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426:620. doi: 10.1038/426620a. [DOI] [PubMed] [Google Scholar]
- Jia K, Chen D, Riddle DL. The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development. 2004;131:3897–3906. doi: 10.1242/dev.01255. [DOI] [PubMed] [Google Scholar]
- Pan KZ, Palter JE, Rogers AN, Olsen A, Chen D, et al. Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. Aging Cell. 2007;6:111–119. doi: 10.1111/j.1474-9726.2006.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson ST, Bonafe M, Johnson TE. daf-16 protects the nematode Caenorhabditis elegans during food deprivation. J Gerontol A Biol Sci Med Sci. 2006;61:444–460. doi: 10.1093/gerona/61.5.444. [DOI] [PubMed] [Google Scholar]
- Syntichaki P, Troulinaki K, Tavernarakis N. eIF4E function in somatic cells modulates ageing in Caenorhabditis elegans. Nature. 2007;445:922–926. doi: 10.1038/nature05603. [DOI] [PubMed] [Google Scholar]
- Huang C, Xiong C, Kornfeld K. Measurements of age-related changes of physiological processes that predict lifespan of Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2004;101:8084–8089. doi: 10.1073/pnas.0400848101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herndon LA, Schmeissner PJ, Dudaronek JM, Brown PA, Listner KM, et al. Stochastic and genetic factors influence tissue-specific decline in ageing C. elegans. Nature. 2002;419:808–814. doi: 10.1038/nature01135. [DOI] [PubMed] [Google Scholar]
- Garigan D, Hsu AL, Fraser AG, Kamath RS, Ahringer J, et al. Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics. 2002;161:1101–1112. doi: 10.1093/genetics/161.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden TR, Beckman KB, Lee AH, Dudek N, Hubbard A, et al. Dramatic age-related changes in nuclear and genome copy number in the nematode Caenorhabditis elegans. Aging Cell. 2007;6:179–188. doi: 10.1111/j.1474-9726.2007.00273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii N, Goto S, Hartman PS. Protein oxidation during aging of the nematode Caenorhabditis elegans. Free Radic Biol Med. 2002;33:1021–1025. doi: 10.1016/s0891-5849(02)00857-2. [DOI] [PubMed] [Google Scholar]
- Gerstbrein B, Stamatas G, Kollias N, Driscoll M. In vivo spectrofluorimetry reveals endogenous biomarkers that report healthspan and dietary restriction in Caenorhabditis elegans. Aging Cell. 2005;4:127–137. doi: 10.1111/j.1474-9726.2005.00153.x. [DOI] [PubMed] [Google Scholar]
- Rea SL, Wu D, Cypser JR, Vaupel JW, Johnson TE. A stress-sensitive reporter predicts longevity in isogenic populations of Caenorhabditis elegans. Nat Genet. 2005;37:894–898. doi: 10.1038/ng1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen E, Bieschke J, Perciavalle RM, Kelly JW, Dillin A. Opposing activities protect against age-onset proteotoxicity. Science. 2006;313:1604–1610. doi: 10.1126/science.1124646. [DOI] [PubMed] [Google Scholar]
- Pinkston JM, Garigan D, Hansen M, Kenyon C. Mutations that increase the life span of C. elegans inhibit tumor growth. Science. 2006;313:971–975. doi: 10.1126/science.1121908. [DOI] [PubMed] [Google Scholar]