Abstract
Background
We evaluated whether early ribavirin pharmacokinetics differ comparing hepatitis C/human immunodeficiency virus coinfected sustained virological responders and nonresponders.
Methods
Twenty-four treatment-naïve coinfected patients received pegylated-interferon alfa-2b (12kD) (1.5μg/kg) once weekly plus daily ribavirin (13.6 mg/kg/d) for up to 48 weeks. Serum HCV RNA, serum alanine aminotransferase, and plasma ribavirin levels were measured frequently during the first 16 days of therapy and monthly thereafter.
Results
Six patients are sustained responders. During the first four weeks of treatment, median plasma ribavirin levels and area under the ribavirin curve were significantly lower (p<0.0001 and p<0.01, respectively) in sustained responders compared with nonresponders. Additionally, ribavirin levels in sustained responders continued to increase significantly until week 8 (p<0.02). At week 4, hemoglobin declines were significantly (p=0.002) greater in sustained responders than nonresponders. At week 1, serum HCV RNA levels and changes in alanine aminotransferase levels relative to baseline could identify likely responders better than plasma ribavirin levels.
Conclusions
We conjecture that intracellular ribavirin accumulation may be enhanced early in treatment in coinfected sustained responders, although this hypothesis should be investigated further. At week 1, serum HCV RNA and changes in alanine aminotransferase levels relative to baseline might identify likely responders.
Keywords: Ribavirin, pharmacokinetics, hepatitis C virus/human immunodeficiency virus coinfection, hepatitis C virus, treatment outcome biomarkers
Introduction
Hepatitis C virus (HCV) and human immunodeficiency virus (HIV) coinfect ∼250,000 individuals in the United States [1]. The number of sustained virological responders (SVRs) to current standard therapy, pegylated-interferon and ribavirin (RBV), is lower in HIV/HCV coinfected than in HCV monoinfected patients. Pegylated-interferon/RBV results in successful therapeutic outcomes only in 27-44% of coinfected patients [2-5] compared with ∼50% of monoinfected patients [6, 7]. Among difficult-to-treat coinfected genotype 1 patients, the percentage of SVRs is even lower, ranging from 8-38% [2-5, 8]. Therefore, understanding the determinants of a successful treatment outcome and the early identification of patients who are likely to be SVRs are important clinical challenges that are particularly relevant to HIV/HCV coinfected patients.
RBV, initially approved in the United States in 1998, principally prevents HCV relapse in patients in whom HCV RNA is undetectable at the end of treatment. Although RBV monotherapy imposes a transient HCV decline early in therapy, the drug has no apparent long-term effect on HCV RNA [9, 10]. Recent studies, however, have suggested that increased RBV doses and increased peripheral blood RBV levels may improve treatment outcomes [11]. To our knowledge, detailed RBV pharmacokinetic analysis early in treatment, in either mono or coinfected patients, has not been performed.
We were interested in evaluating whether early RBV pharmacokinetics differ comparing HIV/HCV coinfected SVRs and nonresponders (NRs) and whether RBV levels early in treatment are predictive of being an SVR. Currently, only failure to achieve ≥2-log10 HCV RNA decline by week 12 is used in the clinic to justify treatment discontinuation [2-5, 12-14] although other parameters, such as alanine aminotransferase (ALT) levels, might also be clinically useful [15]. We measured plasma RBV, serum HCV RNA, and serum ALT levels frequently after the initial three doses of pegylated-interferon alfa-2b (12kD) and RBV in 24 HIV/HCV coinfected treatment naïve patients. We found that early plasma RBV pharmacokinetics differ in SVRs compared with NRs. Although early RBV levels do not discriminate well between likely SVRs and NRs, the linear combination of serum HCV RNA levels at day 7 and the ratio of ALT levels at day 7 to baseline had excellent ability, albeit in this small, coinfected cohort, to identify likely SVRs.
Patients and methods
Study subjects
In brief, 24 HCV/HIV-1 coinfected, pegylated-interferon alfa/RBV treatment naïve patients were recruited from the Hepatitis Clinic of New York/Weill/Cornell Medical Center in a cohort study with convenience sampling to evaluate early RBV pharmacokinetics in coinfected patients. Enrollment was discontinued when we achieved a percentage of end of treatment responders within the range reported previously [5]. Subsequent studies have confirmed that our SVR percentage is in agreement with those of other studies [2-5]. The protocol conforms to the 1975 Helsinki guidelines for the conduct of human research. The Institutional Review Board of Weill Medical College of Cornell University approved the study, and written informed consent was obtained from each patient. Inclusion criteria, as described previously [16], required that patients have detectable HCV RNA, be on a stable course of antiretroviral therapy or no antiretroviral agents for at least four weeks prior to pegylated-interferon/RBV initiation, and to have a CD4+ T cell count ≥ 100 cells/mm3. Pretreatment liver biopsies were obtained on all patients and were assessed using the Scheuer system [17]. Patients were excluded if they had severe depression, immunodeficiency-related opportunistic infections, active substance abuse, or were pregnant or lactating. Subjects were treated with pegylated-interferon alfa-2b 1.5 μg/kg (PEG-INTRON, Schering Plough Corporation, Kenilworth, NJ) once weekly and a total daily dose of RBV (Rebitrol, Schering Plough) 1000-1200 mg, taken every twelve hours, for up to 48 weeks. Patients with HCV RNA below detection (<29 IU/ml) 24 weeks after treatment cessation are SVRs. Of a total of 24 patients enrolled, two patients withdrew during the first week because of treatment-related side effects (hematologic) or noncompliance, and RBV levels were not assayed on these patients.
Sample acquisition
Samples for HCV RNA and RBV determinations were obtained at baseline, after the first pegylated-interferon and RBV doses (6, 12, 24 hours) and on days 2, 3, 5, 6, 7; after the second pegylated-interferon dose (6, 12, 24 hours) and on days 9, 14; after the third pegylated-interferon dose on days 15, 16. Subsequently, patients returned monthly for the first three months, then every six weeks, and three times after treatment discontinuation.
HCV and HIV-1 measurements and HCV genotyping
Total HCV RNA was extracted and quantitated using a validated real-time reverse transcriptase polymerase chain reaction assay with a lower limit of detection of 29 IU/ml (Schering-Plough Research Institute). The amplification target was the HCV 5′-untranslated region. An internal RNA control and appropriate positive and negative controls were added to each run. HIV RNA was measured using the Roche COBAS Amplicor HIV-1 MONITOR assay version 1.5 (Roche Diagnostics, Branchberg, NJ). HCV genotypes were determined by Quest Diagnostics (Teterboro, NJ).
RBV assay
RBV concentration was measured using previously published methods [18-20] in an assay consisting of phenylboronic acid solid phase extraction (PBA SPE) followed by high-performance liquid chromatography (HPLC) with minor modifications. In brief, to 200 μl of previously frozen plasma samples were added 5.0 μl internal standard mixture(0.40 μg/μl each of 3-methylcytidine methosulfate [3MCMS, Sigma, St. Louis, MO] and 5-methylcytidine [5MC, Sigma]), and 800 μl of 250 mM ammonium acetate buffer (pH 8.5). The solutions were then vortexed and loaded onto PBA SPE columns (Bond Elute-PBA [100 mg, 1 ml], Varian Inc, Palo Alto, CA). After washing with five 1 ml aliquots of buffer followed by 1 ml water, the adsorbates were eluted with 1 ml of 100 mM formic acid in methanol. The collected eluent solutions were evaporated to dryness in a 70°C sandbath under nitrogen stream and taken up in 200 μl of mobile phase.
The HPLC apparatus consisted of an “Ultimate” fully integrated micro-HPLC system (LC-Packings, Dionex Corp, Sunnyvale, CA), reverse phase C18 columns, 15 cm, 300 μm internal diameter with a 45 nl volume 10 mm path length 4-wavelength UV detector. The system was controlled by Dionex’s “Chromeleon” software. The HPLC separation was carried out under isocratic conditions. Aliquots of 1.0 μl of sample were injected by an autosampler onto the column at 30°C. The mobile phase composition of 98:2.0:0.15 (v/v) water/methanol/trifluoroacetic acid (Sigma) was chosen to provide acceptable separation (from unidentified component peaks originating in the plasma) and convenient retention times of approximately 7, 11, and 14 minutes for RBV, 3MCMS, and 5MC, respectively. The chromatograms were recorded simultaneously at 207, 215, and 240 nm wavelengths. As the best ratio of RBV signal to background absorption was observed at 215 nm, all results reported here are based on data acquired at that wavelength. Quantitation is based on the area ratios of the RBV peak to the peak of the primary internal standard (3MCMS). The calibration curve was constructed using plasma aliquots from a one-time blood draw of a consenting donor spiked with known amounts of RBV (Sigma) (5 points in the range 1.4 to 20.0 μg/ml) and carried through the extraction and HPLC steps.
Statistical analysis
We used nonparametric methods in an intention-to-treat analysis to compare baseline characteristics of SVRs vs. NRs (Table 1). In the analysis of longitudinal parameters (e.g., plasma RBV levels, serum log10HCV RNA levels, and changes in ALT levels relative to baseline), two patients were excluded due to treatment discontinuation during the first week. Where appropriate, we used either the Mann-Whitney U or the Wilcoxon signed-rank tests (S-PLUS, V.7.0.2, Seattle, WA). To compare categorical variables, we used the two-tailed Fisher exact test. We used the Spearman test to evaluate the correlation between variables. To determine whether plasma RBV levels differ between SVRs and NRs over time, we used Friedman’s test, the nonparametric analogue of the two-way analysis of variance. To evaluate the direction of the association, we used Page’s test[21]. All multiple comparisons were adjusted using the Bonferoni correction and the overall level of significance was set at p=0.05. The RBV area under the curve was calculated by summing the rectangle areas in a given time period. Each area was measured by multiplying the average of two consecutive RBV concentrations by the time between the corresponding points (in days) [22].
Table 1.
Baseline characteristics
| All (n = 24) | SVRs (n=6) | NRs (n=18) | P value | |
|---|---|---|---|---|
| Male: Female, n | 21:3 | 6:0 | 15:3 | 0.54 |
| Median age in years, (IQR) | 46.5 (42.5-48.5) | 47.0 (37.0-55.0) | 46.5 (43.0-48.0) | 0.64 |
| Race, n (%) | ||||
| Caucasian | 7 (29) | 3 (50) | 4 (22) | 0.48 |
| African American | 12 (50) | 2 (33) | 10 (56) | |
| Hispanic | 5 (21) | 1 (17) | 4 (22) | |
| Median weight in kilograms, (IQR)* | 83.0 (78.5-88.5) | 81.6 (65.8-82.6) | 84.6 (79.8-89.8) | 0.04 |
| Median body mass index (kg/m2), (IQR) | 25.5 (23.5-29.0) | 24.3 (22.2-25.2) | 26.7 (23.6-30.0) | 0.10 |
| Median CD4 (cells/mm3), (IQR) | 467.0 (326.5-502.0) | 361.5 (209.0-493.0) | 477.5 (434.0-506.0) | 0.13 |
| Patients with plasma HIV RNA <400 copies/ml, (%) | 16 (67) | 5 (83) | 11 (61) | 0.62 |
| Median serum HCV RNA (log10 IU/ml)*, (IQR) | 6.3 (5.4 -6.7) | 5.3 (4.7-5.5) | 6.6 (6.1-6.8) | 0.003 |
| HCV genotype, (%) | ||||
| HCV 1a/b | 21 (87.5) | 5 (83) | 16 (89) | 1.0 |
| HCV 2 or 3a | 3 (12.5) | 1 (17) | 2 (11) | |
| Median hemoglobin level (g/dl), (IQR) | 13.5 (12.5-15.0) | 14.5 (13.3-15.3) | 13.1 (12.4-14.5) | 0.20 |
| Median ALT (U/l), (range) | 85.5 (40.0 - 119.0) | 203.0 (34.0 - 299.0) | 65.0 (42.0 - 113.0) | 0.14 |
| Inflammatory grade, n (%) | ||||
| 0 - 2 | 14 (58) | 3 (50) | 11 (61) | 0.67 |
| 3 - 4 | 10 (42) | 3 (50) | 7 (39) | |
| Fibrosis stage, n (%) | ||||
| 0 - 2 | 11 (46) | 2 (33) | 9 (50) | 0.65 |
| 3 - 4 | 13 (54) | 4 (67) | 9 (50) |
Abbreviations: SVRs, sustained virological responders; NRs, nonresponders; IQR, interquartile range; IU, international units; ALT, alanine aminotransferase; HCV, hepatitis C virus; HIV, human immunodeficiency virus.
p<0.05 for a difference between SVRs and NRs
To determine the discriminatory ability to identify likely SVRs of plasma RBV levels early in treatment, serum log10HCV RNA, and the ratio between ALT levels at day 7 and at baseline (ALTd7/ALTd0), we computed the area under the receiver operator characteristic curve at day 7 (STATA, Version 8.0, STATA Corp, College Station, TX). The area under the receiver operator characteristic curve is a measure of the probability that in a randomly selected pair of SVR and NR patients, the marker permits correct identification. We also report the positive and negative predictive values, which quantify the clinical value of a marker. To increase the discriminatory ability of these parameters to identify likely SVRs, we combined multiple parameters and nonparametrically assessed [23] whether the linear combination improved the discriminatory ability. The method is based on the observation that the area under the receiver operator characteristic curve for a continuous score S is interpreted as P (SNR ≥ SSVR), where SNR and SSVR are scores for independent, randomly selected subjects for the NRs and SVRs respectively. It uses the Mann-Whitney U-statistic estimator of the area under the curve associated with the linear combination score, and one chooses the coefficients that maximize the Mann-Whitney estimator of the area under the curve. The linear combination has the form:
where the coefficients (α1, α2, α3) can be interpreted as the relative weight of each parameter.
Results
Patient characteristics and virological responses
Six (25%) patients were SVRs (5 HCV genotype-1, one genotype-3). Among SVRs, five patients (four genotype -1) reached undetectable HCV RNA (<29 IU/ml) during the first 10 days of treatment. Among baseline characteristics, only HCV RNA was significantly (p = 0.003) lower in SVRs than NRs (Table 1). ALT levels correlated (r=0.44, p=0.04) with necroinflammation but not with fibrosis.
Early plasma RBV levels are lower in SVRs compared with NRs
Longitudinal RBV levels were obtained on all patients except two who withdrew during the first week. For all patients, the median RBV concentration was 2.0 (range, 0.6 - 4.6) μg/ml at week 2, and it continued to increase until a median concentration of 3.2 (range, 1.1 - 7.3) μg/ml was reached at week 8.
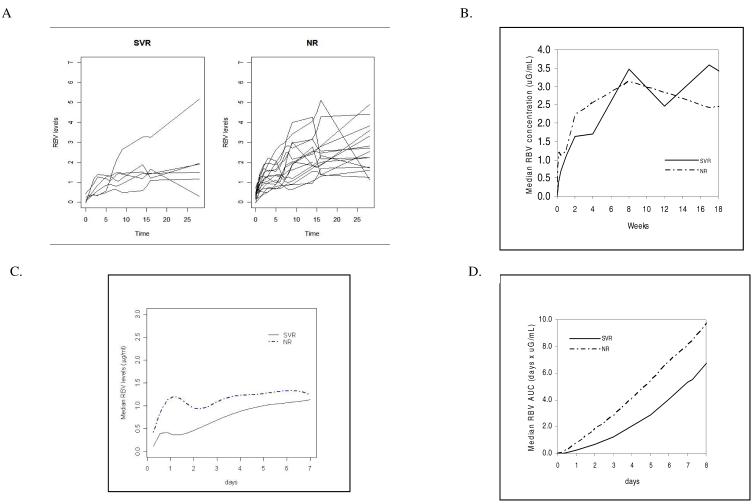
Stratification by treatment outcome revealed interesting differences between SVRs and NRs in early plasma RBV levels and in the rate of RBV accumulation. Median plasma RBV levels were significantly decreased in SVRs compared with NRs during the first two weeks of treatment (p<0.0001, Fig 1A-1C). Similarly, median RBV area under the curve levels were lower in SVRs compared with NRs at days 3, 7, 14, and 28 (p<0.01, Fig 1D).
Fig. 1.
A) Plasma ribavirin levels during the first 28 days of treatment for individual patients stratified according to treatment outcome, sustained virological responders (SVRs) and nonresponders (NRs). B) Differences in median ribavirin (RBV) concentration in plasma during the first 18 weeks of treatment in SVRs and NRs. C) Cubic spline fit of median RBV levels during the first week for SVRs and NRs. An exact cubic spline was fit using the R spline function according to the Forsythe, Malcolm, and Moler method [35]. D) Differences in median RBV area under the curve during the first 8 days of treatment between SVRs and NRs. RBV area under the curve was significantly lower in SVRs compared with NRs for the first 28 days of treatment (p<0.01).
In SVRs, median RBV levels continued to increase significantly until week 8 (p<0.02 comparing median RBV levels at both weeks 2 and 4 with levels at week 8, median slope 0.012 μg/ml day-1). In contrast, in NRs median RBV levels increased only modestly between weeks 2 and 8 (2.3 μg/ml and 2.7 μg/ml, respectively; median slope 0.0012 μg/ml day-1). In SVR patients, RBV slope tended to increase (p=0.078) between weeks 4 and 8 in comparison with NR patients.
We also evaluated hemoglobin declines stratified by treatment outcome. Although median baseline hemoglobin levels were roughly equivalent in SVRs and NRs (p=0.20), hemoglobin decline during the first month of therapy was significantly greater in SVRs than NRs (3.9 vs. 1.4 gm/dl, p=0.002). Since renal function is a major determinant of RBV accumulation [24], we estimated the glomerular filtration rate using the Cockroft-Gault equation [25]. Neither median baseline serum creatinine (p=0.5) nor median estimated glomerular filtration rate (p=0.3) differed significantly between SVRs and NRs. Although not the primary study objective, graphical analysis indicates that RBV levels differ by race (Caucasian versus African-American) in both SVRs and NRs (data not shown).
Early single predictors of successful treatment response
Because of differences in early RBV pharmacokinetics between SVRs and NRs, we evaluated RBV performance capacity by calculating the area under the receiver operator characteristic curve at week 1. We found that early plasma RBV levels had poor discriminatory ability (area under the receiver operator characteristic curve ± standard error: 0.57 ± 0.13, 95% confidence interval [0.30,0.83]) to identify likely SVR patients very early in treatment.
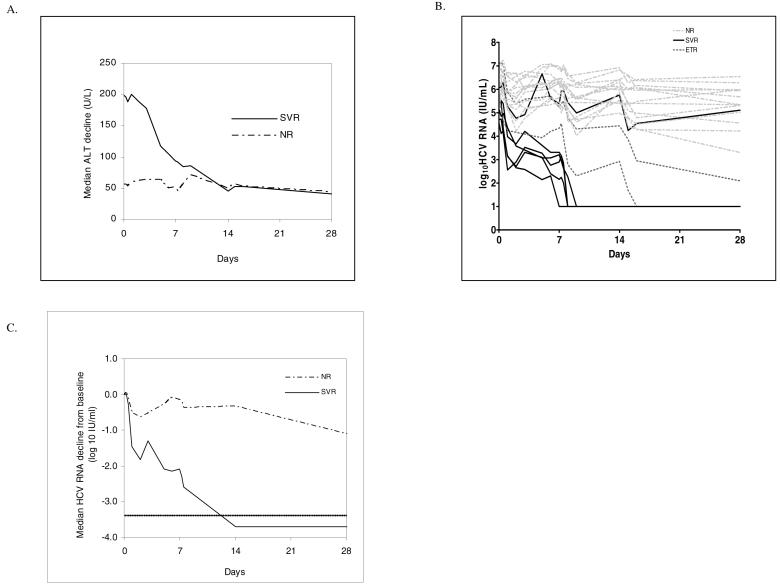
Because of the poor discriminatory ability of early RBV levels, we evaluated other parameters, particularly ALT (Fig 2A) and HCV RNA (Fig 2B-C), for their ability to identify SVRs early in treatment. In SVRs, median baseline ALT values were 3-fold higher, ALT decline during the first 14 days tended to be significantly faster (p=0.04), and, consequently, day 14 levels were equivalent when compared with NRs (Fig 2A). The ALT ratio at day 7 to baseline (ALTd7/ALTd0) had reasonably good discriminatory ability (0.80 ± 0.11, [0.58, 1.0]) to identify likely SVRs very early in treatment. We chose to evaluate the ratio of ALT at day 7 to baseline instead of absolute day 7 ALT values (0.40 + 0.19, [0.04, 0.77]) because of the improved performance characteristics, namely increased sensitivity over specificity ranges 0.4 to 1. We also computed the positive and negative predictive values for ALTd7/ALTd0. Using an ALTd7/ALTd0 cutoff value of 0.7, we calculated positive predictive value = 71% and negative predictive value = 93% (Table 2).
Fig. 2.
A) Median alanine aminotransferase (ALT) decline from baseline in sustained virological responders (SVRs) and nonresponders (NRs) during the first four weeks of pegylated interferon and ribavirin treatment. B) Changes in log10HCV RNA in SVRs, end of treatment responders (ETRs) and NRs during the first 28 days of pegylated interferon and ribavirin treatment. C) Median HCV RNA decline from baseline in SVRs and NRs during the first 28 days of pegylated interferon and ribavirin treatment. Dotted line indicates level of assay detection for HCV RNA (29 IU/ml).
Table 2.
Predictive values of week 1 log10 HCV RNA, the ratio between baseline and week 1 alanine aminotransferase levels, and the linear combination with the best discriminatory ability (ALTd7/ALTd0 + 0.7 log10 HCV RNA).
| log10HCV RNA | ALTd7/ALTd0 | log10HCV RNAd7 - 0.67 ALTd7/ALTd0 + 0.01 RBVd7. | |
|---|---|---|---|
| Cutoff | ≤ 5.5 | ≤ 0.7 | ≤ 4.7 |
| PPV | 60 | 71 | 67 |
| NPV | 100 | 93 | 100 |
| Sn | 100 | 83 | 100 |
| Sp | 75 | 87 | 81 |
| AUROC ± SE | 0.96 ± 0.03 | 0.80 ± 0.11 | 0.98 ± 0.02 |
Formulas and abbreviations: ALTd7/ALTd0, ratio of day 7 to baseline alanine aminotransferase levels; PPV, positive predictive value; NPV, negative predictive value; Sn, sensitivity; Sp, specificity; AUROC, area under the receiver operator characteristic curve; SE, standard error.
Serum log10HCV RNA had good discriminatory ability (0.96 ± 0.03, [0.90, 1.0]) to identify likely SVR patients very early in treatment. Using a week 1 serum log10HCV RNA cutoff value of 5.5, positive predictive value = 60% and negative predictive value = 100% for the detection of a successful therapeutic outcome (Table 2).
Combinations of parameters improve diagnostic accuracy of successful treatment response
To improve on the discriminatory ability of each parameter individually, we examined the ability of linear combinations of the parameters to detect likely SVRs early in treatment. The linear combination that produced the maximum area under the receiver operator characteristic curve (0.98 ± 0.02, [0.93, 1.0]) is:
In fact, this linear combination simultaneously maximizes the specificity at high ranges of sensitivity. Based upon the area under the receiver operator characteristic curve, the combination of day 7 HCV RNA levels and the ratio of day 7 to baseline ALT levels have the best discriminatory ability to identify likely SVRs in comparison with each individual parameter. The addition of plasma RBV levels as early as day 7 does not improve the diagnostic accuracy.
A cutoff value of the score ≤ 4.7 has positive predictive value = 67%, negative predictive value = 100%, sensitivity = 100% and specificity = 81%.
Discussion
In this study, we found that early plasma RBV levels and RBV area under the curve early in treatment were significantly lower in SVRs compared with NRs. Moreover, the median hemoglobin decline over this period was significantly larger in SVRs. We conjecture that decreased early plasma RBV levels in SVRs and enhanced hemoglobin declines early in treatment may reflect higher intracellular RBV concentration in SVRs, although this hypothesis will need to be investigated further. Early studies demonstrated that RBV concentration in erythrocytes can vary from 9 to 60 times more than the plasma concentration following multiple twice daily dosing [26, 27]. Higher intracellular RBV concentration may translate into increased mutagenic effect, a proposed mechanism of action for RBV, which may act by lowering HCV infectivity [28] thereby leading to an SVR.
Increased intracellular RBV concentration could also lead to increased anemia, the most frequent adverse effect of RBV therapy. In anucleate cells that also lack dephosphorylation enzymes such as erythrocytes, RBV is irreversibly phosphorylated intracellularly [29]. In these cells, RBV triphosphate accumulates resulting in competitive inhibition of ATP-dependent oxidative respiration, shortened erythrocyte survival and hemolytic anemia [30, 31]. We found that week 4 hemoglobin declines were significantly greater in SVRs compared with NRs. In agreement with our results, two recent studies found that erythrocyte RBV levels correlate with hemoglobin reductions at weeks 2 - 4 in IFN alfa-2b/RBV treated monoinfected patients [32, 33]. In contrast, another recent study did not observe differences in RBV levels comparing SVRs and NRs [34]. That study, however, used a substantially reduced RBV dose (600-800) than was used in this study. We speculate that, early in treatment, SVRs might have enhanced intracellular RBV transport resulting in increased intracellular RBV concentration and greater hemoglobin reductions. Additional studies with measurement of intracellular ribavirin levels will be required to determine whether this is in fact the case.
Early prediction of a successful treatment outcome to pegylated-interferon/RBV is particularly important in difficult-to-treat HIV/HCV coinfected patients [2-5, 8] to permit early treatment discontinuation in those who are unlikely to respond. In a recent publication, we identified several pegylated-interferon pharmacodynamic parameters that might identify likely SVR patients early in treatment [16]. Here, in order to identify other promising early predictors of successful treatment outcome, we evaluated the predictive value of plasma RBV levels, changes in serum ALT levels at week 1 relative to baseline, and serum log10 HCV RNA levels at week 1 for their potential to identify likely SVRs. We found that the ratio between ALT levels at day 7 to baseline and serum log10 HCV RNA levels at week 1 had a negative predictive value of 100% for the identification of likely SVRs at week 1. The high negative predictive value at week 1 minimizes the likelihood of premature treatment discontinuation in patients who might achieve a successful therapeutic outcome. We also found that early RBV pharmacokinetics differ in SVRs compared with NRs, which may be one determinant of treatment outcome. To evaluate whether these parameters can lead to the early identification of likely nonresponders, studies in larger numbers of patients are needed.
Acknowledgments
We thank Drs. Marshall Glesby and Ira Jacobson for critical review of the manuscript, and Queenie Brown M.S. for assistance with sample procurement. This research was supported, in part, by a General Clinical Research Center Grant (M01-RR00047) from the National Center for Research Resources, National Institutes of Health, RR06555 (ASP), RR18754 (RMR), the Greenberg Foundation for Medical Research, Schering-Plough Research Institute, a fellowship from the Fulbright Foundation (HD) and the Speaker’s Fund for Public Health Research awarded by the city of New York (AHT). A portion of this work was performed under the auspices of the US Department of Energy under contract W-7405-ENG-36.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sherman KE, Rouster SD, Chung RT, Rajicic N. Hepatitis C virus prevalence among patients infected with human immunodeficiency virus: a cross-sectional analysis of the US Adult AIDS Clinical Trials Group. Clin Infect Dis. 2002;34:831–837. doi: 10.1086/339042. [DOI] [PubMed] [Google Scholar]
- 2.Chung RT, Andersen J, Volberding P, Robbins GK, Liu T, Sherman KE, et al. Peginterferon Alfa-2a plus ribavirin versus interferon alfa-2a plus ribavirin for chronic hepatitis C in HIV-coinfected persons. N Engl J Med. 2004;351:451–459. doi: 10.1056/NEJMoa032653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Torriani FJ, Rodriguez-Torres M, Rockstroh JK, Lissen E, Gonzalez-Garcia J, Lazzarin A, et al. Peginterferon Alfa-2a plus ribavirin for chronic hepatitis C virus infection in HIV-infected patients. N Engl J Med. 2004;351:438–450. doi: 10.1056/NEJMoa040842. [DOI] [PubMed] [Google Scholar]
- 4.Carrat F, Bani-Sadr F, Pol S, Rosenthal E, Lunel-Fabiani F, Benzekri A, et al. Pegylated interferon alfa-2b vs standard interferon alfa-2b, plus ribavirin, for chronic hepatitis C in HIV-infected patients: a randomized controlled trial. Jama. 2004;292:2839–2848. doi: 10.1001/jama.292.23.2839. [DOI] [PubMed] [Google Scholar]
- 5.Ballesteros AL, Franco S, Fuster D, Planas R, Martinez MA, Acosta L, et al. Early HCV dynamics on Peg-interferon and ribavirin in HIV/HCV co-infection: indications for the investigation of new treatment approaches. Aids. 2004;18:59–66. doi: 10.1097/00002030-200401020-00007. [DOI] [PubMed] [Google Scholar]
- 6.Manns MP, McHutchison JG, Gordon SC, Rustgi VK, Shiffman M, Reindollar R, et al. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001;358:958–965. doi: 10.1016/s0140-6736(01)06102-5. [DOI] [PubMed] [Google Scholar]
- 7.Fried MW, Shiffman ML, Reddy KR, Smith C, Marinos G, Goncales FL, Jr., et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002;347:975–982. doi: 10.1056/NEJMoa020047. [DOI] [PubMed] [Google Scholar]
- 8.Laguno M, Murillas J, Blanco JL, Martinez E, Miquel R, Sanchez-Tapias JM, et al. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for treatment of HIV/HCV co-infected patients. Aids. 2004;18:F27–36. doi: 10.1097/00002030-200409030-00003. [DOI] [PubMed] [Google Scholar]
- 9.Dusheiko G, Main J, Thomas H, Reichard O, Lee C, Dhillon A, et al. Ribavirin treatment for patients with chronic hepatitis C: results of a placebo-controlled study. J Hepatol. 1996;25:591–598. doi: 10.1016/s0168-8278(96)80225-x. [DOI] [PubMed] [Google Scholar]
- 10.Pawlotsky JM, Dahari H, Neumann AU, Hezode C, Germanidis G, Lonjon I, et al. Antiviral action of ribavirin in chronic hepatitis C. Gastroenterology. 2004;126:703–714. doi: 10.1053/j.gastro.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 11.Lindahl K, Stahle L, Bruchfeld A, Schvarcz R. High-dose ribavirin in combination with standard dose peginterferon for treatment of patients with chronic hepatitis C. Hepatology. 2005;41:275–279. doi: 10.1002/hep.20563. [DOI] [PubMed] [Google Scholar]
- 12.Alberti A, Clumeck N, Collins S, Gerlich W, Lundgren J, Palu G, et al. Short statement of the first European Consensus Conference on the treatment of chronic hepatitis B and C in HIV co-infected patients. J Hepatol. 2005;42:615–624. doi: 10.1016/j.jhep.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 13.Davis GL, Wong JB, McHutchison JG, Manns MP, Harvey J, Albrecht J. Early virologic response to treatment with peginterferon alfa-2b plus ribavirin in patients with chronic hepatitis C. Hepatology. 2003;38:645–652. doi: 10.1053/jhep.2003.50364. [DOI] [PubMed] [Google Scholar]
- 14.Ferenci P, Fried MW, Shiffman ML, Smith CI, Marinos G, Goncales FL, Jr., et al. Predicting sustained virological responses in chronic hepatitis C patients treated with peginterferon alfa-2a (40 KD)/ribavirin. J Hepatol. 2005;43:425–433. doi: 10.1016/j.jhep.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 15.Martinez-Bauer E, Crespo J, Romero-Gomez M, Moreno-Otero R, Sola R, Tesei N, et al. Development and validation of two models for early prediction of response to therapy in genotype 1 chronic hepatitis C. Hepatology. 2006;43:72–80. doi: 10.1002/hep.21002. [DOI] [PubMed] [Google Scholar]
- 16.Talal AH, Ribeiro RM, Powers KA, Grace M, Cullen C, Hussain M, et al. Pharmacodynamics of PEG-IFN a differentiate HIV/HCV coinfected sustained viral responders from non-responders. Hepatology. 2006;43:943–953. doi: 10.1002/hep.21136. [DOI] [PubMed] [Google Scholar]
- 17.Scheuer PJ. Classification of chronic viral hepatitis: a need for reassessment. J Hepatol. 1991;13:372–374. doi: 10.1016/0168-8278(91)90084-o. [DOI] [PubMed] [Google Scholar]
- 18.Granich GG, Krogstad DJ, Connor JD, Desrochers KL, Sherwood C. High-performance liquid chromatography (HPLC) assay for ribavirin and comparison of the HPLC assay with radioimmunoassay. Antimicrob Agents Chemother. 1989;33:311–315. doi: 10.1128/aac.33.3.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Homma M, Jayewardene AL, Gambertoglio J, Aweeka F. High-performance liquid chromatographic determination of ribavirin in whole blood to assess disposition in erythrocytes. Antimicrob Agents Chemother. 1999;43:2716–2719. doi: 10.1128/aac.43.11.2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larrat S, Stanke-Labesque F, Plages A, Zarski JP, Bessard G, Souvignet C. Ribavirin quantification in combination treatment of chronic hepatitis C. Antimicrob Agents Chemother. 2003;47:124–129. doi: 10.1128/AAC.47.1.124-129.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gibbons JD. Nonparametric methods for quantitative analysis. 3rd edition ed. American Sciences Press, Inc.; Columbus, OH: 1997. pp. 348–350. [Google Scholar]
- 22.Press WH, Teukolsky SA, Vetterling WT, Flannery BP. Numerical Recipes in C++ Cambridge University Press; Cambridge, U.K.: 2002. p. 131. [Google Scholar]
- 23.Pepe MS, Thompson ML. Combining diagnostic test results to increase accuracy. Biostatistics. 2000;1:123–140. doi: 10.1093/biostatistics/1.2.123. [DOI] [PubMed] [Google Scholar]
- 24.Bruchfeld A, Lindahl K, Schvarcz R, Stahle L. Dosage of ribavirin in patients with hepatitis C should be based on renal function: a population pharmacokinetic analysis. Ther Drug Monit. 2002;24:701–708. doi: 10.1097/00007691-200212000-00004. [DOI] [PubMed] [Google Scholar]
- 25.Cockroft DW, Gault MH. Prediction of creatinine clearance from serum creatinine. Nephron. 1976;16:31–41. doi: 10.1159/000180580. [DOI] [PubMed] [Google Scholar]
- 26.Laskin OL, Longstreth JA, Hart CC, Scavuzzo D, Kalman CM, Connor JD, et al. Ribavirin disposition in high-risk patients for acquired immunodeficiency syndrome. Clin Pharmacol Ther. 1987;41:546–555. doi: 10.1038/clpt.1987.70. [DOI] [PubMed] [Google Scholar]
- 27.Lertora JJ, Rege AB, Lacour JT, Ferencz N, George WJ, VanDyke RB, et al. Pharmacokinetics and long-term tolerance to ribavirin in asymptomatic patients infected with human immunodeficiency virus. Clin Pharmacol Ther. 1991;50:442–449. doi: 10.1038/clpt.1991.162. [DOI] [PubMed] [Google Scholar]
- 28.Dixit NM, Layden-Almer JE, Layden TJ, Perelson AS. Modelling how ribavirin improves interferon response rates in hepatitis C virus infection. Nature. 2004;432:922–924. doi: 10.1038/nature03153. [DOI] [PubMed] [Google Scholar]
- 29.Glue P. The clinical pharmacology of ribavirin. Semin Liver Dis. 1999;19(Suppl 1):17–24. [PubMed] [Google Scholar]
- 30.De Franceschi L, Fattovich G, Turrini F, Ayi K, Brugnara C, Manzato F, et al. Hemolytic anemia induced by ribavirin therapy in patients with chronic hepatitis C virus infection: role of membrane oxidative damage. Hepatology. 2000;31:997–1004. doi: 10.1053/he.2000.5789. [DOI] [PubMed] [Google Scholar]
- 31.Lafeuillade A, Hittinger G, Chadapaud S. Increased mitochondrial toxicity with ribavirin in HIV/HCV coinfection. Lancet. 2001;357:280–281. doi: 10.1016/S0140-6736(00)03618-7. [DOI] [PubMed] [Google Scholar]
- 32.Homma M, Matsuzaki Y, Inoue Y, Shibata M, Mitamura K, Tanaka N, et al. Marked elevation of erythrocyte ribavirin levels in interferon and ribavirin-induced anemia. Clin Gastroenterol Hepatol. 2004;2:337–339. doi: 10.1016/s1542-3565(04)00064-3. [DOI] [PubMed] [Google Scholar]
- 33.Inoue Y, Homma M, Matsuzaki Y, Shibata M, Matsumura T, Ito T, et al. Erythrocyte ribavirin concentration for assessing hemoglobin reduction in interferon and ribavirin combination therapy. Hepatol Res. 2006;34:23–27. doi: 10.1016/j.hepres.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 34.Saito H, Tada S, Ebinuma H, Ishii H, Kashiwazaki K, Takahashi M, et al. Role of erythrocytes as a reservoir for ribavirin and relationship with adverse reactions in the early phase of interferon combination therapy for chronic hepatitis C virus infections. J Clin Microbiol. 2006;44:3562–3568. doi: 10.1128/JCM.00079-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Forsythe GE, Malcolm MA, Moler CB. Computer methods for Mathematical Computations. Prentice-Hall; Englewood Cliffs, NJ: 1977. [Google Scholar]