Abstract
The outcome of dopaminergic signaling and effectiveness of dopaminergic drugs depend on the relative preponderance of each of the five dopamine receptors in a given brain region. The separate contribution of each receptor to overall dopaminergic tone is difficult to establish at a functional level due to lack of receptor subtype specific pharmacological agents. A surrogate for receptor function is receptor protein or mRNA expression. We examined dopamine receptor mRNA expression by quantitative reverse transcription real-time PCR in the striatum, globus pallidus, frontal cortex and cingulate cortex of embryonic and postnatal mice. Samples of each region were collected by laser capture microdissection. D1- and D2-receptor mRNAs were the most abundant in all the regions of the mature brain. The D1-receptor was predominant over the D2-receptor in the frontal and cingulate cortices whereas the situation was reversed in the striatum and globus pallidus. In the proliferative domains of the embryonic forebrain, D3-, D4- and D5- receptors were predominant. In the corpus striatum and cerebral cortex, the D3- and D4-receptors were the only receptors that showed marked developmental regulation. By analyzing D1 receptor protein expression, we show that developmental changes in mRNA expression reliably translate into changes in protein levels, at least for the D1-receptor.
1. Introduction
Dopamine receptors are associated with a variety of cellular functions. In the mammalian central nervous system, dopamine receptor activation is critical for regulation of mood, motivation and motor function. Dopamine receptors are coupled to G-proteins. Typically, dopamine receptor activation leads to changes in intracellular cAMP levels [31,40,43] and triggers a signaling cascade that culminates in gene transcription [10]. Based on G-protein partners and intracellular signaling mechanisms, two classes of dopamine receptors are recognized [31,40,43]. The D1-like receptors are Gs/olf coupled and their activation results in increased intracellular cAMP. The D2-like receptors are Gi/o coupled and their activation decreases intracellular cAMP. Genes encoding five dopamine receptors have been cloned and based on genetic evidence, the D1-like receptors are further divided into D1- and D5- receptors (D1R and D5R, respectively) and the D2-like receptors into D2-, D3- and D4-receptors (D2R, D3R and D4R, respectively) [47]. Although signaling via the adenylyl cyclase-cAMP system is the principal mode of action, dopamine receptors also activate phospho-lipase C via the Gq/11 system and increase intracellular calcium levels [16,39]. Dopamine receptors also interact with glutamate receptors [4,21,44] and mobilize intracellular Ca2+ stores [23,51].
Activation of the D1-like and D2-like receptors typically produces opposite effects on intracellular signaling cascades and cell physiology [50]. A physiological balance between the activities of the two receptors is critical for normal neurological function. In fact, perturbation of the physiological balance appears to underlie a variety of neurological and neuro-psychiatric conditions such as Parkinson’s disease, depression, schizophrenia, attention deficit hyperactivity, anxiety and drug addiction. Many of the pharmacological agents used in the management of these conditions produce their effects by selectively activating or antagonizing the D1-like or D2-like receptors. Therefore, understanding the relative abundance of each of the five dopamine receptors in a given brain region is helpful for understanding the overall impact of dopaminergic signaling during normal development as well as in disease states.
Accurate estimates of dopamine receptor levels and function in specific brain regions is hampered by low abundance of some of the receptor subtypes, lack of subtype-specific agonists or antagonists, and unreliable antibodies. Low receptor abundance and antibody non-specificity is a particular problem in the developing brain. A surrogate marker for receptor function is receptor mRNA expression. We have used real-time quantitative PCR to compare mRNA levels for each of the five dopamine receptors in different regions of the embryonic and postnatal brain. Samples of the different brain regions were collected by using laser capture microdissection (LCM). Using western blots, we estimated D1-receptor protein levels in the developing and mature striatum and cerebral cortex and compared the protein levels to mRNA levels. Our data show that all five receptor mRNAs are expressed in the proliferative and postmitotic domains as early as embryonic day 12 (E12). The mRNA levels change dynamically during development and each receptor and each brain region show independent patterns of developmental regulation. Furthermore, developmental changes in mRNA levels reliably translate into changes in protein levels, at least for the D1-receptor, validating the usefulness of mRNA measurement as a surrogate for receptor expression.
2. Results
We collected samples of the embryonic and postnatal brains by laser capture microdissection (LCM; Fig. 1A – E). Samples of proliferative and postmitotic regions were collected from embryonic day 12 (E12) and E15 mice. The proliferative regions analyzed were the lateral ganglionic eminence (LGE), medial ganglionic eminence (MGE), caudal ganglionic eminence (CGE) and neuroepithelium of the dorsal cerebral wall. The postmitotic regions were differentiating fields of the striatum and cerebral wall (E15 only). From postnatal brains, we collected samples of the striatum, globus pallidus, frontal cortex and cingulate cortex from postnatal day 0 (P0; day of birth), P21 and P60 mice (Fig. 1C, D, E). Anatomical landmarks based on atlases of prenatal rodent brains [1,18] for E12, E15 and P0 samples or a mouse brain stereotaxic atlas [35] for P21 and P60 samples were used to identify the brain regions. Additional details about the anatomical landmarks used in sample collection are described in the Methods section.
Figure 1.
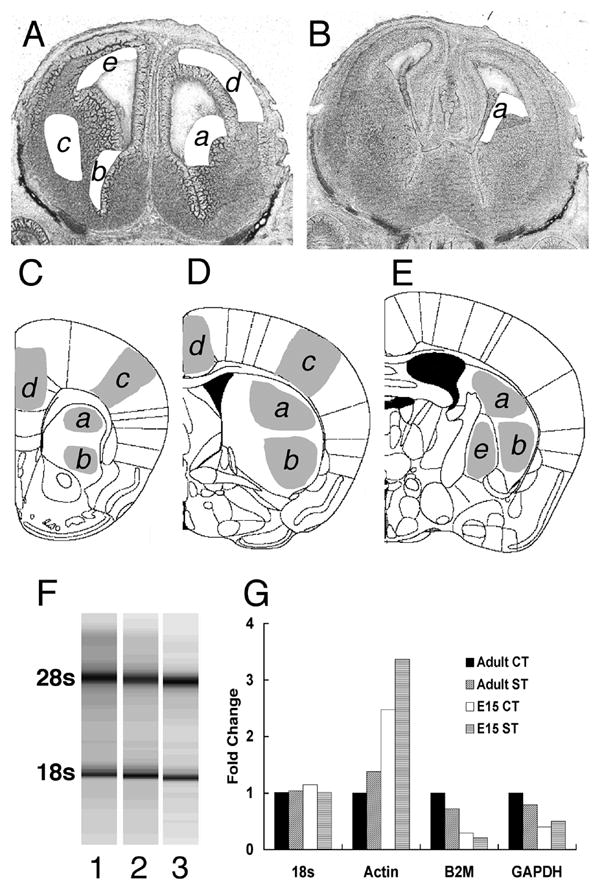
Collection of tissue samples from sections of the embryonic (A, B) and adult (C, D, E) brains by laser capture microdissection. Two cryostat sections from an E15 brain are shown in A and B after sample collection. A section from rostral levels of the forebrain (A) is shown after samples of the lateral ganglionic eminence (a), medial ganglionic eminence (b) and developing striatum (c) were collected from the ventral forebrain. From the same section, samples of postmitotic (d) and proliferative (e) domains of the cerebral wall were collected from the dorsal forebrain. A section from the caudal forebrain (B) is shown after samples of the caudal ganglionic eminence (a) were collected. Diagrams three representative coronal sections taken from the adult mouse brain atlas [35] are shown in C, D and E. Samples of the striatum were collected from sections intervening between those shown in C and E. Samples taken from sections between C and D represent the rostral striatum and samples taken from sections beginning with D and up E represent the caudal striatum. From each section, samples from the dorsal (a) and ventral (b) striatum were collected separately. In addition to striatal samples, samples of the cingulate cortex (d), frontal sensorimotor cortex (c) and globus pallidus (e) were also collected. Every section between C and E was used from every brain. Qualitative analysis of RNA isolation (F) was performed by nondenaturing capillary gel electrophoresis. A representative gel shows intact 28s (upper bands) and 18s (lower bands) rRNA, without degradation bands or smears (lanes 1, 2, and 3 represent E15 samples from lateral, medial and caudal ganglionic eminences, respectively). (G). Selection of endogenous RNA control for real time reverse transcription quantitative PCR. We used 2μg of total RNA from cerebral cortex (CT) and striatum (ST) each, collected from embryonic day 15 (E15) or adult (P60) mice to assess which generally accepted endogenous control was superior for normalizing expression of mRNA for each region at each age Our results demonstrate that only 18s rRNA levels remained steady between E15 and P60 in the cortex and striatum prompting its choice as the endogenous control in all our experiments. Actin, B2M and GAPDH mRNA levels showed variation between E15 and P60 and between cortex and striatum.
We confirmed the quality of RNA isolated from the LCM samples to verify that high quality RNA was used to produce cDNA (Fig. 1F). In addition to standardizing the amount of RNA used for reverse-transcription, we evaluated four endogenous genes as controls to normalize dopamine receptor gene expressions across the various regions and ages. Our results demonstrated that when the same quantity of total RNA was analyzed from all age groups and all brain regions included in this study, 18s rRNA had the least variability compared to other traditional endogenous controls such as actin, β2-microglobulin or GAPDH (Fig. 1G). Thus, 18s rRNA was found to be the most reliable indicator of the sample size. Since 18s rRNA is abundantly and stably expressed under a variety of conditions, standardizing our samples based on 18s levels permitted us to standardize average number of cells in each sample and therefore compare our data across the different brain regions and age groups.
Differences in the expression levels of each dopamine receptor mRNA at each age in each brain region as well as across the pre- and postnatal ages were compared by Student’s t-test or analysis of variance (ANOVA). A t-test was used when only two groups of data were compared and ANOVA was used when more than two groups were compared. Whenever ANOVA indicated significant differences among the groups, a post hoc Bonferroni test was performed for pair-wise comparisons. A total of 67 comparisons (43 ANOVAs and 24 t-tests) were performed in this study. The large number of comparisons increased the probability of occurrence of type 1 statistical error (i.e. false positives). To reduce the probability of a type 1 error we mathematically corrected the probability of rejecting the null hypothesis from the customary 5% to a more stringent 0.1%. Thus, differences among/between groups were considered significant only when p ≤ 0.001. Since the Bonferroni post hoc test itself is a correction for multiple comparisons and since we used it only when ANOVA produced a p ≤ 0.001, statistical significance for the Boinferroni test was set at the customary p ≤ 0.05.
Receptor mRNA expression in the proliferative domains
We analyzed mRNA expression in the proliferative zones of the dorsal cerebral wall (presumptive neocortex) and the ganglionic eminences of the basal forebrain on E12 and E15. Initially, mRNA expression was examined separately for the lateral, medial and caudal ganglionic eminences. However, no significant differences were found among the 3 regions, for any of the five receptor mRNAs on E12 or E15. Therefore, we pooled the data from the lateral, medial and caudal ganglionic eminences.
Neuroepithelium of the dorsal cerebral wall
D1R and D5R mRNAs were expressed at very low levels on E12 and E15 (Fig. 2A). D2R, D3R and D4R mRNAs were expressed at high levels at both the ages (Fig 2A). Thus, D2-like mRNAs (D2, D3 and D4) were significantly more abundant than D1-like mRNAs (D1 and D5) in the neuroepithelium of the dorsal cerebral wall at the early developmental stages of E12 and E15 (t(10)=6.363; p<0.0001). Over the E12 to E15 interval the receptor mRNA levels did not change significantly (Fig 2A).
Figure 2.
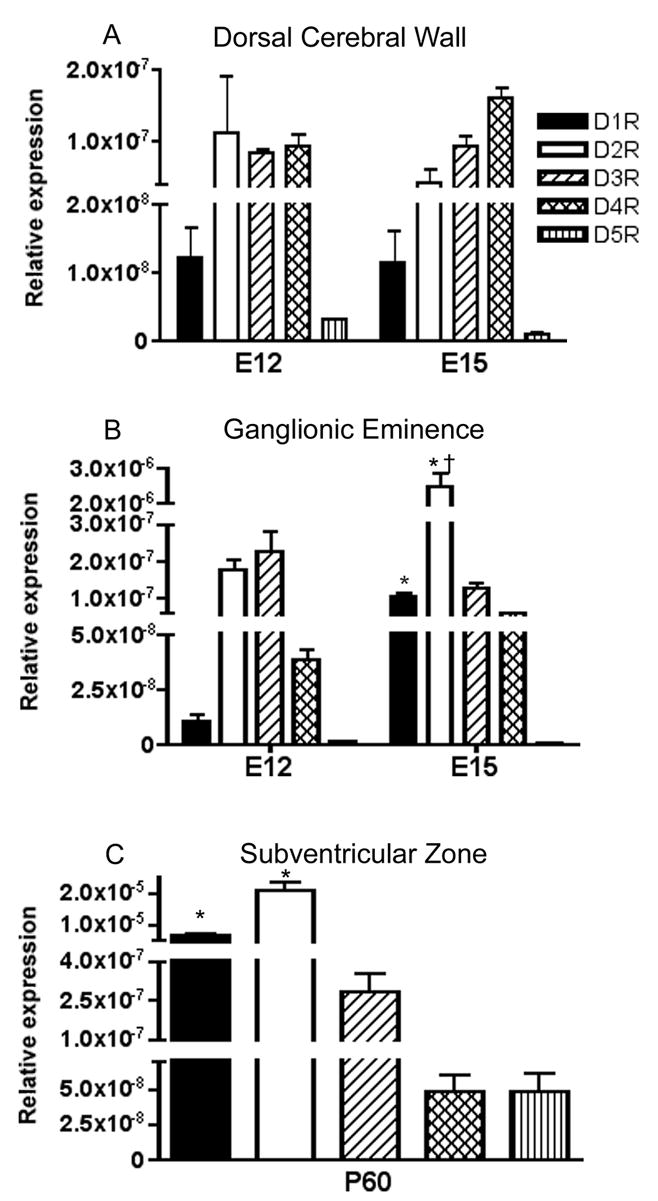
Expression level of dopamine receptor mRNAs in the proliferative neuroepithelium of the dorsal cerebral wall (A) and the ganglionic eminence (B) on E12 and E15 and the subventricular zone on P60 (C). Since there were no statistically significant differences in the expression level of any of the five mRNAs between the lateral, medial and caudal ganglionic eminences at E12 or E15, we combined the data from the 3 subdivisions. (A) Dorsal cerebral wall demonstrates greater D2-like (DR2, DR3 and DR4) than D1-like receptor expression (t(10)=6.363; p<0.0001). (B) * = D1R and D2R are significantly higher at E15 than at E12 (t(18)=6.382 or 5.38, respectively, p<0.0001 for both), † = D2R expression significantly greater than all other dopamine receptors at E15 (p<0.0001 for all post-hoc comparisons). (C) * = D1R and D2R expression lavels are significantly different between each other and compared to all other receptors (F(4,25)=42.8; p<0.0001; Bonferroni p<0.05, for all comparisons) Mean±SEM values from 3–6 individual brains.
Ganglionic eminence
In this region, D1R and D5R mRNA expression was the lowest and D2R and D3R was the highest on E12 (Fig 2B). Over the E12 to E15 interval D1R and D2R mRNA increased significantly (D1R t(18)=6.382; p<0.0001 and D2R t(18)=5.38; p<0.0001) while D3R, D4R and D5R mRNA did not show significant changes (Fig 2B). At E15, D2R mRNA was expressed at significantly higher levels than all the other mRNAs (F(4,49)=38.04; p<0.0001; Bonferroni p<0.001 for all comparisons).
Forebrain subventricular zone at P60
Since the subventricular zone continues to be a proliferative region even in the adult forebrain, we included this region in our analyses. All five dopamine receptor mRNAs were detected in this region (Fig. 2C) with significant differences among the expression levels (F(4,25)=42.8; p<0.0001). D1R and D2R mRNA expression was significantly higher than that of the others (Bonferroni; p<0.05, for all comparisons). While the expression levels of D3R, D4R and D5R did not significantly differ from each other, D2R mRNA level was higher than that of D1R mRNA (Bonferroni; p<0.05) (Fig 2C).
Receptor mRNA expression in the postmitotic regions
Striatum
All five receptor mRNAs were expressed in the striatum at all ages examined from E12 to P60 (Fig 3A, B). D2R mRNA levels were significantly higher than all the other mRNAs at each age (statistical analyses in Table 2), except at E15 when D2R and D1R mRNA levels were not significantly different (Fig. 3A; Table 2). D1R mRNA levels were the second highest. Thus, D1R and D2R mRNAs were the most abundant at all ages examined. Both D1R and D2R mRNAs increased significantly between E12 and P60 (Fig. 3A; statistics in Table 1). The D1R mRNA showed the greatest increase between E12 to E15 (Fig. 3A), which was statistically significant (Table 1) and the expression level did not change significantly after E15 during the postnatal period (Fig. 3A; Table 1). D2R mRNA showed a steady and significant increase from E12 to P60 (Fig. 3A; Table 1). D3R, D4R and D5R mRNA expression showed different profiles compared to those of D1R and D2R mRNAs (Fig. 3B). Thus, D3R mRNA expression had a “U” shaped pattern: the expression declined from E12 to P0 and then returned to pre-existing levels during P0 to P60. The changes were not statistically significant (Table 1). In contrast to D3R, D4R mRNA had an inverted “U” pattern with significant increases from E12 to P0 and a subsequent decline (Fig. 3B; Table 1). Interestingly, D5R, which showed the lowest expression of all the mRNAs in the striatum at E12, significantly increased during later development to reach virtually identical expression levels as D3R at P60 (Fig. 3B; Tables 1 and 2).
Figure 3.
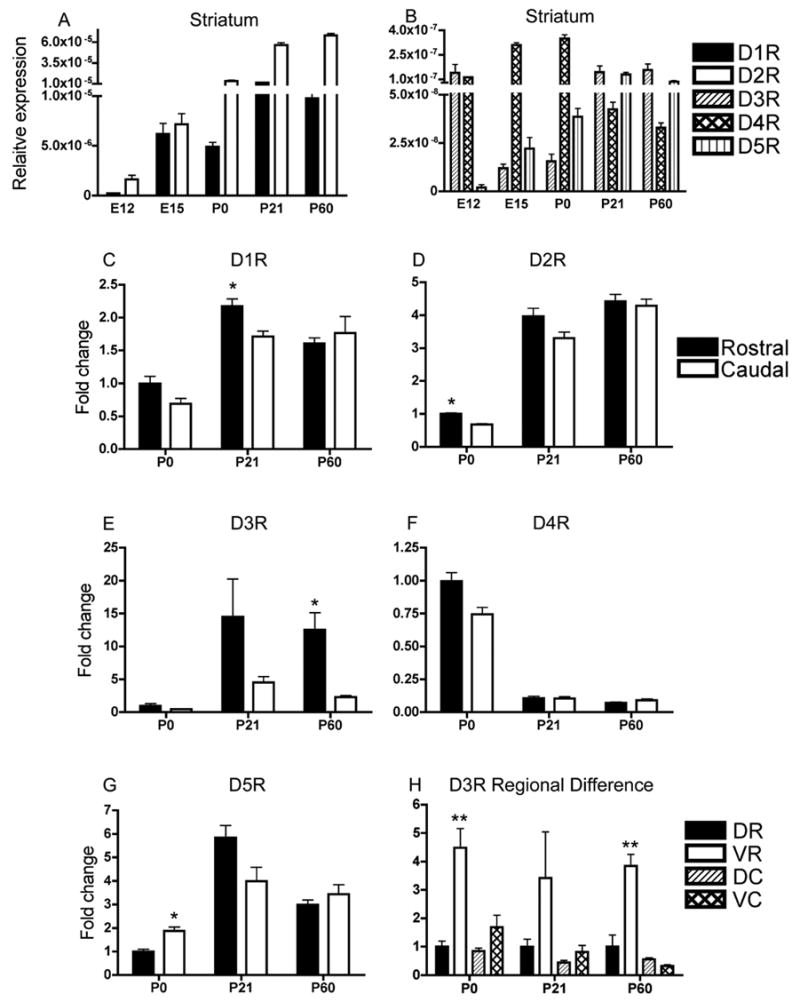
Dopamine receptor mRNA expression in the striatum from E12 to P60 (A, B). D1R and D2R mRNA levels are significantly higher than the other mRNAs (Table 2). Therefore, D1R and D2R expression levels are shown separately in A and D3R, D4R and D5R mRNA are shown in B. Since the striatum is a rostro-caudally elongated structure, we analyzed each mRNA separately for rostral and caudal striatum on P0, P21 and P60 (C, D, E, F, G) (C, * = t(13)=3.394; p<0.005 at P21 rostral versus caudal) (D, * = t(10)=7.673; p<0.0001 at P0 rostral vs. caudal) (E, * = t(22)=3.851; p<0.001 at P60 rostral vs. caudal). We also analyzed mRNA expression separately for dorsal and ventral regions of rostral and caudal divisions. Only D3R mRNA showed significant differences among the four sub-regions with the ventral portion of the rostral striatum showing the highest expression at each (H) (** = P0, F(3,8)=16.58; p<0.001, Bonferroni p<0.01 for all ventro-rostral comparisons; P60, F(3,20)=30.52; p<0.0001, Bonferroni p<0.001 for all ventro-rostral comparisons. Mean±SEM values from 3–6 individual brains. DR = dorsal-rostral; VR = ventral-rostral; DC = dorsal-caudal; VC = ventral-caudal.
Table 2. Results of statistical analyses of dopamine receptor expression levels at each of E12 to P60 in each brain region.
Statistical analyses of differences among dopamine receptor expression levels at each age. For each brain region and age, expression levels of the five receptors (D1R, D2R, D3R, D4R and D5R) were analyzed by ANOVA. Differences between a given receptor pair were analyzed by Bonferroni post-test. NS = not significant, for ANOVA p>0.001 for Bonferroni p>0.05.
| Striatum | Globus Pallidus | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| E12 | E15 | P0 | P21 | P60 | P0 | P21 | P60 | ||
| ANOVA (all 5 receptors) F(dfbtw, dfw) | NS(p=0.0022) 11.41 (4,8) | p<0.0001 27.95 (4,15) | p<0.0001 183.4 (4,55) | p<0.0001 394.8 (4,69) | p<0.0001 767.5 (4,115) | ANOVA (all 5 receptors) F(dfbtw, dfw) | p<0.0001 64.75 (4,10) | p<0.0001 87.95 (4,15) | p=0.0001 9.035 (4,25) |
| D1R vs D2R | NS | p< 0.001 | p< 0.001 | p< 0.001 | D1R vs D2R | p< 0.001 | p< 0.001 | p< 0.01 | |
| D1R vs D3R | p< 0.001 | p< 0.001 | p< 0.001 | p< 0.001 | D1R vs D3R | NS | NS | NS | |
| D1R vs D4R | p< 0.001 | p< 0.001 | p< 0.001 | p< 0.001 | D1R vs D4R | NS | p< 0.05 | NS | |
| D1R vs D5R | p< 0.001 | p< 0.001 | p< 0.001 | p< 0.001 | D1R vs D5R | NS | p< 0.05 | NS | |
| D2R vs D3R | p< 0.001 | p< 0.001 | p< 0.001 | p< 0.001 | D2R vs D3R | p< 0.001 | p< 0.001 | p< 0.001 | |
| D2R vs D4R | p< 0.001 | p< 0.001 | p< 0.001 | p< 0.001 | D2R vs D4R | p< 0.001 | p< 0.001 | p< 0.001 | |
| D2R vs D5R | p< 0.001 | p< 0.001 | p< 0.001 | p< 0.001 | D2R vs D5R | p< 0.001 | p< 0.001 | p< 0.001 | |
| D3R vs D4R | NS | NS | NS | NS | D3R vs D4R | NS | NS | NS | |
| D3R vs D5R | NS | NS | NS | NS | D3R vs D5R | NS | NS | NS | |
| D4R vs D5R | NS | NS | NS | NS | D4R vs D5R | NS | NS | NS | |
| Frontal cortex | Cingulate cortex | ||||||||
| E15 | P0 | P21 | P60 | P0 | P21 | P60 | |||
|
| |||||||||
| ANOVA (all 5 receptors) F(dfbtw, dfw) | p<0.0001 68.8 (4,25) | p<0.0001 44.02 (4,9) | p<0.0001 31.63 (4,15) | p<0.0001 33.89 (4,24) | ANOVA (all 5 receptors) F(dfbtw, dfw) | p<0.0001 72.25 (4,10) | p<0.0001 45.11 (4,14) | p<0.0001 13.02 (4,21) | |
|
| |||||||||
| D1R vs D2R | p< 0.001 | p< 0.001 | p< 0.01 | p< 0.001 | D1R vs D2R | NS | p< 0.01 | p< 0.01 | |
| D1R vs D3R | p< 0.001 | p< 0.001 | p< 0.001 | p< 0.001 | D1R vs D3R | p< 0.001 | p< 0.001 | p< 0.001 | |
| D1R vs D4R | p< 0.001 | p< 0.001 | p< 0.001 | p< 0.001 | D1R vs D4R | p< 0.001 | p< 0.001 | p< 0.001 | |
| D1R vs D5R | p< 0.001 | p< 0.001 | p< 0.001 | p< 0.001 | D1R vs D5R | p< 0.001 | p< 0.001 | p< 0.001 | |
| D2R vs D3R | NS | NS | p< 0.01 | p< 0.01 | D2R vs D3R | p< 0.01 | p< 0.01 | NS | |
| D2R vs D4R | NS | NS | p< 0.01 | p< 0.05 | D2R vs D4R | p< 0.001 | p< 0.01 | NS | |
| D2R vs D5R | NS | NS | p< 0.05 | p< 0.05 | D2R vs D5R | p< 0.01 | p< 0.01 | NS | |
| D3R vs D4R | NS | NS | NS | NS | D3R vs D4R | p< 0.001 | NS | NS | |
| D3R vs D5R | NS | NS | NS | NS | D3R vs D5R | NS | NS | NS | |
| D4R vs D5R | NS | NS | NS | NS | D4R vs D5R | p< 0.001 | NS | NS | |
Table 1. Results of statistical analyses of dopamine receptor expression levels from E12 to P60.
Statistical analyses of developmental change in dopamine receptor mRNA expression. For each brain region changes in expression levels of the five receptors (D1R, D2R, D3R, D4R and D5R) during development (developmental interval indicated in parentheses for each region) was analyzed by ANOVA. Differences in the expression levels between specific developmental intervals were analyzed by Bonferroni test. NS = not significant, for ANOVA p>0.001 for Bonferroni p>0.05.
| Striatum (E12-P60) | |||||
|---|---|---|---|---|---|
| D1R | D2R | D3R | D4R | D5R | |
| ANOVA (E12 to P60) F(dfbtw, dfw) | p<0.0001 17.11 (4,53) | p<0.0001 113.9 (4,53) | NS | p<0.0001 182.8 (4,52) | p<0.0001 20.91 (4,52) |
| E12 vs E15 | NS | NS | p< 0.001 | NS | |
| E12 vs P0 | NS | NS | p< 0.001 | NS | |
| E12 vs P21 | P< 0.001 | p< 0.001 | NS | p< 0.001 | |
| E12 vs P60 | P< 0.001 | p< 0.001 | NS | p< 0.01 | |
| E15 vs P0 | NS | NS | NS | NS | |
| E15 vs P21 | P< 0.05 | p< 0.001 | p< 0.001 | p< 0.001 | |
| E15 vs P60 | NS | p< 0.001 | p< 0.001 | p< 0.01 | |
| P0 vs P21 | P< 0.001 | p< 0.001 | p< 0.001 | p< 0.001 | |
| P0 vs P60 | P< 0.001 | p< 0.001 | p< 0.001 | p< 0.001 | |
| P21 vs P60 | NS | p< 0.01 | NS | p< 0.01 | |
| Globus Pallidus (P0–P60) | |||||
| D1R | D2R | D3R | D4R | D5R | |
|
| |||||
| ANOVA (P0 to P21) F(dfbtw, dfw) | NS | NS | p=0.0001 24.83 (2, 10) | p<0.0001 73.07 (2, 10) | NS |
|
| |||||
| P0 vs P21 | p< 0.01 | p< 0.001 | |||
| P0 vs P60 | p< 0.001 | p< 0.001 | |||
| P21 vs P60 | NS | NS | |||
| Frontal cortex (E15-P60) | |||||
| D1R | D2R | D3R | D4R | D5R | |
|
| |||||
| ANOVA (E15 to P60) F(dfbtw, dfw) | P<0.0001 | NS(p=0.0046) 6.834 (3, 14) | NS | p<0.0001 37.19 (3, 15) | p=0.0003 11.79 (3, 15) |
|
| |||||
| E15 vs P0 | NS | NS | NS | ||
| E15 vs P21 | P< 0.01 | p< 0.001 | p< 0.001 | ||
| E15 vs P60 | P< 0.001 | p< 0.001 | NS | ||
| P0 vs P21 | P< 0.001 | p< 0.001 | p< 0.01 | ||
| P0 vs P60 | P< 0.001 | p< 0.001 | NS | ||
| P21 vs P60 | NS | NS | p< 0.05 | ||
| Cingulate cortex (P0–P60) | |||||
| D1R | D2R | D3R | D4R | D5R | |
|
| |||||
| ANOVA (P0 to P60) F(dfbtw, dfw) | NS | NS | NS | p<0.0001 392.4 (2, 10) | NS |
|
| |||||
| P0 vs P21 | p< 0.001 | ||||
| P0 vs P60 | p< 0.001 | ||||
| P21 vs P60 | NS | ||||
Thus, D1R, D2R and D5R mRNA significantly increased over the E12 to P60 period (Table 1). In contrast, D3R and D4R mRNA fluctuated in inverse relationship to each other over the same period. Only D2R and D5R levels significantly changed between P21 and P60 (Table 1).
Since the striatum is a rostro-caudally elongated structure and since inputs to the striatum (e.g. cortico-striate inputs) are topographically organized along the rostral-caudal axis [19,22,30,37,55], we examined mRNA levels separately for rostral and caudal striatum. This analysis was performed only for the postnatal ages because the sizes of the rostral and caudal samples on E12 and E15 were not large enough for such analyses. We standardized P0 rostral expression level to 1.0 and compared it to caudal expression levels. This permitted us to illustrate not only rostral versus caudal differences but also developmental changes in mRNA expression between the 2 regions. Although there was no significant difference in D1R expression between rostral and caudal striatum at any of the ages, we observed a non-significant (p<0.05) trend by the standards of the present statistical analyses (where significant differences are registered only at p<0.001) of higher D1R expression in the rostral compared to the caudal striatum at P21 (t(13)=3.394; p<0.005 asterisk in Fig. 3C). D2R expression was significantly higher in the rostral striatum at P0 (t(10)=7.673; p<0.0001, asterisk in Fig. 3D). By P60, rostral versus caudal differences were only present for D3R (t(22)=3.851; p<0.001, asterisk in Fig. 3E). D4R mRNA levels showed no significant rostral-caudal differences on P0, P21 or P60 (Fig. 3F). In contrast to the other receptors, D5R mRNA was significantly higher in the caudal striatum at P0 (t(10)=4.384; p=0.001, asterisk in Fig. 3G).
Striatal neurogenesis progresses in a ventro-lateral to dorso-medial gradient [2,7,9,29,49,52]. Dopaminergic innervation and appearance of striosome-matrix compartments also progresses in ventro-lateral to dorso-medial gradient [8,9,11,12,49]. Therefore, we reasoned that postnatal expression patterns of dopamine receptor mRNAs could show regional differences and further subdivided rostral and caudal samples into dorsal and ventral halves. Thus, we analyzed mRNA expression levels separately for each of the resulting 4 regions. Only the D3R mRNA showed significant differences among the 4 regions. At each of the 3 ages examined, the ventral portion of the rostral striatum showed the highest D3R mRNA level at P0 and P60 (P21 not significant) compared to the other 3 regions analyzed (P0, F(3,8)=16.58; p<0.001, Bonferroni p<0.01 for all ventro-rostral comparisons; P60, F(3,20)=30.52; p<0.0001, Bonferroni p<0.001 for all ventro-rostral comparisons; see asterisks in Fig 3H).
Globus pallidus
Reliable identification of this region for LCM from embryonic brains was difficult in our preparations; therefore, we examined the globus pallidus at postnatal ages P0, P21 and P60. As in the case of the striatum, D2R mRNA expression was significantly higher than all the other mRNAs at each of the ages examined (Fig. 4A, Tables 1 and 2). D1R mRNA levels were the second highest, although statistical analyses did not show this to be significant (Table 1). D1R and D2R mRNA did not significantly change from P0 to P60 (Fig. 4A, Table 1). However, D3R mRNA levels increased significantly from P0 to P60 (Fig. 4B, asterisk; Table 1). Also, D4R mRNA level was the highest on P0; declined from P0 to P21 and remained low at P60 (Fig. 4B, Table 1). D5R mRNA levels did not show significant changes during the P0 to P60 period (Fig. 4B, Table 1).
Figure 4.
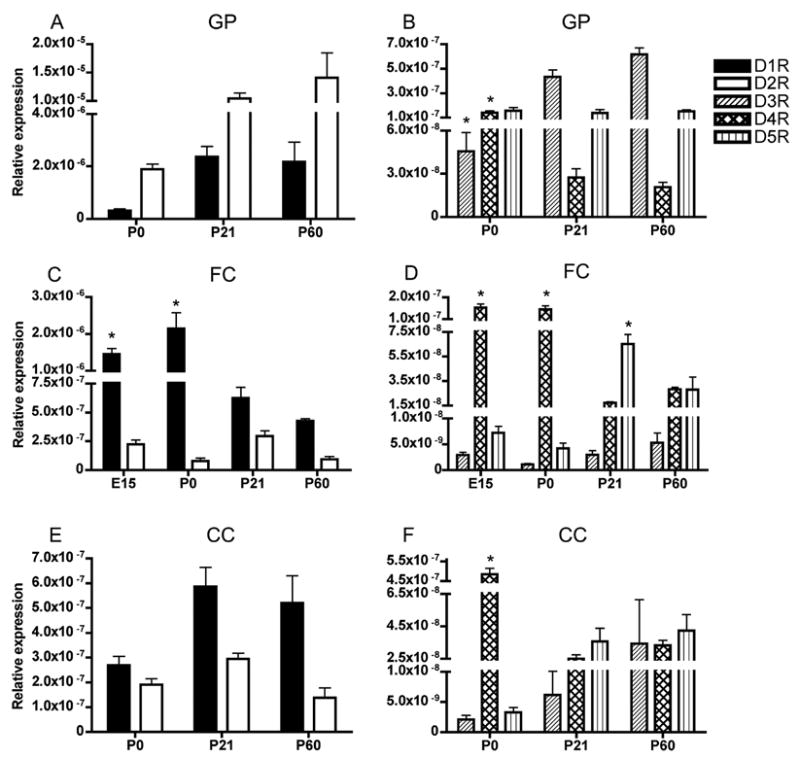
Developmental profiles of dopamine receptor mRNAs in the globus pallidus (A, B), frontal sensory-motor cortex (C, D) and cingulate cortex (E, F). D1R and D2R mRNA levels are higher than the other mRNAs; therefore, they are shown separately for each region (A, C, E). D3R, D4R and D5R mRNA are also shown for each region (B, D, F). The data from embryonic mice are shown only for the frontal cortex, because in the other regions we could not identify postmitotic differentiating fields consistently in the sections of the embryonic brain during LCM. In the globus pallidus (A, B) D2R mRNA was significantly higher than all the other mRNA at each age (Table 2) and (B) * = D3R mRNA significantly increases from P0 to P60 while * = D4R mRNA significantly decreases from P0 to P60 (Table 1). In frontal cortex (C, D) D1R mRNA was significantly higher than all the other mRNAs at every age (Table 2) and declined during development (C, * = Significantly different from P21 and P60, see Table 1). D4R mRNA decreased during development (D, * = Significantly different from P21 and P60, see Table 1). D5R mRNA was the highest at P21(D, * = see Table 1). In cingulate cortex (E, F) D1R mRNA was significantly higher than all the other mRNAs at all ages except P0 (Table 2). In the cingulate cortex, only D4R mRNA showed significant changes during development (F, * = different from P21 and P60, see Table 1). Mean±SEM values from 3–6 individual brains.
Frontal cortex
We analyzed mRNA expression in the frontal cortex on E15, P0, P21 and P60 (Figs. 4C, D). Unlike the striatum and globus pallidus, D1R mRNA showed the highest expression of all the mRNAs in the frontal cortex (Fig. 4C, D; Table 2). D1R mRNA did not change significantly between E15 and P0 (Fig 4C; table 1). However, it significantly declined from P0 to P60 (Fig. 4C, asterisk; Table 1). D2R and D3R mRNA levels did not show significant changes over this time period (Figs. 4C, D; Table 1). D4R mRNA was high on E15 and P0 (as high as D2R mRNA) and decreased significantly from P0 to P21 and remained low at P60 (Fig. 4D, Table 1). D5R mRNA also showed significant developmental changes with its peak expression occurring at P21 (Fig. 4D, Table 1).
Cingulate cortex
We analyzed mRNA expression in the cingulate cortex during postnatal development: on P0, P21 and P60 (Fig. 4E, F). The cingulate cortex, located near the medial edge of the cerebral wall is developmentally less advanced than the more lateral regions such as the presumptive frontal cortex. Therefore, we could not reliably distinguish between proliferative and postmitotic regions of the presumptive cingulate cortex at E12 or E15 in our LCM preparations and decided not to include these age groups in the analysis. D1R mRNA was the most abundant of all the mRNAs in the cingulate cortex, except at P0 when D1R and D2R were expressed at comparable levels (Fig. 4E, F; Table 2). D1R, D2R, D3R and D5 mRNA expression did not vary significantly during development (Figs. 4E and 4F, Table 1). However similar to the frontal cortex, D4R mRNA expression showed significant decreases between P0 and P60 (Fig 4F, Table 1).
Sex differences
There were no statistically significant differences in the expression level of any of the 5 mRNAs in any of the regions analyzed between male and female mice at P60 (Fig. 5). We did not analyze the differences at any of the other age groups.
Figure 5.
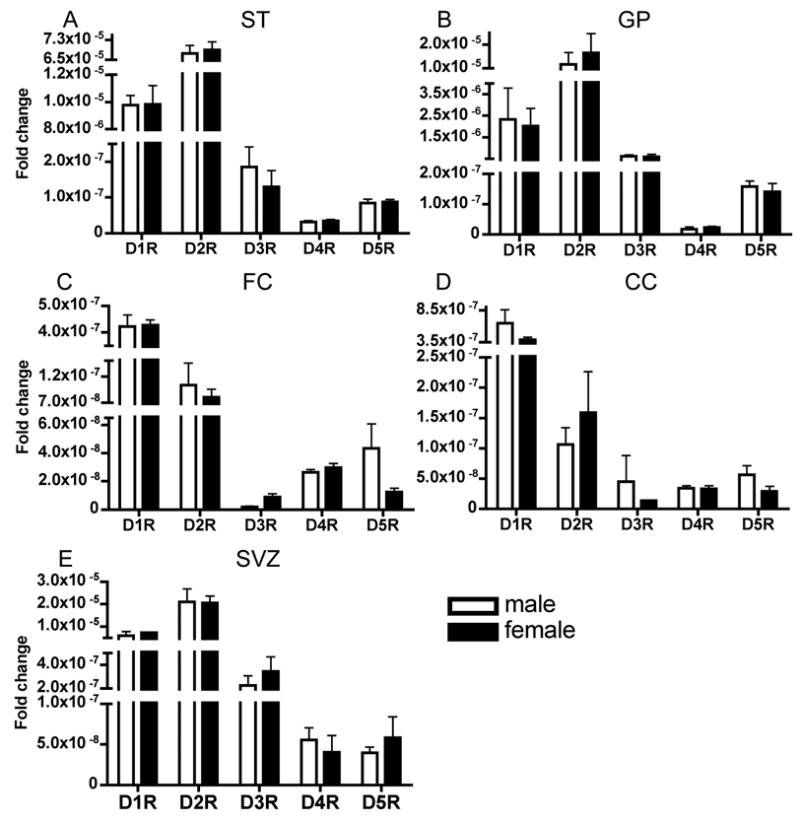
Sex differences in the expression levels of dopamine receptor mRNAs at P60 in the striatum (A), globus pallidus (B), frontal cortex (C), cingulate cortex (D) and the subventricular zone (E). There were no statistically significant differences by t-test between male and female mice in any mRNA in any region. Mean±SEM values from 3–6 individual brains.
D1-like to D2-like mRNA ratios
We re-analyzed mRNA levels for D1-like (D1R and D5R) and D2-like (D2R, D3R and D4R) receptors and calculated D1-like to D2-like receptor ratios (Fig. 6A). In the striatum, at all ages examined, D2-like receptor expression was higher than D1-like receptor expression. The ratio approached its highest value (~0.8) at E15 indicating that D1- and D2-like receptor mRNA levels were closely balanced at that age. At all other ages, D2-like mRNA was more abundant than D1-like mRNA in the striatum. In the globus pallidus D1-to-D2 ratio was consistently smaller than 1.0 at all the ages examined, indicating the preponderance of the D2-like receptors. In the frontal cortex, the ratio was consistently greater than 1.0 indicating the preponderance of the D1-like receptor at all ages examined. The ratio reached its highest value on P0 and declined from P0 to P21, indicating fluctuations in the receptor mRNA levels during postnatal development. In the cingulate cortex, the ratio was <1.0 at P0 indicating the predominance of the D2-like receptors at this age. The ratio rose above 1.0 at P21 and P60, indicating a reversal of the D2-like receptor mRNA predominance during the postnatal period. The increase in the ratio at P21 was due to an increase in D1R mRNA as well as a significant decline in D4R mRNA over the P0 to P21 period (Tables 1 and 2).
Figure 6.
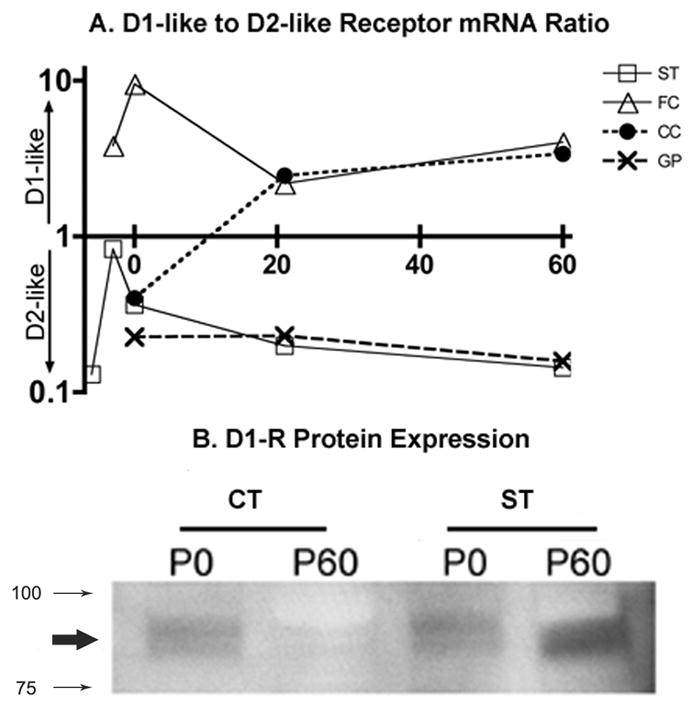
An overview of the ratio between D1-like receptors (summation of D1R and D5R) and D2-like receptors (summation of D2R, D3R and D4R) is shown for each region during development (A; age along the abscissa; 0, 21 and 60 = P0, P21 and P60, respectively). Only the striatum at E15 approximates equipoise (ratio 0.8) between the two dopamine receptor classes. The ventral forebrain represented by the striatum (ST) and globus pallidus (GP) show D2-like dominance in receptor expression, whereas the dorsal forebrain represented by frontal (FC) and cingulate (CC) cortex show D1-like dominance. An immunoblot showing D1R protein expression (B). Membrane fractions from postnatal days 0 and 60 (P0 and P60, respectively) cortex and striatum were analyzed for D1R protein expression. The D1R band (bold arrow) appears between ~ 100 and 75 KD (smaller arrows). The highest levels of protein are seen in the adult striatum and the lowest levels in the adult cortex with intermediate levels at P0 in the cortex and striatum.
D1-receptor protein expression
We examined D1-receptor protein in membrane preparations from P0 and P60 frontal cortex and striatum (Fig. 6B). Our goal was to determine if mRNA levels corresponded with membrane associated protein levels. Since the relative expression of the D1R mRNA was different between the frontal cortex and the striatum (striatum > frontal cortex) and since developmental regulation of D1R mRNA over the P0 to P60 interval was also different (decline in frontal cortex versus increase in the striatum), we chose these two brain regions and developmental stages. Consistent with the mRNA expression level, D1R protein expression in membrane fractions declined from P0 to P60 in the frontal cortex and increased in the striatum. The expression level was higher in the striatum than frontal cortex at P0 and P60, in agreement with mRNA expression levels.
3. Discussion
Our data show that dopamine receptor mRNA expression in the mouse forebrain begins early in the embryonic period and that each mRNA shows a unique, region-specific developmental profile. D1R and D2R mRNAs predominate over the other three mRNAs during development and at maturity (Fig. 7). The embryonic neuroepithelial domains show a preponderance of D2-like (D2R, D3R and D4R) receptor mRNA. Interestingly, although all five mRNA levels change during development, D3R and D4R are the only mRNAs that are up- and down-regulated during postnatal development (Fig. 7). D3R mRNA shows striking developmental variation in the ventral forebrain, especially in the striatum, whereas D4R mRNA shows striking variation in the dorsal forebrain. Membrane-associated receptor protein levels correspond with mRNA levels, at least for D1R in the frontal cortex and striatum and at least at P0 and P60, suggesting that mRNA level is good indicator of protein expression.
Figure 7.
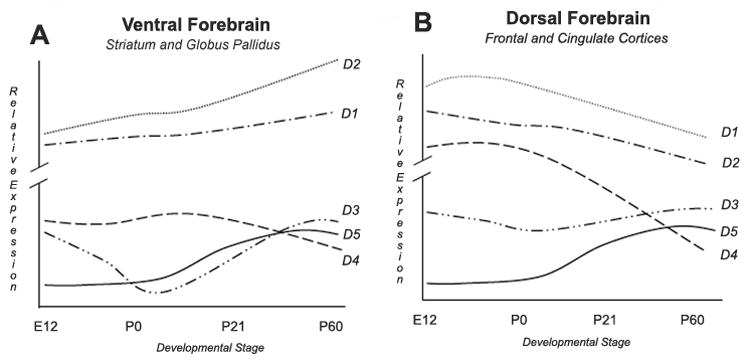
A summary of the salient differences between ventral (A) and dorsal (B) forebrain with regard to dopamine receptor mRNA expression. For the sake of simplicity, the striatum and globus pallidus are grouped together under ventral forebrain and frontal and cingulate cortices under dorsal forebrain, overlooking differences between each of the 2 regions within a group. Data on prenatal expression are taken from the striatum (E12 and E15) and frontal cortex (E15) only. In both the forebrain regions, D1R and D2R mRNA are by far the most abundant. However, both D1R and D2R mRNA expression is significantly higher in the ventral than dorsal forebrain. D2R mRNA levels are higher than D1R mRNA in the ventral forebrain whereas the opposite is the case in the dorsal forebrain. Whereas both D1R and D2R mRNA increase during development in the ventral forebrain, both show decline in the dorsal forebrain. D3R mRNA shows the most striking developmental change in the ventral forebrain with two peaks separated by a trough anchored around the time of birth. D4R mRNA shows the most striking developmental change in the dorsal forebrain with its high expression level early in development and a rapid decline in the postnatal period. D5R mRNA shows comparable developmental changes in both the regions.
The ontogeny of dopamine receptors has been examined previously by using receptor binding, in situ RNA hybridization or RT-PCR in the brains of rodents and primates [13,14,17,26,38,41,42,45,46]. With the exception of the in situ hybridization studies, the mRNA expression was not analyzed in anatomically defined regions of the embryonic and postnatal brain. Although in situ hybridization analyses offer exquisite anatomical resolution, often at the single cell level, the data can be difficult to quantify due to the low signal to noise ratios in the autoradiograms, especially in the embryonic brain. Our study offers quantitative data on relative expression level of each of the five dopamine receptor mRNAs from E12 to P60 in samples of the forebrain collected by LCM based on anatomical landmarks. Thus, our data not only complement earlier studies but also offer novel information on expression of the dopamine receptors during brain development.
Receptor mRNA expression begins in the ventral and dorsal forebrain prior to the arrival of dopaminergic axons from the midbrain. Tyrosine-hydroxylase positive axons enter the mouse basal forebrain by E13 and dorsal forebrain by E14, coincident with increases in dopamine content of these two regions [11,33,36]. D1-like and D2-like receptor binding sites appear in the mouse basal forebrain by E13 [34], at least a day after the appearance of the mRNA. Similar findings were reported earlier by others [42]. It is possible that developmental regulation of mRNA and binding site expression occurs by independent mechanisms. Perhaps receptor binding site expression occurs upon exposure to dopamine, the endogenous ligand, and mRNA expression occurs earlier, independent of exposure to dopamine [42].
Although all five receptor mRNAs were expressed in the embryonic proliferative domains of the dorsal and ventral forebrain, D2-like (D2R, D3R and D4R) mRNA was the most abundant in the proliferative domains. The predominance of the D2-like receptor did not change significantly over the E12 to E15 interval. Among the D2-like mRNAs, the ganglionic eminence expressed high levels of D3R mRNA. In fact D3R was the most abundant mRNA in this region on E12. This is consistent with earlier reports of high levels of D3R mRNA in the ganglionic eminence illustrated by in situ hybridization methods [6]. In general, expression of dopamine receptor mRNAs in forebrain neuroepithelial regions is consistent with the ability of dopamine or its receptor agonists to modulate cell proliferation [33,36,56,57]. The subventricular zone of the P60 forebrain also expressed all five receptor mRNAs. Although D2R mRNA was the most abundant, D1R and D3R mRNAs were also expressed prominently. D5R mRNA was significantly higher in this region compared to the embryonic proliferative domains. Once again, expression of dopamine receptor mRNAs is consistent with the reported effects of dopamine receptor agonists and antagonists on cell proliferation in the subventricular zone [15,20,53,54].
Each brain region showed unique characteristics with respect to developmental regulation of the five mRNAs. D1R and D2R mRNA increased more or less in tandem during development in the striatum and the globus pallidus (Fig. 7). In sharp contrast to the D1R, D2R and D5R mRNAs, which increased from E12 to P60, D3R and D4R mRNA expression showed up- and down-regulation during development, suggesting that these two mRNAs may play significant roles in brain development. Other reports also emphasize the notion that D3R mRNA is markedly regulated during brain development. For example, D3R mRNA and binding site distribution changed dramatically within the different layers of the developing rodent somastosensory cortex (whisker barrel field) suggesting a role for this dopamine receptor in cortical pattern formation [14]. In the present study, D3R mRNA was the only mRNA that showed region-specific expression patterns and significant developmental changes along dorso-ventral and rostro-caudal axes of the striatum.
Recently we showed that dopamine receptor activation modulates migration of GABA neurons from the embryonic mouse striatum to the cerebral cortex [5]. Activation of the D1R promoted the migration whereas activation of the D2R decreased it suggesting that the ability of the neurons to migrate to the cortex may be influenced by the balance of expression between D1-like and D2-like receptors. Thus, if a neuron in the embryonic striatum expressed predominantly D1-like receptors, it may be more likely to migrate to the cortex whereas a neuron expressing predominantly D2-like receptors may be more likely to remain within the striatum. The present data support this speculation by showing that the D2-like receptors predominate in the embryonic and postnatal striatum (Fig. 3) whereas D1-like receptors predominate in the embryonic and postnatal cerebral cortex (Fig. 4).
The mRNA expression patterns in the frontal and cingulate cortices in the dorsal forebrain were significantly different than the ventral forebrain regions (Fig. 7A, B). The overall mRNA expression levels in the dorsal forebrain were significantly lower than those in the ventral forebrain. Another major difference was that D1R mRNA was more abundant than D2R mRNA in the dorsal forebrain. In addition, in the frontal cortex, the D1R mRNA declined during development. This is the only region in which a decline in D1R mRNA was seen. Another significant difference was that D4R mRNA expression was the highest of all the mRNAs in the cingulate cortex on P0 – even higher than D1R or D2R mRNA. Whether D4R mRNA plays a significant role in the development of this cortical region remains unknown. D4R is expressed on GABA interneurons in the mature cerebral cortex and is implicated in the pathophysiology of neuro-psychiatric disorders [32]. The differences in receptor mRNA expression between dorsal and ventral forebrain (Fig. 7A, B) may underlie the differences in the response of these regions to antipsychotic medication [24,25].
Our data also show that expression levels of the mRNA and membrane-associated protein changes correspond with each other, at least for D1R. We were unable to perform similar analyses for the other receptors because the commercially available antibodies for the other receptors did not identify receptor-specific bands in our hands. Although limited to only the D1R, the mRNA-protein correspondence is strong in two brain regions and during two stages of postnatal development. Therefore, we suggest that mRNA expression is a reliable marker of receptor protein expression in the developing and mature brain. Similar correlations do not exist between mRNA and receptor binding site expression for D1 or D2 family of receptors during development or at maturity [17,38,42]. Previous reports show that D1-like binding sites predominate over D2-like binding sites in the embryonic and postnatal striatum [17,38,42]. This is in contrast to the higher D2-like mRNA expression in the striatum during pre- and postnatal development reported in this study (Fig. 7). Whether the discrepancy is due to differential sensitivities of the techniques for detection of mRNA versus binding sites or if it is due to differences in the processing or trafficking of the receptor protein remains to be determined. Finally, it should be noted, that our method is unable to distinguish between receptor mRNA expression patterns at pre- or post-synaptic regions or between neurons and glial cells.
In summary, our data show that dopamine receptor mRNA expression shows region-specific developmental profiles (Fig. 7). D3R and D4R are the only mRNAs that are up- and down-regulated during postnatal development. D3R mRNA shows developmental variation in the ventral forebrain, whereas D4R mRNA shows developmental variation in the dorsal forebrain. The ratio between D1-like and D2-like mRNAs illustrates the predominance of the D1R in the dorsal forebrain and the predominance of the D2R in the ventral forebrain. Although mRNA expression may correlate with protein expression, the functional significance of developmental modulation of mRNA levels remains unclear because of the lack of correlation between mRNA and binding site expression. The potential for formation of D1R – D2R heteromers, especially in the young-adult brain [39] adds to the complexity involved in correlating mRNA expression and receptor function.
4. Experimental Procedures
Animals
Timed-pregnant CD1 mice were purchased from Charles River Laboratories (Wilmington, MA). Female mice housed with a male for the previous 15 – 17 hours were examined for the presence of vaginal plugs at 9:00 AM. Presence of the plug was taken to indicate conception and the day of plug discovery was designated E0. The day of birth was designated postnatal day 0 (P0). The experimental procedures were approved by Institutional Animal Care and Use Committee and were consistent with the NIH guidelines for the care and use of laboratory animals.
Laser capture microdissection (LCM)
We collected samples of telencephalic proliferative and postmitotic domains from E12 and E15 mice by using LCM. Embryos were removed one at a time from dams deeply anesthetized by an intraperitoneal injection of a mixture of Ketamine (50 mg/kg) and Xylazine (10 mg/kg). The embryos were decapitated; their heads were frozen in liquid nitrogen vapor and stored at −80°C. The frozen heads were sectioned on a cryostat at 70μm thickness in the coronal plane. The sections were mounted on RNase-free, MembraneSlides® (Molecular Machines and Industries, Glattbrugg, Switzerland), dried at room temperature and stored at −80°C. The sections were viewed under a Nikon TE2000 inverted microscope connected to CellCut® LCM equipment (Molecular Machines and Industries). The lateral ganglionic eminence (LGE), medial ganglionic eminence (MGE), caudal ganglionic eminence (CGE), neuroepithelium of the dorsal cerebral wall, striatal differentiating field and postmitotic regions of the cerebral cortex (from E15 only) were identified and dissected (UVCut software package; Molecular Machines and Industries) (Fig. 1A, B). Immediately upon dissection, the sample was picked up on adhesive lids of 500μl mini IsolationCaps® (Molecular Machines and Industries) (Fig. 1A, B). Each region of interest was collected from both hemispheres and pooled. We collected samples from 3–6 brains for each of the two embryonic stages.
P0, P21and P60 mice were anesthetized by isoflurane inhalation and decapitated. Brains were removed and frozen in liquid nitrogen vapor and stored at −80°C. Coronal sections were cut and mounted as described above. From these sections, we collected samples of the striatum, globus pallidus, frontal cortex and cingulate cortex by LCM as described above (Fig. 1C, D, E). The different regions were identified with the help of an adult mouse brain stereotaxic atlas [35]. Since the striatum is a rostro-caudally elongated structure and striatal inputs an outputs are topographically organized [19,22,30,37,55], we collected separate samples from rostral and caudal striatum. The junction between the anterior and posterior limbs of the anterior commissure was considered to be the boundary between rostral and caudal subdivisions of the striatum. The sections at the junction and rostral to it were considered to represent rostral striatum and those caudal to the junction were considered the caudal striatum. From each subdivision, we collected separate samples from the dorsal and ventral portions. Thus, from each brain, we collected four samples of the striatum: dorsal-rostral, ventral-rostral, dorsal-caudal and ventral-caudal. Samples of each brain region collected from the two hemispheres were pooled. Each region of interest sample was pooled from both hemispheres. Samples from male and female mice were collected and analyzed separately for each brain region from the P60 age group. We collected samples from 3–6 brains for each of the two postnatal age groups.
The stereotaxic coordinates for the different regions in the P60 brains were as follows. Rostral striatum from 5.34 mm inter-aural, 1.54 mm bregma to 4.06 mm inter-aural, 0.26 mm bregma; caudal striatum from 3.94 mm inter-aural, 0.14 mm bregma to 3.22 mm inter-aural, −0.58 mm bregma. Globus pallidus: 3.82 mm inter-aural, 0.02 mm bregma to 2.22 mm inter-aural, −1.58 mm bregma. The frontal and cingulated cortices were collected from the same sections that were used for collection of striatal samples.
We also collected samples of the cerebral cortex and striatum from E15 and P60 brains by micro-dissection for preparation of RNA samples for use as endogenous reference genes. We did not use LCM for this purpose because regional specificity was not necessary for validation of an endogenous control.
RNA isolation
RNA from the LCM samples was extracted with PicoPure RNA isolation kit (Arcturus-Molecular Devices, Mountain View, CA), and additional DNase digestion (Qiagen, Valencia, CA), according to manufacture’s instructions. RNA, for validation of endogenous controls, collected by microdissection from cerebral cortex and striatum E15 and P60, was isolated with RNAqueous-4PCR kit (Applied Biosystems, Foster City, CA) with DNase treatment according to manufacture’s instructions.
Reverse transcription and quantitative real-time PCR
After validation of RNA sample quality and quantity with a Bioanalyzer 2100 (Agilent Technologies, Foster City, CA) (Fig. 1F), 2μg of RNA were converted to cDNA by RT reaction using SuperScript II and random hexamer primers (Invitrogen, Carlsbad, CA). We used 0.5μl of cDNA per well in a 20μl reaction volume for PCR. Proprietary primers and fluorescent probe sequences were designed by Applied Biosystems TaqMan Gene expression assays. The following genes were analyzed, each is followed by its corresponding catalogue number D1R (Mm01353211_m1), D2R (Mm00438541_m1), D3R (Mm00432887_m1), D4R (Mm00432893_m1), D5R (Mm00658653_s1), and reference genes 18s rRNA (Hs Hs99999901_s1), β-actin (4352933E), β-2-microglobulin (B2M) (Mm00437762_m1), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (4352932E). TaqMan Universal master PCR mix was used for all PCR reactions (Applied Biosystems). Real-time PCR was performed in a Prism 7000 (Applied Biosystems) using the following thermal settings: 1 cycle of 2 min at 50°C, 1 cycle of 10 min at 95°C and 40 cycle of 15 sec at 95°C and 1min at 60°C. Relative mRNA expression for endogenous control genes was calculated by using the 2−ΔΔCt method[27]. Cycle thresholds for all genes were selected 2 cycles past baseline amplification within the linear range on log scaling. All dopamine receptor target gene expressions were normalized to the endogenous control and the mean normalized expression was analyzed using Q-gene software [48]. The data were analyzed by a Student’s t-test or ANOVA using Prism 4 for Macintosh analysis package (Graphpad Software Inc., San Diego, CA).
Isolation of cell membrane fractions and immunoblotting
We collected samples of cerebral cortex and striatum from P0 and P60 mice by micro-dissection. Membrane fractions from the micro-dissected samples were prepared as described previously [3]. Briefly, tissue samples were homogenized by hand with a teflon/glass homogenizer on ice with cold homogenization buffer (250mM sucrose, 10mM Tris, 1mM dithiothreitol (DTT), 1mM MgCl2, complete mini protein inhibitor (Roche Applied Science, Indianapolis, IN, pH 7.4), and centrifuged at 600 X g for 10 min. The membrane fraction was precipitated by centrifugation of the supernatant at 200,000 X g (70.1 Ti rotor at 50,000 rpm for 30 min) and resuspended in resuspension buffer (10mM Tris, 1mM EDTA, 1mM DTT and complete mini protein inhibitor). Protein concentration of the membrane fractions was measured by BAC kit (Pierce, Rockford, IL), and the samples were solubilized in sample buffer (63mM Tris-HCl, 2% SDS, 10% Glycerol, 2% β-Mercaptoethanol, 0.01% Bromophenol blue). Solubilized membrane fraction protein (20μg) without heat denaturation was fractionated by SDS-PAGE 20-4% gradient gel and transferred to PVDF (Millipore, Billerica, MA). The blots were incubated with 5% skim milk in TBS (20mM Tris, 137mM NaCl, pH 7.6) for 1 hr at room temperature, followed by incubation with monoclonal D1R antibody (1:500) (Chemicon, Temecula, CA) in TBS containing 1.5% BSA overnight at 4°C [28]. Blots were washed with TBS, 0.05% Tween 20 three times, and incubated with horseradish peroxidase-conjugated donkey anti-mouse secondary antibody (1:20,000) (GE Healthcare, Piscataway, NJ), for 1 hr. Immunoreactivity was detected by using ECL plus (GE Healthcare), and exposed to autoradiogrphic film (Kodak, Rochester, NY).
Statistical Analysis
All statistical analyses were performed with Prism v4.0a for Macintosh (GraphPad Software, Inc., San Diego, CA). Analysis of variance was performed for dopamine receptor expression levels across ages and within age for each region. Post-hoc analysis was performed by using Bonferroni correction for multiple comparisons. Student’s t test was performed for analysis of male and female differences and in other cases involving comparison between only 2 groups. Due to multiple comparisons and the risk of a Type I error, statistical significance for ANOVA and t-tests was set at p<0.001.
Acknowledgments
Supported by USPHS grants NS43426, DA020796 and P30NS45776 and American Heart SDG 0535138N. We thank Ms. Luba Zagachin of the Neuroscience Center real-time PCR Core facility for her assistance with this work. We also gratefully acknowledge assistance from Dr. Charles Vanderburg of the Advanced Tissue Resource Center at the Harvard Center for Neurodegeneration and Repair.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Altman J, Bayer SA. Atlas of prenatal rat brain development. CRC Press; Boca raton, Fl., USA: 1995. [Google Scholar]
- 2.Bayer SA. Neurogenesis in the rat neostriatum. International Journal of Developmental Neuroscience. 1984;2:163–175. doi: 10.1016/0736-5748(84)90008-X. [DOI] [PubMed] [Google Scholar]
- 3.Bergson C, Mrzljak L, Smiley JF, Pappy M, Levenson R, Goldman-Rakic PS. Regional, cellular, and subcellular variations in the distribution of D1 and D5 dopamine receptors in primate brain. J Neurosci. 1995;15:7821–36. doi: 10.1523/JNEUROSCI.15-12-07821.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cepeda C, Levine MS. Dopamine and N-Methyl-D-Aspartate Receptor Interactions in the Neostriatum. Dev Neurosci. 1998;20:1–18. doi: 10.1159/000017294. [DOI] [PubMed] [Google Scholar]
- 5.Crandall JE, McCarthy DM, Araki KY, Sims JR, Ren JQ, Bhide PG. Dopamine receptor activation modulates GABA neuron migration from the basal forebrain to the cerebral cortex. J Neurosci. 2007;27:3813–3822. doi: 10.1523/JNEUROSCI.5124-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diaz J, Ridray S, Mignon V, Griffon N, Schwartz JC, Sokoloff P. Selective expression of dopamine D3 receptor mRNA in proliferative zones during embryonic development of the rat brain. J Neurosci. 1997;17:4282–4292. doi: 10.1523/JNEUROSCI.17-11-04282.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fentress JC, Stanfield BB, Cowan WM. Observations on the development of the striatum in mice and rats. Anatomy and Embryology. 1981;163:275–298. doi: 10.1007/BF00315705. [DOI] [PubMed] [Google Scholar]
- 8.Fishell G, Van der Kooy D. Pattern formation in the striatum: developmental changes in the distribution of striatonigral neurons. J Neurosci. 1987;7:1969–1978. doi: 10.1523/JNEUROSCI.07-07-01969.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fishell G, Van der Kooy D. Pattern formation in the striatum: Neurons with early projections to the substantia nigra survive the cell death period. J Comp Neurol. 1991;312:33–42. doi: 10.1002/cne.903120104. [DOI] [PubMed] [Google Scholar]
- 10.Gerfen CR. Molecular effects of dopamine on striatal-projection pathways. Trends Neurosci. 2000;23:S64–70. doi: 10.1016/s1471-1931(00)00019-7. [DOI] [PubMed] [Google Scholar]
- 11.Golden GS. Prenatal development of the biogenic amine systems of the mouse brain. Dev Biol. 1973;33:300–311. doi: 10.1016/0012-1606(73)90139-5. [DOI] [PubMed] [Google Scholar]
- 12.Graybiel AM, Hickey TL. Chemospecificity of ontogenetic units in the striatum: demonstration by combining [3H]thymidine neuronography and histochemical staining. Proc Natl Acad Sci USA. 1982;79:196–202. doi: 10.1073/pnas.79.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guennoun R, Bloch B. D2 dopamine receptor gene expression in the rat striatum during ontogeny: an in situ hybridization study. Dev Brain Res. 1993;60:79–87. doi: 10.1016/0165-3806(91)90157-e. [DOI] [PubMed] [Google Scholar]
- 14.Gurevich EV, Joyce JN. Dopamine D(3) receptor is selectively and transiently expressed in the developing whisker barrel cortex of the rat. J Comp Neurol. 2000;420:35–51. doi: 10.1002/(sici)1096-9861(20000424)420:1<35::aid-cne3>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 15.Hoglinger GU, Rizk P, Muriel MP, Duyckaerts C, Oertel WH, Caille I, Hirsch EC. Dopamine depletion impairs precursor cell proliferation in Parkinson disease. Nat Neurosci. 2004;7:726–35. doi: 10.1038/nn1265. [DOI] [PubMed] [Google Scholar]
- 16.Jin LQ, Goswami S, Cai G, Zhen X, Friedman E. SKF83959 selectively regulates phosphatidylinositol-linked D1 dopamine receptors in rat brain. J Neurochem. 2003;85:378–86. doi: 10.1046/j.1471-4159.2003.01698.x. [DOI] [PubMed] [Google Scholar]
- 17.Jung AB, Bennett JP., Jr Development of striatal dopaminergic function. 1. Pre- and postnatal development of mRNAs and binding sites for striatal D1 (D1a) and D2 (D2a) receptors. Dev Brain Res. 1996;94:109–120. doi: 10.1016/0165-3806(96)00033-8. [DOI] [PubMed] [Google Scholar]
- 18.Kaufman MH. The Atlas of Mouse Development. 2. Academic Press; New York: 1992. [Google Scholar]
- 19.Kincaid AE, Wilson CJ. Corticostriatal innervation of the patch and matrix in the rat neostriatum. J Comp Neurol. 1996;374:578–592. doi: 10.1002/(SICI)1096-9861(19961028)374:4<578::AID-CNE7>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 20.Kippin TE, Kapur S, van der Kooy D. Dopamine specifically inhibits forebrain neural stem cell proliferation, suggesting a novel effect of antipsychotic drugs. J Neurosci. 2005;25:5815–23. doi: 10.1523/JNEUROSCI.1120-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee FJ, Xue S, Pei L, Vukusic B, Chery N, Wang Y, Wang YT, Niznik HB, Yu XM, Liu F. Dual regulation of NMDA receptor functions by direct protein-protein interactions with the dopamine D1 receptor. Cell. 2002;111:219–30. doi: 10.1016/s0092-8674(02)00962-5. [DOI] [PubMed] [Google Scholar]
- 22.Lévesque M, Gagnon S, Parent A, Deschênes M. Axonal arborizations of corticostriatal and corticothalamic fibers arising from the second somatosensory area in the rat. Cerebral Cortex. 1996;6:759–770. doi: 10.1093/cercor/6.6.759. [DOI] [PubMed] [Google Scholar]
- 23.Lezcano N, Bergson C. D1/D5 dopamine receptors stimulate intracellular calcium release in primary cultures of neocortical and hippocampal neurons. J Neurophysiol. 2002;87:2167–75. doi: 10.1152/jn.00541.2001. [DOI] [PubMed] [Google Scholar]
- 24.Lidow MS, Elsworth JD, Goldman-Rakic PS. Down-regulation of the D1 and D5 dopamine receptors in the primate prefrontal cortex by chronic treatment with antipsychotic drugs. J Pharmacol Exp Ther. 1997;281:597–603. [PubMed] [Google Scholar]
- 25.Lidow MS, Goldman-Rakic PS. Differential regulation of D2 and D4 dopamine receptor mRNAs in the primate cerebral cortex vs. neostriatum: effects of chronic treatment with typical and atypical antipsychotic drugs. J Pharmacol Exp Ther. 1997;283:939–46. [PubMed] [Google Scholar]
- 26.Lidow MS, Rakic P. Scheduling of monoaminergic neurotransmitter receptor expression in the primate neocortex during postnatal development. Cereb Cortex. 1992;2:401–16. doi: 10.1093/cercor/2.5.401. [DOI] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Luedtke RR, Griffin SA, Conroy SS, Jin X, Pinto A, Sesack SR. Immunoblot and immunohistochemical comparison of murine monoclonal antibodies specific for the rat D1a and D1b dopamine receptor subtypes. J Neuroimmunol. 1999;101:170–87. doi: 10.1016/s0165-5728(99)00142-3. [DOI] [PubMed] [Google Scholar]
- 29.Marchand R, Lajoie L. Histogenesis of the striopallidal system in the rat. Neurogenesis of its neurons. Neuroscience. 1986;17:573–590. doi: 10.1016/0306-4522(86)90031-x. [DOI] [PubMed] [Google Scholar]
- 30.McGeorge AJ, Faull RLM. The organization and collateralization of corticostriate neurones in the motor and sensory cortex of the rat brain. Brain Res. 1987;423:318–324. doi: 10.1016/0006-8993(87)90855-9. [DOI] [PubMed] [Google Scholar]
- 31.Monsma FJ, Mahan LC, McVittie LD, Gerfen CR, Sibley DR. Molecular cloning and expression of a D1 dopamine receptor linked to adenylyl cyclase activation. Proc Natl Acad Sci USA. 1990;87:6723–6727. doi: 10.1073/pnas.87.17.6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mrzljak L, Bergson C, Pappy M, Huff R, Levenson R, Goldman-Rakic PS. Localization of dopamine D4 receptors in GABAergic neurons of the primate brain. Nature. 1996;381:245–248. doi: 10.1038/381245a0. [DOI] [PubMed] [Google Scholar]
- 33.Ohtani N, Goto T, Waeber C, Bhide PG. Dopamine modulates cell cycle in the lateral ganglionic eminence. J Neurosci. 2003;23:2840–50. doi: 10.1523/JNEUROSCI.23-07-02840.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohtani N, Mitsuhashi T, Bhide PG. Effects of dopamine on cell cycle parameters in the lateral ganglionic eminence. Soc Neurosci Abstr. 1999;25:254. [Google Scholar]
- 35.Paxinos G, Franklin KBJ. The mouse brain in stereotaxic coordinates. 2. Academic Press; San Diego, CA, USA: 2001. [Google Scholar]
- 36.Popolo M, McCarthy DM, Bhide PG. Influence of dopamine on precursor cell proliferation and differentiation in the embryonic mouse telencephalon. Dev Neurosci. 2004;26:229–244. doi: 10.1159/000082140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ragsdale CW, Jr, Graybiel AM. A simple ordering of neocortical areas established by the compartmental organization of their striatal projections. Proc Natl Acad Sci USA. 1990;87:6196–6199. doi: 10.1073/pnas.87.16.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rao PA, Molinoff PB, Joyce JN. Ontogeny of dopamine D1 and D2 receptor subtypes in rat basal ganglia: a quantitative autoradiographic study. Brain Res Dev Brain Res. 1991;60:161–77. doi: 10.1016/0165-3806(91)90045-k. [DOI] [PubMed] [Google Scholar]
- 39.Rashid AJ, So CH, Kong MM, Furtak T, El-Ghundi M, Cheng R, O’Dowd BF, George SR. D1–D2 dopamine receptor heterooligomers with unique pharmacology are coupled to rapid activation of Gq/11 in the striatum. Proc Natl Acad Sci U S A. 2007;104:654–9. doi: 10.1073/pnas.0604049104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson SW, Caron MG. Chimeric D2/D3 dopamine receptors efficiently inhibit adenylyl cyclase in HEk 293 cells. J Neurochem. 1996;67:212–219. doi: 10.1046/j.1471-4159.1996.67010212.x. [DOI] [PubMed] [Google Scholar]
- 41.Sales N, Martres MP, Bouthenet ML, Schwartz JC. Ontogeny of dopaminergic D2 receptors in the rat nervous system: characterization and detailed autoradiographic mapping with [125I]iodosulpiride. Neuroscience. 1989;28:673–700. doi: 10.1016/0306-4522(89)90014-6. [DOI] [PubMed] [Google Scholar]
- 42.Schambra UB, Duncan GE, Breese GR, Fornaretto MG, Caron MG, Fremeau RT., jr Ontogeny of D1A and D2 dopamine receptor subtypes in rat brain using in situ hybridization and receptor binding. Neuroscience. 1994;62:65–85. doi: 10.1016/0306-4522(94)90315-8. [DOI] [PubMed] [Google Scholar]
- 43.Schinelli S, Paolillo M, Corona Gl. Opposing actions of D1- and D2-dopamine receptors on arachidonic acid release and cyclic AMP production in striatal neurons. J Neurochem. 1994;62:944–949. doi: 10.1046/j.1471-4159.1994.62030944.x. [DOI] [PubMed] [Google Scholar]
- 44.Schoffelmeer AN, Vanderschuren LJ, De Vries TJ, Hogenboom F, Wardeh G, Mulder AH. Synergistically interacting dopamine D1 and NMDA receptors mediate nonvesicular transporter-dependent GABA release from rat striatal medium spiny neurons. J Neurosci. 2000;20:3496–503. doi: 10.1523/JNEUROSCI.20-09-03496.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seeman P, Bzowej NH, Guan HC, Bergeron C, Becker LE, Reynolds GP, Bird ED, Riederer P, Jellinger K, Watanabe S, et al. Human brain dopamine receptors in children and aging adults. Synapse. 1987;1:399–404. doi: 10.1002/syn.890010503. [DOI] [PubMed] [Google Scholar]
- 46.Shearman LP, Zeitzer J, Weaver DR. Widespread expression of functional D1-dopamine receptors in fetal rat brain. Dev Brain Res. 1997;102:105–15. doi: 10.1016/s0165-3806(97)00091-6. [DOI] [PubMed] [Google Scholar]
- 47.Sibley DR, Monsma FJ., Jr Molecular biology of dopamine receptors. Trends Pharmacol Sci. 1992;13:61–9. doi: 10.1016/0165-6147(92)90025-2. [DOI] [PubMed] [Google Scholar]
- 48.Simon P. Q-Gene: processing quantitative real-time RT-PCR data. Bioinformatics. 2003;19:1439–40. doi: 10.1093/bioinformatics/btg157. [DOI] [PubMed] [Google Scholar]
- 49.Song DD, Harlan RE. Genesis and migration patterns of neurons forming the patch and matrix compartments of the rat striatum. Dev Brain Res. 1994;83:233–245. doi: 10.1016/0165-3806(94)00144-8. [DOI] [PubMed] [Google Scholar]
- 50.Stoof JC, Kebabian JW. Two dopamine receptors: biochemistry, physiology and pharmacology. Life Sci. 1984;35:2281–96. doi: 10.1016/0024-3205(84)90519-8. [DOI] [PubMed] [Google Scholar]
- 51.Tang TS, Bezprozvanny I. Dopamine receptor-mediated Ca(2+) signaling in striatal medium spiny neurons. J Biol Chem. 2004;279:42082–94. doi: 10.1074/jbc.M407389200. [DOI] [PubMed] [Google Scholar]
- 52.Van der Kooy D, Fishell G. Neuronal birthdate underlies the development of striatal compartments. Brain Res. 1986;401:155–161. doi: 10.1016/0006-8993(87)91176-0. [DOI] [PubMed] [Google Scholar]
- 53.Van Kampen JM, Hagg T, Robertson HA. Induction of neurogenesis in the adult rat subventricular zone and neostriatum following dopamine D3 receptor stimulation. Eur J Neurosci. 2004;19:2377–2387. doi: 10.1111/j.0953-816X.2004.03342.x. [DOI] [PubMed] [Google Scholar]
- 54.Van Kampen JM, Robertson HA. A possible role for dopamine D(3) receptor stimulation in the induction of neurogenesis in the adult rat substantia nigra. Neuroscience. 2005 doi: 10.1016/j.neuroscience.2005.07.054. [DOI] [PubMed] [Google Scholar]
- 55.Wilson CJ. Morphology and synaptic connections of crossed corticostriatal neurons in the rat. J Comp Neurol. 1987;263:567–580. doi: 10.1002/cne.902630408. [DOI] [PubMed] [Google Scholar]
- 56.Zhang L, Bai J, Undie AS, Bergson C, Lidow MS. D1 dopamine receptor regulation of the levels of the cell-cycle-controlling proteins, cyclin D, P27 and Raf-1, in cerebral cortical precursor cells is mediated through cAMP-independent pathways. Cereb Cortex. 2005;15:74–84. doi: 10.1093/cercor/bhh110. [DOI] [PubMed] [Google Scholar]
- 57.Zhang L, Lidow MS. D1 dopamine receptor regulation of cell cycle in FGF- and EGF-supported primary cultures of embryonic cerebral cortical precursor cells. Int J Dev Neurosci. 2002;20:593–606. doi: 10.1016/s0736-5748(02)00104-1. [DOI] [PubMed] [Google Scholar]


