Abstract
By using matrix-assisted laser desorption/ionization time-of-flight MS, individual peptidergic neurons from Aplysia are assayed. A semiquantitative method is developed for comparing single-cell profiles by using spectral normalization, and peptides are localized to specific cells by mass spectrometric cell mapping. In addition to all previously identified products of the egg-laying hormone (ELH) gene, other peptides are formed from proteolytic hydrolysis of Leu-Leu residues within ELH and acidic peptide (AP). AP exhibits further processing to yield AP1–20 and AP9–27. These peptides appear to be colocalized in vesicles with ELH, transported to specific neuronal targets, and released in a Ca2+-dependent manner. A differential peptide distribution is observed at a specific target cell, and a low-frequency variation of AP, [Thr21]AP, is detected in a single animal.
Proteolytic cleavage of neuropeptide precursors is a well-regulated process in vertebrate and invertebrate neurons, where distinct products from a single gene can have diverse functions (1). For example, egg laying in the marine mollusk Aplysia is controlled by a cluster of peptidergic cells located in the rostral margin of the abdominal ganglion (2, 3). Posttranslational processing of the egg-laying hormone (ELH) precursor (3) results in multiple products that are differentially packaged (4, 5) and transported to multiple targets (6). Several of these peptides appear to act locally on the abdominal ganglion (7, 8), whereas others are released into the vasculature to act on distant sites. Although this peptide family is well-characterized, in terms of DNA (9), processing (10), and release (11), the functions of many of these peptides are not well understood.
Often, as in the case of the opioid peptides, different bioactive products derived from the same prohormone can have diverse effects (12, 13). Furthermore, the extent of prohormone processing can vary with environment, such as the influence of osmotic stress on the pro-oxytocin and pro-dynorphin systems (14–16). The 271-residue ELH prohormone (pELH) undergoes an early cleavage at a tetrabasic site, and the resulting peptides are sorted into different vesicles. In addition to conventional processing, such as cleavage at basic sites and C-terminal amidation, further modification can greatly affect biological activity of the final product peptides. In the bag cells, the nine-residue α-bag cell peptide (BCP) is further cleaved into the more potent α1–8 and α1–7 forms (17). Acidic peptide (AP) is stored in the same vesicles with ELH and ɛ-BCP (5). Although AP was first reported more than 20 years ago (18, 19), no specific physiological roles have been determined. We report that AP undergoes further processing to yield two major peptides that are released from the bag cell clusters on electrical stimulation. Also present are at least five modified forms of ELH, somewhat analogous to those in the atrial gland (20).
Cellular peptide identification can be a difficult task. Most methods require concentration and purification of the individual peptides before sequencing and/or compositional analysis. Matrix-assisted laser desorption/ionization (MALDI) with time-of-flight MS (TOF MS) provides an alternative for highly accurate and precise molecular weight determination. When combined with knowledge of the prohormone, identification of most, if not all, of the peptides present in complex biological samples is possible. Previous studies have used MALDI to study gene expression within identified neurons from the pulmonate snail Lymnaea stagnalis (21, 22). Additional MALDI-based methods detect novel in vitro processing of neuropeptide Y (NPY) in human cerebrospinal fluid (23) and monitor peptide changes in the neurointermediate lobe of rats (24). High salt concentrations associated with cells from Aplysia make such assays difficult because of variable salt tolerances encountered with MALDI. However our recently reported method enables cells from marine species such as Aplysia and Pleurobranchaea californica to be directly assayed for peptides (25).
By examining the peptides found in bag cell clusters and in several identified neurons, we observe: (i) three forms of α-BCP (α1–9, α1–8, and α1–7) in cells, (ii) novel processing of ELH and AP by cleavage at Leu-Leu residues, (iii) AP undergoing additional N- and C-terminal processing with one form, AP9–27, often being the most intense mass peak observed, (iv) greatly enhanced C-terminal peptides (ELH, AP) versus N-terminal peptides near specific target abdominal cells compared with ratios found in bag cells, (v) the additional AP-related peptides being released from bag cell neurons in a Ca2+-dependent manner, and (vi) Aplysia NPY, although previously reported to be localized in the bag cells (26), being localized to the abdominal RG cluster.
MATERIALS AND METHODS
Animals.
Animals less than 200 g were obtained from Aplysia Research Facility (Miami, FL); those from 200 to 500 g were from Pacific Biomarine (Venice, CA) and Marinus (Long Beach, CA).
Cellular Sample Preparation.
MALDI mass spectra were obtained from individual cells and cluster sections as previously described (25). The abdominal ganglion was removed, the physiological saline was replaced with the MALDI matrix solution [10 mg/ml of 2,5-dihydroxybenzoic acid (DHB) in water], and electrochemically sharpened tungsten rods were used to isolate and transfer the cell(s) onto a MALDI sample plate containing ≈0.5 μl of DHB. Note that with roughly half of the preparations, freshly isolated ganglia were treated with a protease (27) to soften connective tissues before cell dissection.
Multiple sampling protocols were investigated. Immediately after dissection, several abdominal ganglia were placed into either artificial seawater (ASW) or DHB containing two different peptidase inhibitor mixtures (17, 28). The first inhibitor mixture contained 62 μg/ml each of leupeptin, antipain, diprotin A, Phe-Ala-Ala (University of Illinois Biotechnology Center), Ser-Leu, and Leu-Arg, as well as 250 μg/ml each of bacitracin and trypsin inhibitor types II-L, II-O, III-O, and I-S. The second was a saturated solution of 1,10-phenanthroline, 4-(chloromercuri)benzenesulfonic acid, (−)-1-chloro-3-tosylamido-7-amino-2-heptanone hydrochloride, and α-toluenesulfonyl fluoride. As an alternative approach, abdominal ganglia were placed into DHB containing 0.1% (vol/vol) trifluoroacetic acid (TFA) immediately after animal dissection. To test for matrix-dependent effects, an alternate MALDI matrix was used consisting of a 0.1:30:70 TFA/acetonitrile/water solution saturated with α-cyano-4-hydroxycinnamic acid. Unless specified, chemicals were purchased from Sigma or Aldrich.
Release Experiments.
To reduce the volume of extracellular solution and still collect samples of that solution, a small volume release chamber was constructed, as in Fig. 1A. ASW (with or without peptidase inhibitors) flowed from the chamber through the sample and bath outlet capillaries (50 μm i.d. and 150 μm i.d., respectively) via gravimetric pressure. Heights of the inlet and outlet capillaries were adjusted for ≈2-min residence time, with a flow rate of ≈2 μl/min. Different peptidase inhibitor mixtures in ASW were used: (i) 62 μg/ml each of leupeptin, Ser-Leu, Leu-Arg, and Phe-Ala-Ala, (ii) 250 μg/ml each of trypsin inhibitors types II-L, II-O, III-O, and I-S in ASW, and (iii) a combined set of i and ii.
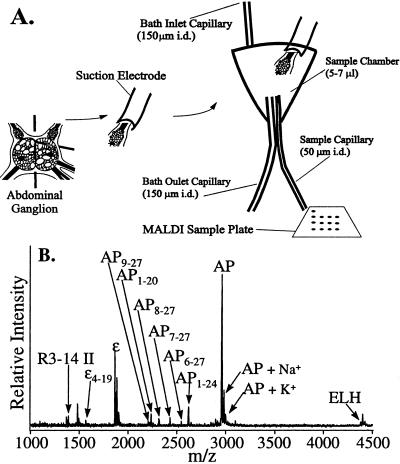
Figure 1.
(A) Schematic diagram showing the peptide release experimental details. (B) A mass spectrum of the extracellular matrix after stimulating an afterdischarge. Because of the high levels of cations in the media, the peak intensities for this experiment cannot be compared with spectra obtained from isolated cells.
Bag cell clusters were cut away from the abdominal ganglion with a 5-mm section of the pleural-abdominal connective. A pulled piece of polyethylene tubing was placed over the connective and used as a suction pipette electrode. The bag cell cluster then was moved to the release chamber, a Ag/AgCl electrode was placed in the bath as a reference electrode, and another in the suction pipette near the tissue, for recording and stimulation. An A-M Systems Differential AC Amplifier, model 1700 was used both to record from and to stimulate the tissue in the chamber. Extracellular media sampling was carried out by placing the drop at the end of the sample capillary (50–100 nl) onto a MALDI sample probe followed by the addition of 0.5 μl of DHB.
MS of Synthetic Peptide Standards.
α-BCP, α1–8, α1–7, β-BCP, γ-BCP, and ELH (American Peptide) as well as AP, AP9–27, and AP1–20 (University of Illinois Biotechnology Center) were mixed in 0–100% (vol/vol) ASW. In each case, 50 nl–1 μl of peptide solution was combined with ≈0.5 μl of DHB on a MALDI sample probe.
MS Instrumentation.
Two MALDI-TOF mass spectrometers were used, namely a TofSpec (Micromass, Manchester, U.K.) and a Voyager Elite with delayed ion extraction (PerSeptive Biosystems, Framingham, MA). Each instrument used a pulsed nitrogen laser (337 nm) as the desorption/ionization source, and mass spectra were acquired by using both linear and reflectron modes with a 20-kV acceleration potential. Each unsmoothed mass spectrum shown is the average of 20–100 laser pulses, with mass calibration performed internally by using two peaks corresponding to identified peptides.
RESULTS
Peptides in Bag Cell Neurons.
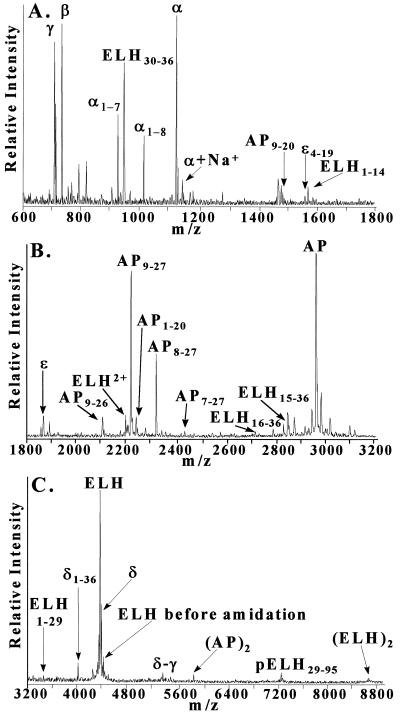
During a 2-year period, more than 500 MALDI mass spectra from over 100 animals were obtained. Fig. 2 is a mass spectrum acquired from an individual bag cell, with the known as well as previously unreported peptides labeled. Essentially all peaks in the mass spectra presented correspond to the protonated molecular weights of peptides. One exception is the occasional detection of peaks distributed over m/z 600–800, which likely correspond to phospholipids in the cellular membranes.
Figure 2.
Representative mass spectrum of a single bag cell. The mass scale is divided into (A) low, (B) middle, and (C) high m/z regions. Mass assignment for each peak is based on internal calibration with α-BCP and ELH. Peaks generally correspond to [M+H]+, where M is the molecular weight of each peptide. Note that in A, the [M+Na]+ peak for α-BCP is indicated; in B, ELH2+ is the [M+2H]2+ peak for ELH; and in C (AP)2 and (ELH)2 are the [2M+H]+ peaks for AP and ELH, respectively. δ-γ is a processing intermediate.
We need to determine whether the detection of unknown peptides is the result of sample preparation, cellular processing, or extracellular degradation. Previous studies with Lymnaea have demonstrated that the acidity (≈pH 2) of the DHB matrix, combined with short sample handling times, is likely to inactivate endogenous proteolytic enzymes (21, 22). However, because gradual peptide degradation is reported to occur in bag cell cluster extracts, even when incubated in 0.2 M HCl for 2–3 hr (29), we investigated a number of other sample preparation paradigms. With ganglion incubation and analysis in the presence of two independent, yet widely used regimens reported to prevent degradation in bag cell neurons (17, 28) and by dissecting the cells without protease pretreatment, we observe the same peptides as in Fig. 2. Although additional peaks are found, these peaks also are observed in controls and are attributed to the added inhibitors. Similarly, all peptides are detected when acidified DHB or α-cyano-4-hydroxycinnamic acid matrix is used, however, cell lysis encountered with these approaches is severe and interferes with isolating single identified cells. Finally, the same peptide profile is observed when the cells are dissected from the animal in seawater and immediately dried on the sample target so that the elapsed time from dissection to spectral acquisition is only several minutes (25).
Peptide Heterogeneity.
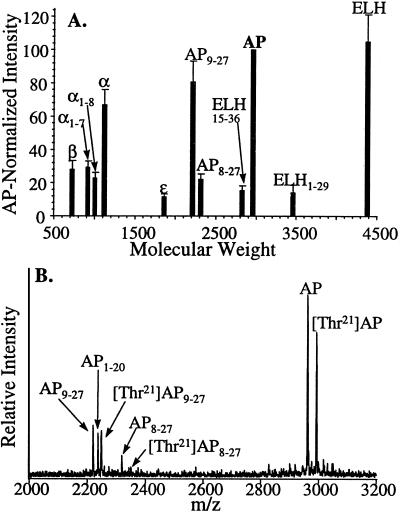
With the ability to screen individual bag cells and cluster sections, the peptide profiles from multiple animals were compared. Eleven major peaks spanning m/z 700–4,500 were analyzed in 34 representative mass spectra that were obtained from small groups of bag cells containing all 11 peaks of interest. Furthermore, each selected mass spectrum was acquired from a different animal, with at least one from each month of the year. The intensities of 11 major peaks were normalized to the intensity of AP and then averaged, to account for differences in the two mass spectrometers, different sampling conditions, and variability in cell size from different animals. Fig. 3A shows average peak intensity versus mass-to-charge ratio, with errors expressed as the SEM. This composite resembles a typical bag cell mass spectrum, and the associated errors for each peak average 20%, reflecting both the inter-animal variance and the reproducibility of the instrumentation and preparation. Although most single-cell spectra are similar to the composite, approximately 20% of the individual somas profiled within a bag cell cluster appear different and usually have greatly suppressed amino-terminal peptides.
Figure 3.
(A) A composite mass spectrum of bag cells from 34 different animals. For each mass spectrum, the peaks were normalized (IntensityAP = 100). Errors are given as the SEM. (B) Representative mass spectrum of the AP region from an animal expressing a [Thr21]AP variation. Both wild-type and variant AP-related peptides are seen. The absence of two peaks for AP1–20 is evidence that the amino acid substitution is in the residues 21–27, whereas the mass difference between each wild-type/variant pair enables the substituted residue to be identified.
Of the animals surveyed, two contained additional peptides not observed in other animals. As one example, Fig. 3B is representative of 13 mass spectra obtained from the left and right bag cell clusters of a single animal. The typical peaks for AP-related peptides are present with the exception of a unique second series. This higher mass can be attributed to a low-frequency variation that has resulted in a substituted AP that is 30.01 mass units heavier.
Peptide Distribution.
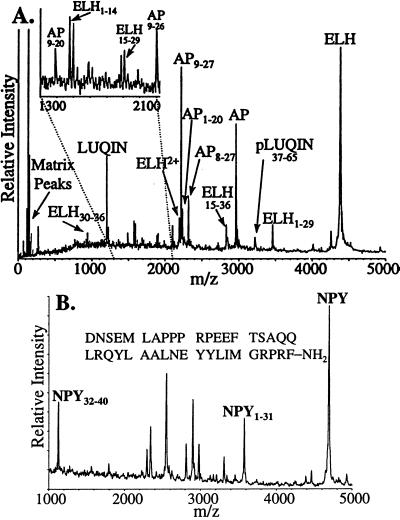
The differences between relative amounts of peptides from different cells and even cellular targets is investigated. Beyond differences within a bag cell cluster, there are differences in the complement of the bag cell peptides found at presumed target neurons. For example, Fig. 4A is a mass spectrum of an individual L3 neuron. Still attached are bag cell terminals containing detectable amounts of the bag cell peptides. Clearly seen are the L3 peptide LUQIN and the bag cell peptides ELH, AP, and AP9–27, but no amino-terminal pELH peptides.
Figure 4.
(A) A mass spectrum of an individual L3 neuron (identified on the basis of position, size, and color) with the major peptide peaks identified. Baseline analysis revealed no peptide peaks corresponding to N-terminal peptides of pELH. (Inset) An expanded mass scale for the region between the dotted lines. (B) Aplysia NPY is one of the peptides identified in a single cell from the abdominal RG cluster.
Aplysia NPY was isolated from pooled abdominal ganglia and, based on immunoreactivity, has been reported to reside in the bag cells (26). We have never detected NPY (Mr 4,687) in the bag cells, however, a survey of morphologically identified abdominal neurons reveals that NPY, as well as peptides corresponding to truncated forms, are stored in the RG cluster (Fig. 4B).
Peptide Release.
The peptides released from cells are found in the extracellular media containing high alkali salt concentrations, so the technique of removing excess salts from intact cells cannot be used. No peptides are seen because of the high salts when 0.5–1 μl of extracellular media is sampled directly. A recently reported method (30) for joining MALDI MS with capillary electrophoresis was adapted for use in our system. A small sample (50–100 nl) of extracellular media is deposited onto the MALDI sample plate followed by deposition of a large excess (0.5 μl) of the DHB matrix. This method reduces the absolute amount of salts deposited onto the sample plate, thus reducing the alkali ion concentration on the target. However, measuring the peptides present in nanoliter volume samples is difficult as the total amount of peptide is small. Therefore, a small volume chamber was designed to decrease the volume of extracellular solution around the cluster and thus decrease dilution of released peptides. By using this chamber with 50–100 nl samples, peptides can be detected, as shown in the mass spectrum in Fig. 1B. Present in the mass spectrum are ELH, AP, and novel AP peptides.
As the appearance of the mass spectrum is crucially dependent on the presence and amounts of salts (25), the effects of different salt concentrations on the appearance of mass spectra from different synthetic peptides were explored. MALDI-TOF MS of equimolar synthetic α-BCP, AP, AP9–27, AP1–20, and ELH in various dilutions of ASW in water shows that α-BCP and AP1–20 retain high peak intensity in up to 100% ASW, whereas AP9–27, and ELH are less salt tolerant, with drastically decreased signals. Thus, direct comparison of the intensity of peptide peaks from intact cells and cellular releasates cannot be made. The novel ELH-related peptides are not observed in release data. However, full-length ELH is the most salt-sensitive peptide investigated. Hence, it is likely that ionization of the shortened forms is suppressed, preventing detection in releasate.
DISCUSSION
MALDI-TOF MS is inherently different from conventional methods in that all peptides present at significant concentrations are detected. We do not have to preselect a particular peptide as in immunotechniques nor are we limited to peptides containing a particular amino acid residue as in radiolabeling. We observe the same complement and intensity of peptides regardless of sampling strategy even though more than 10 different combinations of cell isolation/peptidase inhibitor/sample matrices have been investigated. This observation strongly argues that intracellular processing is responsible for the observed peptides, so these peptides are examined by using the known pELH sequence.
pELH Processing.
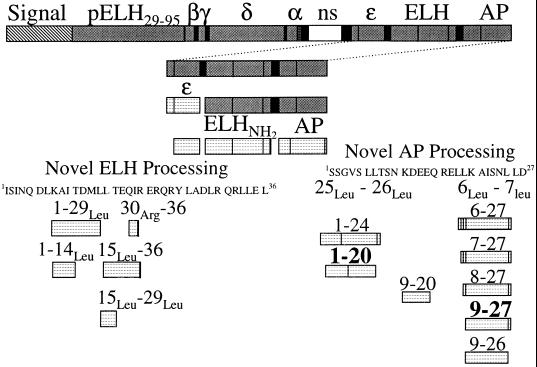
Different signal sequences using amino acid analysis have been reported previously (10, 29). However, MALDI-TOF MS unambiguously determines that signal sequence cleavage occurs after residue 28 of pELH, because, based on mass, we observe a peptide between Ser29 and α-BCP, designated pELH29–95 in Fig. 5. Although this signal sequence length is different than that previously reported, it does fall within their margin of error (10, 29).
Figure 5.
A schematic diagram of pELH processing based on our data. Light coloring indicates the peptide is detected by MS. Solid vertical lines represent cleavage sites and black bars are entirely cleaved. The white bar, labeled ns, is not seen in this or previous studies (10). The residue numbers are designated to describe the novel peptide processing. Residue numbers in bold indicate major products.
A large number of peptides are detected in the bag cells, most of which are assigned to previously identified peptides. Although expected from the gene sequence, the peptide product located between α-BCP and ɛ-BCP is seen neither by extraction followed by HPLC (29) nor in the present study. MALDI provides a more complete picture of the peptides present in individual bag cells than conventional techniques. As one example, unamidated ELH (containing a C-terminal Gly) is observed (Fig. 2C), although the amidated peak is more intense.
We detect three forms of α-BCP (i.e., α1–9, α1–8, and α1–7). Full-length α-BCP has been proposed to be intracellularly processed (17), with bioactivity depending on the relative amounts of the products, as α1–8 and α1–7 are 30 and 10 times more potent, respectively, than α1–9. The three forms previously were recovered from cellular homogenates (17, 31). However, in the presence of peptidase inhibitors, α1–7 was not observed in releasates within the reported 1-pmol detection limit (28). Our data suggests that α1–9 is intracellularly processed into both α1–8 and α1–7, as all three peptides are detected in intact cells. After release, α1–9 is known to quickly degrade into α1–5, α3–9, α6–9, and others, whereas extracellular peptidases presumably degrade α1–8 similarly (32). None of these expected degradants are detected in our experiments. This finding is additional evidence that the DHB cell rinsing protocol prevents extracellular degradation.
Peptides in the bag cell neurons are identified based on experimentally observed masses combined with knowledge of the pELH sequence, and all known peptides in Fig. 2 are identified with an average mass accuracy of 0.005%. Furthermore, peaks corresponding to previously unidentified peptides are assigned with the same mass accuracy. As one example, AP9–27 has a calculated and observed protonated monoisotopic mass of 2,215.15 Da and 2,215.26 Da, respectively. Significantly, the only sequence along pELH that has a protonated mass of 2,215.26 ± 0.75 Da is AP9–27.
The measurement of the mass spectra from low-frequency variants provides independent confirmation of our assignment of AP9–27. Fig. 3B shows an unusual mass spectrum where there are paired peaks for AP, AP9–27, and AP8–27, but not for AP1–20. Whereas several amino acid substitutions result in a 30.0 ± 0.5 Da difference, comparison of these modifications with the prohormone sequence shows that only an Ala21 to Thr21 switch (a single base substitution) accounts for the observed peaks. The fact that the novel forms of AP also have corresponding variant forms strongly supports the mass assignments.
Because the peptides detected in the bag cell neurons do not result from nonspecific degradation or instrumental artifacts, an in vivo processing scheme is strongly suggested. The pattern of processing for AP and ELH-derived peptides is shown schematically in Fig. 5. Although we observe a large number of shortened forms of ELH and AP, all of the novel peptides appear to have the same first processing step, cleavage at Leu-Leu sites (or in one case, Leu-Arg), similar to the action of the enzyme renin. This renin-like activity has been reported for Aplysia atrial gland processing of A-ELH and [Ala27]A-ELH (20). Because of the high degree of homology between ELH, A-ELH, and [Ala27]A-ELH, similar Leu14-Leu15 processing in the bag cells is not surprising. The similarity between the atrial gland and the bag cell neurons continues in that subsequent cleavage of the N-terminal Leu15 is reported for A-ELH and also is observed here. However, in the bag cells there is a second ELH cleavage observed at Leu29-Arg30 that is not seen in the atrial gland, and in the atrial gland there is a cleavage at Leu33-Arg34 that is not observed in the bag cells. This finding could be attributed to the disulfide bond as well as sequence differences in the C-terminal region of the atrial gland forms (20). As further evidence that the Leu-Leu processing is not caused by extracellular degradation, the atrial gland forms were observed even when atrial glands were boiled before extraction (20). In the bag cells, the doubly cleaved peptide (ELH15–29) is observed. Although these shortened forms of ELH have not been identified or sequenced previously, shortened ELH forms have been postulated based on immunoassay results (10, 33).
AP is reported to be stored in the same vesicle as ELH (5), and so is expected to be processed similarly. In AP, two of three Leu-Leu sites are cleaved and processed, yielding a large number of final products. In addition, several observed peptides require removal of additional amino acid residues in a manner similar to the carboxypeptidase processing that follows the conversion of angiotensinogen to angiotensin I by renin. Based on the observed profiles, carboxypeptidase activity removes residues in a step-wise fashion, so that AP1–24 and AP1–20 are observed, and this activity stops at AP1–20. This finding contrasts with the activity seen after cleavage at the N-terminal Leu6–Leu7 cleavage. After Leu6–Leu7 cleavage, there is aminopeptidase activity that stops at the peptide AP9–27 (AP6–27 through AP9–27 are observed). The most intense peak in many bag cell mass spectra is AP9–27. As with ELH processing, the peptide between the two cleavages, AP9–20, also is seen. Interestingly, there is no cleavage at the central Leu18–Leu19. In summary, although a number of AP-derived peptides are observed, AP9–27, AP1–20, and the multiply cleaved AP9–20 represent the most fully intracellularly processed forms.
Previously published results (34, 35) have shown that nonstoichiometric amounts of ELH and AP have been recovered from tissue extracts, and it was suggested that ELH and AP peptide were differentially packaged, even though electron microscopy of bag cells has not demonstrated a new vesicle type. However, those results of a variable ELH/AP ratio also can be explained by multiple forms of AP and ELH described here. As further evidence supporting multiple forms of these peptides, [3H]Leu radiolabeling studies indicate the presence of another component that closely follows levels of AP (35). In addition, it has been proposed that a physiologically inactive—but immunoreactive—form of ELH is released in response to cooler ambient temperatures (33).
Bag Cell Peptide Distribution.
In addition to the assay of cell somas in the bag cell cluster, the assay of presumed target abdominal neurons allows bag cell peptide distribution to be studied. The mass spectrum in Fig. 4A is of the abdominal L3 neuron. Mass spectra of L3 neuron somas (n = 3) contain a number of identifiable peptides. In addition, the N-terminal peptides from pELH are not at detectable levels (<5 fmol) and may be lacking. This finding is surprising considering that the application of α-BCP effects the membrane properties of L3, as well as other left upper quadrant neurons (17, 31, 36). Further studies involving the distribution of the bag cell peptides throughout the abdominal ganglion are underway. The peptides present at bag cell terminals should represent fully processed peptides (peptides ready to be released). From a comparison of the AP9–27: AP intensity ratio in Fig. 4A (terminals) to that in Fig. 2B (somas), a significantly higher normalized intensity of AP9–27 is evident. This result strongly confirms the ability of the bag cell neurons to target specific vesicles, in this case the ELH/AP type, to specific target cells.
The only undetected peptide reported to be in the bag cells is Aplysia NPY (26). Encoded by a different gene, NPY was purified from whole ganglia and localized by staining. Results from screening identified abdominal cells [LUQ (left upper quadrant), R2, R3–14, R15, RB, etc.] indicate that the NPY gene is not expressed in the bag cells, rather in cells of the RG cluster.
Peptide Release.
The measurement of peptides within cells does not imply the novel peptides are released. Peptides such as ELH, α-, β-, and γ-BCPs have been shown to be released during the bag cell afterdischarge (11). Because of the improved mass resolution and desalting techniques, we have used MALDI to follow peptide release, optimizing the system for the detection of the novel peptides.
The profile of released peptides is different from that seen in the isolated cell (compare Figs. 1B and 2). However, the observed peptides agree with the processing scheme in Fig. 5. This discrepancy in peak intensities and detection is, at least partly, caused by a different “salt tolerance” for each peptide. Some peptides are less easily ionized and detected in the presence of salt than others. For example, ionization of AP9–27 and ELH is significantly suppressed in the presence of salts, which contributes to the low intensity of these two peptides in mass spectra of releasate. Thus, we cannot compare mass spectra from isolated cells and releasate. However, spectra from bag cell release can be compared with each other as all peptides are in a similar environment. As we do not observe peptides when the cells are not stimulated electrically, nor when the cells are stimulated in a low Ca2+/high Mg2+ matrix (data not shown), we are observing physiological release. We do observe release of the full complement of shortened AP peptides. Interestingly, we also observe release of the R3–14 peptide II and several additional peptides not observed in bag cell neurons, even though most, if not all, of the abdominal cells have been removed in our preparation.
In summary, we have identified several additional peptides in Aplysia bag cell neurons derived from the known ELH precursor and demonstrated that they are transported to target cells and released in a physiologically dependent manner. We currently are evaluating the biological activity of AP9–27, as this peptide is often the most intense peak in the mass spectrum. From a technological standpoint, we have expanded the role of single-cell MALDI-TOF MS to obtain semiquantitative results by using internal spectral normalization. In addition, we have used MALDI-TOF MS for mapping gene expression, in a manner similar to immunohistochemistry, to find the cells that produce NPY.
Acknowledgments
We thank Juliann Gleeson and Richard Milberg for assisting with the mass spectrometers. R.W.G. acknowledges the support of a Beckman Institute Research Assistantship and an American Chemical Society Analytical Division Fellowship, sponsored by R.W. Johnson Pharmaceutical Research Institute, and J.V.S. acknowledges a Sloan Fellowship. This study is supported by National Institutes of Health Grant 1 R29NS31609 and National Science Foundation Grant CHE 96-22663. The TofSpec mass spectrometer was purchased in part with National Institutes of Health Grant RR07141. A. californica were partially provided by the National Center for Research Resources, National Resource for Aplysia at the University of Miami under National Institutes of Health Grant RR10294.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: AP, acidic peptide; ASW, artificial seawater; BCP, bag cell peptide; DHB, 2,5-dihydroxybenzoic acid; ELH, egg-laying hormone; MALDI, matrix-assisted laser desorption/ionization; pELH, egg-laying hormone prohormone; TOF MS, time-of-flight MS; NPY, neuropeptide Y.
A commentary on this article begins on page 3338.
References
- 1.Sossin W S, Fisher J M, Scheller R H. Neuron. 1989;2:1407–1417. doi: 10.1016/0896-6273(89)90186-4. [DOI] [PubMed] [Google Scholar]
- 2.Conn P J, Kaczmarek L K. Mol Neurobiol. 1989;3:237–273. doi: 10.1007/BF02740607. [DOI] [PubMed] [Google Scholar]
- 3.Wayne N L. J Endocrinol. 1995;147:1–4. doi: 10.1677/joe.0.1470001. [DOI] [PubMed] [Google Scholar]
- 4.Fisher J M, Sossin W, Newcomb R, Scheller R H. Cell. 1988;54:813–822. doi: 10.1016/s0092-8674(88)91131-2. [DOI] [PubMed] [Google Scholar]
- 5.Kreiner T, Fisher J M, Sossin W, Scheller R H. Mol Brain Res. 1989;6:135–142. doi: 10.1016/0169-328x(89)90047-8. [DOI] [PubMed] [Google Scholar]
- 6.Sossin W S, Sweet-Cordero A, Scheller R H. Proc Natl Acad Sci USA. 1990;87:4845–4848. doi: 10.1073/pnas.87.12.4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kauer J A, Fisher T E, Kaczmarek L K. J Neurosci. 1987;7:3623–3632. doi: 10.1523/JNEUROSCI.07-11-03623.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scheller R H, Rothman B S, Mayeri E. Trends Neurosci. 1983;6:340–345. [Google Scholar]
- 9.Scheller R H, Jackson J F, McAllister L B, Rothman B S, Mayeri E, Axel R. Cell. 1983;32:7–22. doi: 10.1016/0092-8674(83)90492-0. [DOI] [PubMed] [Google Scholar]
- 10.Newcomb R, Fisher J M, Scheller R H. J Biol Chem. 1988;263:12514–12521. [PubMed] [Google Scholar]
- 11.Newcomb R, Scheller R H. Brain Res. 1990;521:229–237. doi: 10.1016/0006-8993(90)91547-t. [DOI] [PubMed] [Google Scholar]
- 12.Lewis R V, Stern A S. Annu Rev Pharmacol Toxicol. 1983;23:353–372. doi: 10.1146/annurev.pa.23.040183.002033. [DOI] [PubMed] [Google Scholar]
- 13.Mains R, Eipper B A. J Biol Chem. 1981;256:5683–5688. [PubMed] [Google Scholar]
- 14.Fleminger G, Lahm H, Udenfriend S. Proc Natl Acad Sci USA. 1984;81:3587–3590. doi: 10.1073/pnas.81.11.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lorenz R G, Evans C J, Barchas J D. Life Sci. 1985;37:1523–1528. doi: 10.1016/0024-3205(85)90184-5. [DOI] [PubMed] [Google Scholar]
- 16.Nordmann J J, Cazalis M, Dayanithi G, Castanas E, Giraud P, Legros J J. Cell Tissue Res. 1986;246:177–182. doi: 10.1007/BF00219015. [DOI] [PubMed] [Google Scholar]
- 17.Sigvardt K A, Rothman B S, Brown R O, Mayeri E. J Neurosci. 1986;6:803–813. doi: 10.1523/JNEUROSCI.06-03-00803.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arch S J. Gen Physiol. 1972;59:47–59. doi: 10.1085/jgp.59.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kupfermann I. Am Zool. 1972;12:513–519. [Google Scholar]
- 20.Nagle G T, Painter S D, Blankenship J E, Kurosky A. J Biol Chem. 1988;263:9223–9237. [PubMed] [Google Scholar]
- 21.Li K W, Hoek R M, Smith F, Jiménez C R, van der Schors R C, van Veelen P A, Chen S, van der Greef J, Parish D C, Benjamin P R, Geraerts W P M. J Biol Chem. 1994;269:30288–30292. [PubMed] [Google Scholar]
- 22.de With N D, Li K W, Jiménez C R, Vonk N, Dreisewerd K, Hillenkamp F, Karas M, Geraerts W P M. Peptides. 1997;18:765–770. doi: 10.1016/s0196-9781(97)00020-x. [DOI] [PubMed] [Google Scholar]
- 23.Ekman R, Juhasz P, Heilig M, Ågren H, Costello C E. Peptides. 1996;17:1107–1111. doi: 10.1016/s0196-9781(96)00168-4. [DOI] [PubMed] [Google Scholar]
- 24.Jiménez C R, Ki K W, Dreisewerd K, Mansvelder H D, Brussaard A B, Reinhold B B, Van der Schors R C, Karas M, Hillenkamp F, Burbach J P H, Costello C E, Geraerts W P M. Proc Natl Acad Sci USA. 1997;94:9481–9486. doi: 10.1073/pnas.94.17.9481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garden R W, Moroz L L, Moroz T P, Shippy S A, Sweedler J V. J Mass Spectrom. 1996;31:1126–1130. doi: 10.1002/(SICI)1096-9888(199610)31:10<1126::AID-JMS403>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 26.Rajpara S M, Garcia P D, Roberts R, Eliassen J C, Owens D F, Maltby D, Myers R M, Mayeri E. Neuron. 1992;9:505–513. doi: 10.1016/0896-6273(92)90188-j. [DOI] [PubMed] [Google Scholar]
- 27.Nick T A, Moreira J E, Kaczmarek L K, Carew T J, Wayne N L. J Neurophysiol. 1996;76:3351–3359. doi: 10.1152/jn.1996.76.5.3351. [DOI] [PubMed] [Google Scholar]
- 28.Pulst S M, Rothman B L, Mayeri E. Neuropeptides. 1987;10:249–259. doi: 10.1016/0143-4179(87)90075-8. [DOI] [PubMed] [Google Scholar]
- 29.Newcomb R, Scheller R H. J Neurosci. 1987;7:854–863. doi: 10.1523/JNEUROSCI.07-03-00854.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang H Y, Caprioli R M. J Mass Spectrom. 1996;31:1039–1046. doi: 10.1002/(SICI)1096-9888(199609)31:9<1039::AID-JMS398>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 31.Rothman B S, Mayeri E, Brown R O, Yuan P M, Shively J E. Proc Natl Acad Sci USA. 1983;80:5753–5757. doi: 10.1073/pnas.80.18.5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Squire C R, Talebian M, Menon J G, Dekruyff S, Lee T D, Shively J E, Rothman B S. J Biol Chem. 1991;266:22355–22363. [PubMed] [Google Scholar]
- 33.Wayne N L, Nick T, Block G D. Gen Comp Endocrinol. 1996;102:351–359. doi: 10.1006/gcen.1996.0078. [DOI] [PubMed] [Google Scholar]
- 34.Arch S, Linstedt A, Whitney G, Teal P, Smock T. J Neurosci. 1986;6:1545–1552. doi: 10.1523/JNEUROSCI.06-06-01545.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Molloy S, Bruns C, Arch S. Peptides. 1987;8:829–836. doi: 10.1016/0196-9781(87)90067-2. [DOI] [PubMed] [Google Scholar]
- 36.Brownell P, Mayeri E. Science. 1979;204:417–420. doi: 10.1126/science.35827. [DOI] [PubMed] [Google Scholar]