Summary
Fragile X-associated tremor/ataxia syndrome (FXTAS) is a recently recognized neurodegenerative disorder in fragile X premutation carriers with FMR1 alleles containing 55-200 CGG repeats. Previously, we developed a Drosophila model of FXTAS and demonstrated that transcribed premutation repeats alone are sufficient to cause neurodegeneration, suggesting that rCGG repeat-binding proteins (RBPs) may be sequestered from their normal function by rCGG binding. Here we identify Pur α and hnRNP A2/B1 as RBPs. We show that Pur α and rCGG repeats interact in a sequence-specific fashion that is conserved between mammals and Drosophila. Overexpression of Pur α in Drosophila could suppress rCGG-mediated neurodegeneration in a dose-dependent manner. Furthermore, Pur α is also present in the inclusions of FXTAS patient brains. These findings support the disease mechanism of FXTAS of rCGG repeat sequestration of specific RBPs, leading to neuronal cell death, and implicate that Pur α plays important role in the pathogenesis of FXTAS.
Introduction
Fragile X syndrome is caused primarily by expansion of the CGG trinucleotide repeat in the 5′ untranslated region (5′ UTR) of the fragile X mental retardation 1 (FMR1) gene (Warren, 2001). While normal individuals generally possess between 5 and 54 repeats, fully affected individuals have more than 200 CGG repeats on what are referred to as full mutation alleles. Premutation alleles (55-200 CGG repeats) of the FMR1 gene are precursors of full mutation alleles and they expand, through genetic instability, into full mutation during maternal germline transmission (Sherman, 2002). Fragile X-associated tremor/ataxia syndrome (FXTAS) is a recently identified neurodegenerative disorder found among many male premutation carriers in or beyond their fifth decade of life (Hagerman and Hagerman, 2004). Female carriers may also develop FXTAS, though the incidence is far higher in males (Hagerman et al., 2004). Common features of the syndrome include progressive intention tremor, gait ataxia, parkinsonism, autonomic dysfunction, and cognitive decline (Hagerman et al., 2005). The neuropathological hallmark of FXTAS is the ubiquitin-positive intranuclear inclusion, present in both neurons and astrocytes throughout the brain (Greco et al., 2002). Furthermore, the cerebellum in FXTAS patients displayed marked dropout of Purkinje cells, Purkinje axonal torpedoes and Bergmann gliosis. However, intranuclear inclusions were absent from Purkinje cells, although they were present in a small number of neurons in the dentate nucleus and diffusely in cerebellar astrocytes (Greco et al., 2002).
The molecular pathogenesis of FXTAS remains unclear. However, several lines of evidence have led us, as well as others, to propose an RNA-mediated gain-of-function toxicity model for FXTAS (Hagerman and Hagerman, 2002; Jin et al., 2003). In cells from premutation carriers with a broad range of repeat lengths, the level of FMR1 mRNA was elevated up to 8-fold over normal levels, while the amount of FMR1-encoded protein (FMRP) appeared to remain at, or slightly below, normal levels (Kenneson et al., 2001; Tassone et al., 2000). Indeed, even in the high-end normal range (∼54 repeats), the FMR1 message level was nearly double that found in the most common alleles (∼30 repeats). In a “knock-in” mouse model, in which the endogenous CGG repeats (5 CGG repeats in the wild-type mouse Fmr1 gene) had been replaced with a ∼100 CGG repeat fragment, intranuclear inclusions were found to be present throughout the brain, with the exception of cerebellar Purkinje cells (Willemsen et al., 2003). An increase in both the number and size of the inclusions was observed during the life course, which correlates with the progressive character of the phenotype observed in humans. Neuropathological studies in humans have revealed a highly significant association between length of the CGG tract and frequency of intranuclear inclusions in both neurons and astrocytes, indicating that the CGG repeat is a powerful predictor of neurological involvement clinically (age of death) as well as neuropathologically (number of inclusions) (Greco et al., 2006). Notably, FMR1 mRNA was found in the inclusions associated with FXTAS patients (Tassone, 2004). Furthermore, intranuclear inclusions can be formed in both primary neural progenitor cells and established neural cell lines, as was revealed using a reporter construct with an FMR1 5′ UTR harboring expanded (premutation) CGG repeats (Arocena et al., 2005). Finally, we have described a fly model of FXTAS expressing the FMR1 untranslated-CGG repeats 5′ to the EGFP coding sequence and demonstrated that premutation-length riboCGG (rCGG) repeats are toxic and sufficient to cause neurodegeneration (Jin et al., 2003). These observations encouraged us to propose that transcription of the CGG90 repeats leads to an RNA-mediated neurodegenerative disease. We further posited a mechanism by which rCGG repeat-binding proteins (RBPs) may become functionally limited by their sequestration to lengthy rCGG repeats, mechanistically similar to the pathophysiology of myotonic dystrophy (Ranum and Day, 2004).
To test this mechanism, here we identify two known RNA-binding proteins, Pur α and hnRNP A2/B1, as they are associated with premutation-length rCGG repeats. Pur α is a conserved RNA-binding protein that is expressed in neuronal cytoplasm and involved in dendritic mRNA transport. We show that the interaction between Pur α and rCGG repeats is conserved and sequence-specific. Overexpression of Pur α in Drosophila could suppress rCGG-mediated neurodegeneration in a dose-dependent manner. Further, Pur α is found to be part of inclusions induced by rCGG repeats in both Drosophila and FXTAS patients. These data support the model that fragile X premutation-length rCGG repeats sequester specific RBPs, leading to neuronal cell death, and implicate that Pur α plays important role in the pathogenesis of FXTAS.
Results
Identification of rCGG repeat-binding proteins (RBPs)
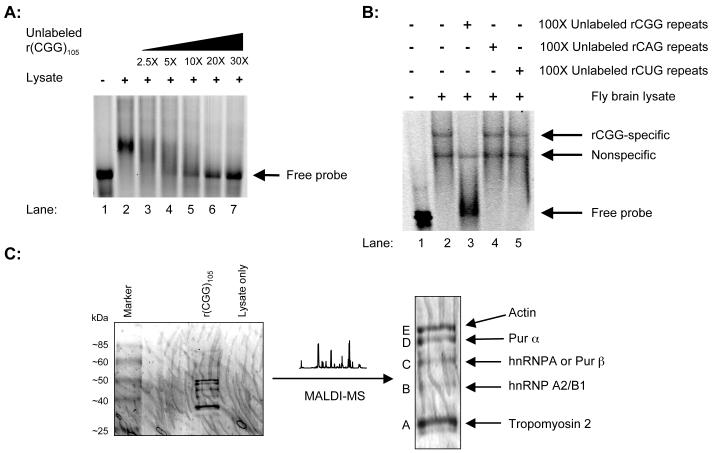
To test the sequestration model, we first questioned whether specific rCGG repeat-binding proteins are present in brain lysates. It is noted that CGG-specific DNA-binding proteins have been identified, but these proteins do not bind RNA (Deissler et al., 1996). We designed an in vitro transcription construct that contained a T7 promoter. The same DNA fragment was used previously to show that transcribed CGG repeats are toxic in Drosophila (Jin et al., 2003). We labeled RNA transcripts by in vitro transcription with fluorescent nucleoside triphosphates (NTPs). The RNA was then used in gel shift assays with both mouse and fly brain lysates. With mouse brain lysates, we observed an RNA-protein complex migrating much slowly than probe alone. This interaction could be competed off by increasing amounts of unlabeled rCGG (Figure 1A). Similarly, a specific RNA-protein complex was also observed with fly brain lysates (Figure 1B). Furthermore, the interaction was rCGG-specific and could not be competed off by the addition of the unlabeled rCAG or rCUG repeats (Figure 1B). These data suggest that specific rCGG repeat-binding protein(s) is present in both mouse and fly brains.
Figure 1. Identification of rCGG repeat-binding proteins (RBPs).
A. Gel shift assay with mouse brain lysates. Lane 1, rCGG probe only; Lane 2, rCGG probe with mouse brain lysates; Lanes 3-7, rCGG probe and mouse brain lysates with the indicated increasing amounts of unlabeled rCGG repeats (molar ratio). B. Gel shift competition assay with fly brain lysates. Lane 1, rCGG probe only; Lane 2, rCGG probe with fly brain lysates; Lanes 3-5, rCGG probe and fly brain lysates in the presence of 100-fold unlabeled triplet repeat RNA, as indicated (molar ratio). C. Identification of RBPs. Coomassie Blue staining gel with RBPs is shown, and distinct bands were cut for protein identification. The identities of those proteins are indicated on the right.
To identify the proteins that bind to rCGG, we scaled up the binding reaction using biotinylated rCGG repeats, which allows the capture of the repeats by binding to streptavidin-coated magnetic beads and the purification of any associated proteins. Given that the phenotype in both human and mouse models is most severe in the cerebellum, mouse cerebellar lysates were used for protein purification (Greco et al., 2006; Willemsen et al., 2003). The putative rCGG repeat-binding proteins were purified by binding to a column of magnetic streptavidin beads. Next, the eluted proteins were separated on a 4-20% gradient SDS-PAGE gel, and specific bands (A-E) were excised for protein identification by automated MALDI-MS (Figure 1C). Initial analysis revealed identities of 3 out of the 5 bands. Bands A and E were tropomyosin and actin, respectively. Band B was revealed as hnRNP A2/B1. Bands C and D were initially unidentifiable and subsequently reanalyzed by peptide sequencing. Band D was determined to be Pur α, and Band C was either hnRNP A or Pur β. Both tropomyosin and actin were later found to be able to interact with other RNAs non-specifically and did not study further (data not shown). Among the RNA-binding proteins that we identified here, given the neurodegenerative phenotype associated with Pur α knockout mice (see below), we focused our following studies on the Pur proteins.
Pur α displays a sequence-specific interaction with rCGG repeats
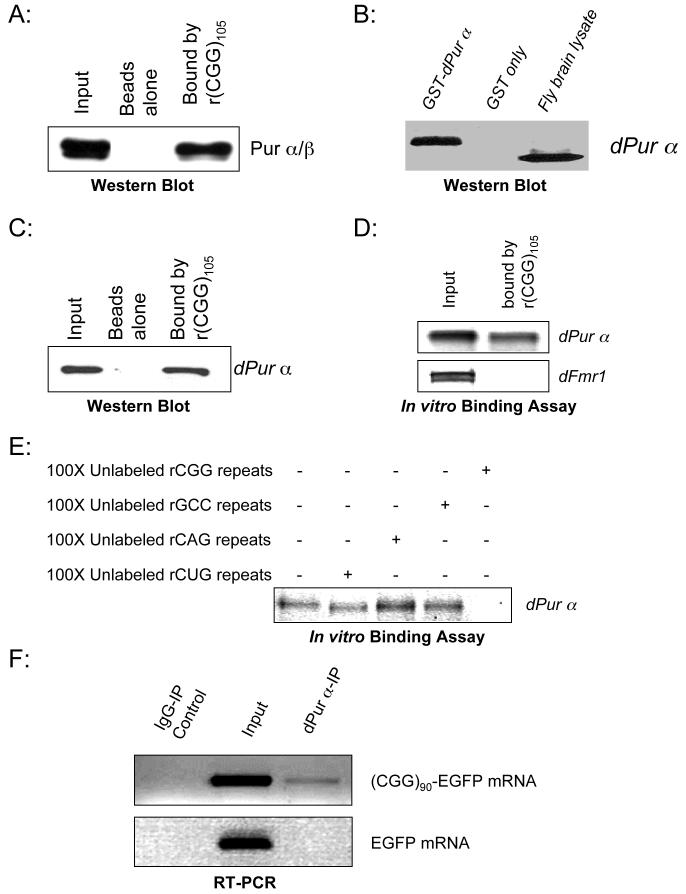
Pur α and Pur β (purine-rich binding proteins) are single-stranded DNA- and RNA-binding proteins that have been implicated in many biological processes, including transcriptional control, initiation of DNA replication, and RNA transport/translation (Gallia et al., 2000; Johnson, 2003). To confirm the interaction between rCGG repeats and the Pur proteins, we repeated the binding reactions using CGG105 biotinylated RNA and cytoplasmic preparations of mouse cerebellar lysates. After binding to the magnetic streptavidin columns, the eluted fractions were used in a Western blot analysis using a specific antibody raised against Pur proteins that was published previously (Khalili et al., 2003). This antibody recognizes both proteins, as demonstrated by the presence of 2 appropriately sized bands (Figure 2A). Cytoplasmic Pur α was found to strongly bind to the rCGG repeats. However, despite similar amounts of Pur α and β in the input, little Pur β protein was captured by the CGG105 repeat RNAs.
Figure 2. Pur α displays a sequence-specific interaction with rCGG repeats.
A. Mouse Pur α protein binds to rCGG repeats. The biotinylated r(CGG)105 repeats were incubated with mouse cerebellar cytoplasmic lysates and captured by DynaBeads. Western blot analysis using antibody against Pur α/β is shown. B. An antibody specifically recognizes Drosophila Pur α protein. Western blot analysis with recombinant proteins (GST-dPur α and GST alone), and wildtype fly brain lysates was shown to test the specificity of this antibody. C. Drosophila Pur α protein also binds to rCGG repeats. The biotinylated rCGG repeats were incubated with fly brain lysates. The captured proteins were eluted and used for Western blot analysis with anti-dPur α antibody. D. Drosophila Pur α protein directly binds to rCGG repeats. The biotinylated rCGG repeats were used for binding reactions with in vitro-translated dPur α or dFmrp. The inputs (10%) and bound fractions are shown. E. Drosophila Pur α protein binds to rCGG repeats specifically. Shown is dPur α protein bound to rCGG repeats in the presence of 100-fold excess (molar ratio) different triplet repeat RNAs. F. Drosophila Pur α protein is associated with rCGG repeat-containing mRNA in vivo. dPur α protein was immunoprecipitated from fly larve expressing either (CGG)90-EGFP or EGFP alone, and the co-immunoprecipitate RNA was isolated. The input (10%) and immunoprecipitated RNAs were used for RT-PCR using the primers specific for EGFP.
We further explored the role of Pur proteins in rCGG-mediated neurodegeneration using the fly model. In the Drosophila genome, there is a single Pur protein ortholog: Pur α. We first tested whether Drosophila Pur protein could bind to the rCGG repeats as well. To characterize the Pur α/rCGG interaction, we developed a specific polyclonal antibody against the Drosophila Pur α protein, as shown in Figure 2B. Similar to the binding assay with mouse cerebellar lysates, we performed binding reactions using CGG105 biotinylated RNA and wild-type fly brain lysates. The captured proteins were used for Western blot analysis with the dPur α antibody. As shown in Figure 2C, endogenous fly dPur α protein could also bind to rCGG repeats. Another RNA-binding protein, Drosophila Fmrp (dFMR1), which is known to bind to G-rich RNAs (G-quartet), could not bind to rCGG repeats (Data not shown). This data suggests that the interaction between Pur α and rCGG repeats is conserved and specific.
To further address whether Pur α directly binds to rCGG repeats, we cloned the full-length cDNA of dPur α by PCR from a fly brain cDNA library. We incubated the in vitro-translated fly dPur α with biotinylated rCGG repeats and our data showed that dPur α could directly bind to rCGG repeats (Figure 2D). Another RNA-binding protein, Drosophila Fmrp (dFMR1), could not bind the rCGG repeats; it was therefore used as a negative control (Wan et al., 2000). To further determine the specificity of the interaction between dPur α and rCGG repeats, we performed RNA-binding assays with or without different unlabeled competitors. The interaction between dPur α and rCGG repeats could not be competed off using unlabeled rCAG repeats, rCUG repeats, or rGCC repeats, which are complementary to rCGG repeats (Figure 2E). Only unlabeled rCGG was able to compete off dPur α from the captured RNA. These data demonstrate that dPur α displays a sequence-specific interaction with rCGG repeats and that this property has been conserved through evolution.
To further determine whether Pur α interacts with rCGG repeats in vivo, using the dPur α antibody, we performed immunoprecipitation experiment. In this experiment, dPur α protein was immunoprecipitated from fly larvae expressing either (CGG)90-EGFP or EGFP alone. The co-immunoprecipitated RNAs were isolated and subjected to RT-PCR using EGFP-specific primers. We found that only the mRNAs containing the rCGG repeats ((CGG)90-EGFP) could be co-immunoprecipitated along with dPur α (Figure 2F). This demonstrate that Pur α is indeed associated with rCGG-repeat-containing mRNA in vivo. However, we noted that only small portion of rCGG-repeat-containing mRNA was co-immunoprecipitated. This is likely due to that a large of portion of dPur α was sequestered into the inclusion by rCGG repeats, and only small amount of dPur α was soluble and could be immunoprecipitated (See below).
Pur α is part of rCGG-induced inclusions
Our previous studies have shown that fragile X premutation rCGG repeats could induce the formation of inclusions, which are Hsp70- and ubiquitin-positive (Jin et al., 2003). Given that Pur α could bind to rCGG repeats both in vitro and in vivo, and FMR1 mRNA was present in the intranuclear inclusions of FXTAS patients’ brains (Tassone, 2004), we examined whether dPur α is also present in the inclusions induced by rCGG repeats in the fly model. Previous studies have shown that the inclusions formed by expanded polyglutamine protein were SDS-insoluble and would remain within the stacking gel after separation on SDS-PAGE gel (Zhou et al., 2001). We performed Western blot analysis using whole head lysates from both wildtype flies and transgenic flies expressing r(CGG)105 repeats in the eye. We observed the SDS-insoluble complexes in the stacking gel with both anti-dPur α and anti-Hsp70 antibodies (Figure 3A). This suggests that dPur α is part of the inclusions induced by the fragile X premutation rCGG repeats (Figure 3A). We did not observe decreased soluble dPur α in the head lysates since the rCGG repeats were only expressed in the eyes of the transgenic flies.
Figure 3. Pur α is present in the rCGG-induced inclusions.
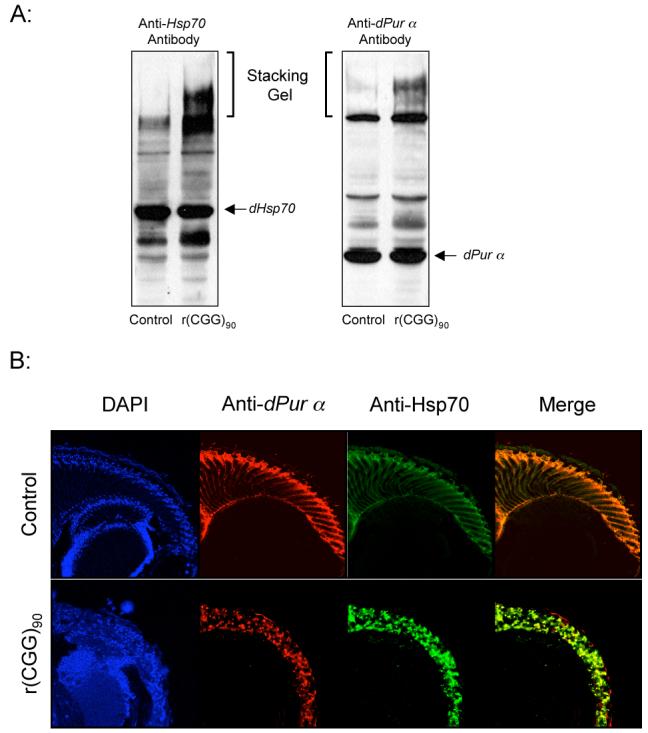
A. Western blot of protein extracted from fly heads, detected with either anti-dPur α antibody or anti-Hsp70 antibody. Arrows indicate migration position of SDS-soluble dPur α and Hsp70 proteins. The aggregated SDS-insoluble protein complex remained in the stacking gel. The genotypes of flies used are w1118; gmr-GAL4 in trans to w1118 (control) and UAS-(CGG)90-EGFP. B. Drosophila Pur α protein is part of rCGG-induced inclusions. Confocal images are shown of the brain transverse sections from 7-day-old flies of either EGFP alone (control) or (CGG)90-EGFP in trans to gmr-GAL4, stained with antibodies against Hsp70 (green) and dPur α protein (red). The nuclei were stained with DAPI (blue).
Furthermore, we performed immunohistochemistry and examined the distribution of dPur α and Hsp70 proteins in both wildtype and rCGG-expressing fly eyes. In wildtype flies, dPur α was ubiquitously expressed and no inclusion was detected. However, in the eyes expressing rCGG repeats, we found that most of the Hsp70-positive inclusions contained dPur α protein as well (Figure 3B). These observations suggest that fragile X premutation rCGG repeats could alter the distribution of dPur α and sequester dPur α into inclusions.
Overexpression of Pur α suppresses rCGG-mediated neurodegeneration in Drosophila
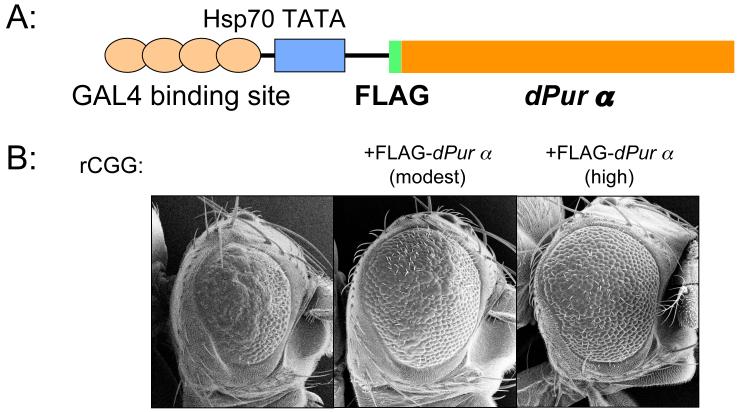
Based on findings from human postmortem samples and animal models, it has been hypothesized that rCGG RBPs may be sequestered from their normal function by binding to lengthy rCGG repeats, leading to neuronal degeneration. If the hypothesis is correct, then we predict that rCGG-mediated neurodegeneration would be suppressed by increasing the expression of RBPs. Accordingly; we generated fly UAS lines that overexpress Drosophila dPur α in the presence of a GAL4 driver. We then crossed these transgenic lines with the CGG-repeat transgenic lines that exhibit photoreceptor neurodegeneration. We discovered that overexpression of Drosophila Pur α could suppress the rCGG-mediated eye neurodegeneration in a dose-dependent manner (Figure 4 and data not shown). This finding strongly supports the mechanistic model proposed above and suggests that Pur α may play an important role in the pathogenesis of FXTAS.
Figure 4. Overexpression of Pur α suppresses rCGG-mediated neurodegeneration in the fly.
A. Schematic representation of pUAST-FLAG-dPur α construct. The full-length cDNA of the Drosophila Pur α gene with FLAG inserted into the downstream of the ATG translational start site was cloned into pUAST plasmid. B. Column 1, flies expressing (CGG)90-EGFP only; Column 2, flies expressing both (CGG)90-EGFP and moderate levels of fly dPur α; Column 3, flies expressing both (CGG)90-EGFP and high levels of fly dPur α. Shown are SEM eye images.
Pur α is present in the inclusions of FXTAS patient brain
Our results in Drosophila model suggest that Pur α could play important role in the pathogenesis of FXTAS. However, in a previous study, the protein composition of the inclusions from post-mortem FXTAS brain tissues was analyzed by mass spectrometry and Pur α was not among the proteins identified (Iwahashi et al., 2006). To determine whether Pur a is part of the inclusion of FXTAS brain tissues as what we observed in our fly model, we performed immunohistochemistry using post-mortem FXTAS brain sections. We found that Pur α is indeed present in the ubiquitin-positive inclusions found in FXTAS brain tissues (Figure 5). This result validates our finding in Drosophila model and suggests that Pur α could play important role in the pathogenesis of FXTAS in human.
Figure 5. Pur α is present in the inclusions of FXTAS patient brain.
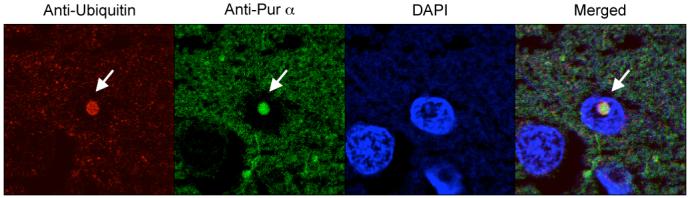
Confocal images are shown of the sup-mid temporal cortex section from a FXTAS patient, stained with antibodies against Ubiquitin (red) and Pur α protein (green). The nuclei were stained with DAPI (blue). Arrow indicates the inclusion containing both ubiquitin and Pur α protein.
Discussion
Fragile X-associated tremor/ataxia syndrome (FXTAS) is a neurodegenerative disorder recently appreciated among adult fragile X syndrome premutation carriers (Hagerman et al., 2005). These patients do not exhibit any clinical features of FMR1-null fragile X syndrome and the full-mutation patients display none of the neurodegenerative features of premutation FXTAS. Thus, it would seem that these variant FMR1 alleles could produce distinctive phenotypes via independent disease mechanisms. While fragile X syndrome is clearly due to the absence of FMR1 expression, previous studies have suggested that FXTAS is RNA-mediated and caused by FMR1 transcripts with premutation-length CGG repeats (Arocena et al., 2005; Jin et al., 2003; Willemsen et al., 2003). One hypothesis is that rCGG repeat-binding proteins (RBPs) could be sequestered from their normal function(s) by binding to these lengthy rCGG repeats, thereby causing neurodegeneration (Hagerman and Hagerman, 2002; Jin et al., 2003). Here we report the identification and characterization of one such RBP, Pur α. We provide both biochemical and genetic data to support the disease mechanism of FXTAS of rCGG repeat sequestration of specific RBPs, leading to neuronal cell death.
rCGG repeat-specific RNA-binding proteins
Specific triplet repeat RNA-binding proteins have previously been implicated in myotonic dystrophy (Miller et al., 2000; Timchenko et al., 1996). There are 2 genetically distinct forms of this neuromuscular disorder, DM1 and DM2, caused by CTG or CCTG repeat expansions in unrelated genes (Ranum and Day, 2004). For both forms, RNA that contains the repeat expansion is central to pathogenesis. CUG repeat RNAs bind to the splicing factor muscleblind (MBNL) in vitro and co-localize with it in vivo, suggesting that MBNL is sequestered by CUG repeats (Jiang et al., 2004; Mankodi et al., 2001; Miller et al., 2000). The fact that loss of MBNL in the mouse leads to phenotypes similar to myotonic dystrophy further supports this notion (Kanadia et al., 2003). Moreover, levels of another splicing factor, CUGBP1, are altered in DM1, and perturbation of normal splicing is observed, which suggests that pathogenesis is caused by the expanded CUG repeat-containing RNA inducing these alterations in splicing via MBNL and CUGBP1 (Timchenko et al., 2002; Timchenko et al., 2001; Timchenko et al., 2004). As in DM1, the repeat-containing transcript present in DM2 affects the levels of CUGBP1 and MBNL, resulting in altered splicing. This RNA-based model of pathogenesis accounts for the remarkably similar multisystemic phenotypes seen in DM1 and DM2, despite the fact that the causative mutations occur in noncoding regions of unrelated genes (Ranum and Day, 2004).
Several lines of evidence, including studies in mouse, Drosophila, and mammalian neuronal cultures, pointed to the critical role of fragile X premutation rCGG repeats in the pathogenesis of FXTAS (Arocena et al., 2005; Jin et al., 2003). To identify rCGG RBPs, we have performed a series of biochemical studies to demonstrate the presence of RBPs. Using both mouse and Drosophila brain lysates, we detected the presence of RBPs in both species and identified RBPs from mouse brain lysates using mass spectrometry. The role of one RBP, hnRNP A2/B1, in rCGG-mediated neurodegeneration was extensively studied in the accompanying paper (Sofola et al., 2007). Here we show that the interaction between rCGG repeats and the other key RBP, Pur α, is specific, in that the interaction cannot be competed off by other repeats, such as rCAG, rCUG, or rGCC. Indeed Pur α was previously found to have a binding preference for the purine-rich single-stranded form of its recognition sequence, which is composed of repeats of NGG (Gallia et al., 2000). Although Pur α has the ability to bind to NGG ribo-repeats, however, the major ribo-repeat binding substrate for Pur α is likely to be rCGG repeats since neither rUGG nor rAGG long repeats (>8) exist in human genome. The identification of Pur α as one of RBPs also validates the approach that we have taken and demonstrates that triplet repeat-specific binding proteins could be identified by combining large-scale in vitro purification and proteomic analysis. Identification of specific RBPs enabled us to test the proposed model of RNA-mediated sequestration.
Pur α and rCGG-mediated neurodegeneration
Pur α is a ubiquitous, sequence-specific DNA- and RNA-binding protein that is highly conserved in eukaryotic cells (Gallia et al., 2000). Pur α has been implicated in diverse cellular functions, including transcriptional regulation and RNA transport/translation (Gallia et al., 2000) (Kanai et al., 2004; Ohashi et al., 2002). As an RNA-binding protein, Pur α is part of an mRNP complex and interacts with multiple proteins, including FMRP, the protein encoded by the FMR1 gene (Ohashi et al., 2002). The normal biological role(s) of the interaction between rCGG repeats and Pur α remains to be determined. However, given the role of Pur α in RNA transport, it is possible that this interaction might be required for FMR1 mRNA transport into dendrites. Of special importance with regard to the data reported above, mice with targeted disruption of the Pur α gene appear normal at birth, but by 2 weeks of age, they develop neurological problems manifested by tremor and gait disturbances, progressing to spontaneous seizures and death by 4 weeks (Khalili et al., 2003). It is remarkable that the early mouse phenotype is reminiscent of that observed in FXTAS patients, which strongly suggests that functionally limiting Pur α, as suggested above, may contribute to the FXTAS phenotype.
In the present study we have identified Pur α as an rCGG repeat-binding protein. This interaction between rCGG repeats and Pur α is specific both in vitro and in vivo. Significantly, overexpression of Pur α could suppress the rCGG-mediated neurodegeneration in the Drosophila model of FXTAS. Furthermore, we demonstrate that Pur α is present in the inclusions of FXTAS brain tissues. These data combined strongly support the hypothesis that the gradual loss of functional Pur α in the cells expressing fragile X premutation rCGG repeats contributes to the FXTAS phenotype. The loss of functional Pur α could be mediated by the sequestration of endogenous Pur α by excessive rCGG repeats, a model supported by our finding that Pur α is part of the inclusions induced by rCGG repeats in both Drosophila and human. In addition, despite the biochemical interaction between Pur α and FMRP, the involvement of Pur α in rCGG-mediated neurodegeneration is independent of FMRP, as appears to be the case in humans, since loss or overexpression of Drosophila Fmrp has no effect on the fly phenotypes caused by fragile X premutation rCGG repeats (Duan and Jin, unpublished data) (Kanai et al., 2004). How might the depletion of Pur α lead to FXTAS, a late adult onset disease? It is likely that the amount of Pur α binding to rCGG repeats increases with expanded repeats. Thus in post-mitotic neurons, the FMR1 mRNAs containing fragile X premutation rCGG repeats could accumulate and titrate Pur α from soluble fraction over the time, which lead to the depletion of functional Pur α eventually. Given the importance of Pur α as part of an mRNP complex involved in mRNA transport, the depletion of Pur α could lead to dysfunction of mRNA transport in the neuron and eventual neuronal cell death (Kanai et al., 2004; Ohashi et al., 2002). Consistent with this idea, patchy axonal loss is indeed observed in FXTAS postmortem brain tissue (Greco et al., 2006; Greco et al., 2002), and previous studies have implicated axonal transport in the pathogenesis of neurodegenerative diseases, such as Alzheimer’s and Huntington’s disease (Gunawardena and Goldstein, 2005; Gunawardena et al., 2003; Stokin et al., 2005). However, the precise mechanism by which Pur α is involved in the pathogenesis of FXTAS requires further study.
In summary, through biochemical purification, we have identified Pur α as an rCGG repeat binding protein. We show that Pur α and rCGG repeats interact in a sequence-specific fashion that is conserved between mammals and Drosophila. Overexpression of Pur α in Drosophila could suppress rCGG-mediated neurodegeneration in a dose-dependent manner. Furthermore, Pur α is also present in the inclusions of FXTAS patient brains. These findings strongly suggest that the RNA-mediated neurodegeneration observed in human FXTAS is mediated by the rCGG repeat sequestration of specific RBPs and implicate that Pur α as one of RBPs plays important role in the pathogenesis of FXTAS. Our data also further support a general mechanistic consequence of transcribed RNA repeats in human disease.
Experimental Procedures
RNA-binding assays
To prepare brain lysates, both mouse and Drosophila brains were washed twice with phosphate-buffered saline (PBS) and homogenized in lysis buffer (10 mM Tris-HCl, pH 7.6, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT). The samples were then centrifuged for 5 min at 10,000 rpm to pellet nuclei and the supernatant (cytoplasmic fraction) was collected and used for RNA-binding assays and RBP purifications. Radioactive-, biotin-, or fluorescent-labeled CGG repeat RNAs were synthesized using the RNAMaxx High Yield Transcription Kit (Stratagene). The RNA probe (100ng) was incubated with 20 μg brain lysates (both mouse and Drosophila) or with 50 ng in vitro-translated protein at room temperature for 30 minutes in 1X binding buffer (20 mM Tris-HCl, pH 7.6, 100 mM KCl, 5 mM MgCl2, 5 mM DTT and 10% glycerol). The binding reaction was loaded and separated on native polyacrylamide gel, which was analyzed via Storm 840 phosphorimager (Amersham Biosciences Corp). For the binding reaction with biotinylated RNAs, DynaBeads M-280 Streptavidin (Dynal, Invitrogen) were used to capture the rCGG-protein complex. The beads were washed before use and resuspended in binding buffer. For the competition assay, the amount of the molar excess unlabeled RNA probe was added to the binding reaction prior to the addition of the labeled probe.
Identification of rCGG-repeat-associated proteins
To identify the proteins associated with rCGG repeats, a large-scale purification using biotinylated rCGG repeats was performed. 5 μg biotinylated rCGG repeats RNA probe and 1 mg mouse cerebellar lysates were used for the binding reaction. The binding reaction was carried out in 1X binding buffer in the presence of 100-fold molar excess tRNA (20 mM Tris-HCl, pH 7.6, 100 mM KCl, 5 mM MgCl2, 5 mM DTT and 10% glycerol). The biotinylated rCGG repeats were then captured by DynaBeads M-280 Streptavidin (Dynal, Invitrogen) and washed for five times. The captured proteins were separated by SDS-PAGE gel. After Coomassie Blue staining, the distinct bands present from the rCGG repeat-binding reaction were cut, subjected to trypsin in-gel digestion and protein identification analysis at the HHMI Biopolymer Laboratory and W.M. Keck Foundation Biotechnology Resource Laboratory at Yale University.
Generation of antibody specific to Drosophila Pur α protein
Rabbit anti-dPur α polyclonal sera directly against the peptide within dPur alpha (CEKMKKSSDSITAEIN) was raised. The peptide and affinity-purified antisera were prepared by New England Peptide, Inc. The specificity of the antibody was tested by Western blot analysis using recombinant dPur alpha protein and fly brain lysates. Western blot analysis was performed as described (Jin et al., 2003). Antibody was used at a dilution of 1:1000. The horseradish peroxidase-conjugated anti-rabbit secondary antibodies were used (Amersham Biosciences, formerly Amersham Pharmacia Biotech, Inc) and detected by Enhanced ChemiLuminescence (Amersham Biosciences, formerly Amersham Pharmacia Biotech, Inc).
Immunoprecipitation, RNA isolation, and RT-PCR
Fly larve expressing either fragile X premutation rCGG repeats (elav-GAL4; UAS-(CGG)90-EGFP) or EGFP alone (elav-GAL4; UAS-EGFP) were collected and homogenized in 1 mL ice-cold lysis buffer (10 mM Tris, pH 7.4, 150 mM NaCl, 30 mM EDTA, 0.5% Triton X-100) with 2X complete protease inhibitors. All further manipulations of the brain lysates were performed at 4°C or on ice. Nuclei and debris were pelleted at 10,000 X g for 10 minutes; the supernatant was collected and precleared for 1 hour with 100 uL recombinant protein G agarose (Invitrogen). Anti-dPur α antibody was incubated with recombinant protein G agarose at 4°C for 2 hours and washed 3 times with lysis buffer. The precleared lysates were immunoprecipitated with antibody-coated recombinant protein G agarose at 4°C overnight. The precipitated complexes were used for Western blot analysis or RNA isolation. RNA from precleared lysates and IPed complexes were isolated using Trizol (Gibco BRL Life Technologies) and cleaned with the RNeasy (QIAGEN). For RT-PCR, RNA was reverse-transcribed with oligo(dT)12-18 and SuperScript II (Invitrogen). The regular PCRs were carried out using EGFP-specific primers as described previously (Jin et al., 2003).
Drosophila genetics
The pUAST constructs were generated by cloning full-length Drosophila Pur alpha cDNA into the pUAST transformation vectors. The constructs were confirmed by DNA sequencing and then injected in a w1118 strain using standard methods. All the other UAS lines, insertions, and GAL4 lines used in this study were obtained from the Bloomington Drosophila stock center. Fly lines were grown on standard medium with yeast paste added. All the crosses were performed at 25°C.
Microscopy and immunohistochemistry
For scanning electron microscopy (SEM) images, whole flies were dehydrated in ethanol, dried with hexamethyldisilazane (Sigma-Aldrich), and analyzed with an ISI DS-130 LaB6 SEM/STEM microscope. For immunohistochemistry, dissected fly heads were fixed for 1 hour in 4% paraformaldehyde, rinsed in PBS, then saturated in 20% sucrose overnight at 4°C. Tissues were embedded in OCT compound (Tissue-Tek) and frozen by immersion in liquid nitrogen. Sections were cut at -20°C, and then stained with different antibodies. Primary antibodies: mouse anti-Hsp70/hsc70 (1:100; StressGen) and rabbit anti-dPur alpha (1:250). Secondary fluorochrome-conjugated antibodies: Cy3 (1:500; Jackson ImmunoResearch) or Cy5 (1:500; Jackson ImmunoResearch). For human tissues, microtome sectioned slices of paraffin-embedded sup-mid temporal cortex derived from FXTAS patients were subjected to immunofluorescent staining. After standard deparaffinization, the section was double stained using a mouse monoclonal antibody against Pur-alpha (Khalili et al., 2003) and a polyclonal rabbit anti-Ubiquitin (DAKO) with the corresponding fluorescently conjugated secondary antibodies. Chromatin was stained with DAPI (Molecular Probes). Confocal microscopy was performed on a Zeiss LSM 510 NLO system.
Acknowledgements
The authors would like to thank J. Taylor and R. Apkarian of The Integrated Microscopy and Microanalytical Facility for their help with SEM, E. Johnson for the antibodies against mouse Pur alpha and beta proteins, C. Greco and P. Hagerman for providing FXTAS patient brain sections. The authors would also like to thank the members of the Warren and Jin labs for their assistance, and S. Chang and K. Garber for critical reading of the manuscript. P.J. is supported by NIH grants R01 NS051630 and R01 MH076090. P.J. is a recipient of the Beckman Young Investigator Award and the Basil O’Connor Scholar Research Award, as well as an Alfred P Sloan Research Fellow in Neuroscience. S.T.W. is supported by NIH grants R37 HD20521 and P30 HD24064.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arocena DG, Iwahashi CK, Won N, Beilina A, Ludwig AL, Tassone F, Schwartz PH, Hagerman PJ. Induction of inclusion formation and disruption of lamin A/C structure by premutation CGG-repeat RNA in human cultured neural cells. Hum Mol Genet. 2005;14:3661–3671. doi: 10.1093/hmg/ddi394. [DOI] [PubMed] [Google Scholar]
- Deissler H, Behn-Krappa A, Doerfler W. Purification of nuclear proteins from human HeLa cells that bind specifically to the unstable tandem repeat (CGG)n in the human FMR1 gene. J Biol Chem. 1996;271:4327–4334. doi: 10.1074/jbc.271.8.4327. [DOI] [PubMed] [Google Scholar]
- Gallia GL, Johnson EM, Khalili K. Puralpha: a multifunctional single-stranded DNA- and RNA-binding protein. Nucleic Acids Res. 2000;28:3197–3205. doi: 10.1093/nar/28.17.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco CM, Berman RF, Martin RM, Tassone F, Schwartz PH, Chang A, Trapp BD, Iwahashi C, Brunberg J, Grigsby J, et al. Neuropathology of fragile X-associated tremor/ataxia syndrome (FXTAS) Brain. 2006;129:243–255. doi: 10.1093/brain/awh683. [DOI] [PubMed] [Google Scholar]
- Greco CM, Hagerman RJ, Tassone F, Chudley AE, Del Bigio MR, Jacquemont S, Leehey M, Hagerman PJ. Neuronal intranuclear inclusions in a new cerebellar tremor/ataxia syndrome among fragile X carriers. Brain. 2002;125:1760–1771. doi: 10.1093/brain/awf184. [DOI] [PubMed] [Google Scholar]
- Gunawardena S, Goldstein LS. Polyglutamine diseases and transport problems: deadly traffic jams on neuronal highways. Arch Neurol. 2005;62:46–51. doi: 10.1001/archneur.62.1.46. [DOI] [PubMed] [Google Scholar]
- Gunawardena S, Her LS, Brusch RG, Laymon RA, Niesman IR, Gordesky-Gold B, Sintasath L, Bonini NM, Goldstein LS. Disruption of axonal transport by loss of huntingtin or expression of pathogenic polyQ proteins in Drosophila. Neuron. 2003;40:25–40. doi: 10.1016/s0896-6273(03)00594-4. [DOI] [PubMed] [Google Scholar]
- Hagerman PJ, Hagerman RJ. The fragile-X premutation: a maturing perspective. Am J Hum Genet. 2004;74:805–816. doi: 10.1086/386296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman RJ, Hagerman PJ. The fragile X premutation: into the phenotypic fold. Curr Opin Genet Dev. 2002;12:278–283. doi: 10.1016/s0959-437x(02)00299-x. [DOI] [PubMed] [Google Scholar]
- Hagerman RJ, Leavitt BR, Farzin F, Jacquemont S, Greco CM, Brunberg JA, Tassone F, Hessl D, Harris SW, Zhang L, et al. Fragile-X-associated tremor/ataxia syndrome (FXTAS) in females with the FMR1 premutation. Am J Hum Genet. 2004;74:1051–1056. doi: 10.1086/420700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman RJ, Ono MY, Hagerman PJ. Recent advances in fragile X: a model for autism and neurodegeneration. Curr Opin Psychiatry. 2005;18:490–496. doi: 10.1097/01.yco.0000179485.39520.b0. [DOI] [PubMed] [Google Scholar]
- Iwahashi CK, Yasui DH, An HJ, Greco CM, Tassone F, Nannen K, Babineau B, Lebrilla CB, Hagerman RJ, Hagerman PJ. Protein composition of the intranuclear inclusions of FXTAS. Brain. 2006;129:256–271. doi: 10.1093/brain/awh650. [DOI] [PubMed] [Google Scholar]
- Jiang H, Mankodi A, Swanson MS, Moxley RT, Thornton CA. Myotonic dystrophy type 1 is associated with nuclear foci of mutant RNA, sequestration of muscleblind proteins and deregulated alternative splicing in neurons. Hum Mol Genet. 2004;13:3079–3088. doi: 10.1093/hmg/ddh327. [DOI] [PubMed] [Google Scholar]
- Jin P, Zarnescu DC, Zhang F, Pearson CE, Lucchesi JC, Moses K, Warren ST. RNA-mediated neurodegeneration caused by the fragile X premutation rCGG repeats in Drosophila. Neuron. 2003;39:739–747. doi: 10.1016/s0896-6273(03)00533-6. [DOI] [PubMed] [Google Scholar]
- Johnson EM. The Pur protein family: clues to function from recent studies on cancer and AIDS. Anticancer Res. 2003;23:2093–2100. [PubMed] [Google Scholar]
- Kanadia RN, Johnstone KA, Mankodi A, Lungu C, Thornton CA, Esson D, Timmers AM, Hauswirth WW, Swanson MS. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–1980. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Kenneson A, Zhang F, Hagedorn CH, Warren ST. Reduced FMRP and increased FMR1 transcription is proportionally associated with CGG repeat number in intermediate-length and premutation carriers. Hum Mol Genet. 2001;10:1449–1454. doi: 10.1093/hmg/10.14.1449. [DOI] [PubMed] [Google Scholar]
- Khalili K, Del Valle L, Muralidharan V, Gault WJ, Darbinian N, Otte J, Meier E, Johnson EM, Daniel DC, Kinoshita Y, et al. Puralpha is essential for postnatal brain development and developmentally coupled cellular proliferation as revealed by genetic inactivation in the mouse. Mol Cell Biol. 2003;23:6857–6875. doi: 10.1128/MCB.23.19.6857-6875.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankodi A, Urbinati CR, Yuan QP, Moxley RT, Sansone V, Krym M, Henderson D, Schalling M, Swanson MS, Thornton CA. Muscleblind localizes to nuclear foci of aberrant RNA in myotonic dystrophy types 1 and 2. Hum Mol Genet. 2001;10:2165–2170. doi: 10.1093/hmg/10.19.2165. [DOI] [PubMed] [Google Scholar]
- Miller JW, Urbinati CR, Teng-Umnuay P, Stenberg MG, Byrne BJ, Thornton CA, Swanson MS. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. Embo J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi S, Koike K, Omori A, Ichinose S, Ohara S, Kobayashi S, Sato TA, Anzai K. Identification of mRNA/protein (mRNP) complexes containing Puralpha, mStaufen, fragile X protein, and myosin Va and their association with rough endoplasmic reticulum equipped with a kinesin motor. J Biol Chem. 2002;277:37804–37810. doi: 10.1074/jbc.M203608200. [DOI] [PubMed] [Google Scholar]
- Ranum LP, Day JW. Myotonic dystrophy: RNA pathogenesis comes into focus. Am J Hum Genet. 2004;74:793–804. doi: 10.1086/383590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman S. Epidemiology. In: Hagerman RJ, editor. Fragile X Syndrome: Diagnosis, Treatment and Research. The Johns Hopkins University Press; Baltimore, MD: 2002. pp. 136–168. [Google Scholar]
- Sofola OA, Jin P, Duan R, Liu H, de Haro M, Nelson DL, Botas J. RNA binding proteins hnRNP A2/B1 and CUGBP1 supress Fragile X CGG premutation repeat-induced neurodegeneration in a Drosophila model of FXTAS. Neuron. 2007 doi: 10.1016/j.neuron.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokin GB, Lillo C, Falzone TL, Brusch RG, Rockenstein E, Mount SL, Raman R, Davies P, Masliah E, Williams DS, Goldstein LS. Axonopathy and transport deficits early in the pathogenesis of Alzheimer’s disease. Science. 2005;307:1282–1288. doi: 10.1126/science.1105681. [DOI] [PubMed] [Google Scholar]
- Tassone F, Hagerman RJ, Taylor AK, Gane LW, Godfrey TE, Hagerman PJ. Elevated levels of FMR1 mRNA in carrier males: a new mechanism of involvement in the fragile-X syndrome. Am J Hum Genet. 2000;66:6–15. doi: 10.1086/302720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassone F, Iwahashi C, Hagerman PJ. FMR1 RNA within the intranuclear inclusions of fragile X-associated tremor/ataxia syndrome (FXTAS) RNA Biology. 2004;1:103–105. doi: 10.4161/rna.1.2.1035. [DOI] [PubMed] [Google Scholar]
- Timchenko LT, Miller JW, Timchenko NA, DeVore DR, Datar KV, Lin L, Roberts R, Caskey CT, Swanson MS. Identification of a (CUG)n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Res. 1996;24:4407–4414. doi: 10.1093/nar/24.22.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timchenko LT, Tapscott SJ, Cooper TA, Monckton DG. Myotonic dystrophy: discussion of molecular basis. Adv Exp Med Biol. 2002;516:27–45. doi: 10.1007/978-1-4615-0117-6_2. [DOI] [PubMed] [Google Scholar]
- Timchenko NA, Cai ZJ, Welm AL, Reddy S, Ashizawa T, Timchenko LT. RNA CUG repeats sequester CUGBP1 and alter protein levels and activity of CUGBP1. J Biol Chem. 2001;276:7820–7826. doi: 10.1074/jbc.M005960200. [DOI] [PubMed] [Google Scholar]
- Timchenko NA, Patel R, Iakova P, Cai ZJ, Quan L, Timchenko LT. Overexpression of CUG triplet repeat-binding protein, CUGBP1, in mice inhibits myogenesis. J Biol Chem. 2004;279:13129–13139. doi: 10.1074/jbc.M312923200. [DOI] [PubMed] [Google Scholar]
- Wan L, Dockendorff TC, Jongens TA, Dreyfuss G. Characterization of dFMR1, a drosophila melanogaster homolog of the fragile X mental retardation protein [In Process Citation] Mol Cell Biol. 2000;20:8536–8547. doi: 10.1128/mcb.20.22.8536-8547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren ST, Sherman SL. The fragile X syndrome. In: Scriver CR, Beaudet AL, Valle D, Childs B, Kinzler KW, Vogelstein B, editors. The Metabolic & Molecular Bases of Inherited Disease. McGraw-Hill Companies; 2001. pp. 1257–1290. [Google Scholar]
- Willemsen R, Hoogeveen-Westerveld M, Reis S, Holstege J, Severijnen LA, Nieuwenhuizen IM, Schrier M, van Unen L, Tassone F, Hoogeveen AT, et al. The FMR1 CGG repeat mouse displays ubiquitin-positive intranuclear neuronal inclusions; implications for the cerebellar tremor/ataxia syndrome. Hum Mol Genet. 2003;12:949–959. doi: 10.1093/hmg/ddg114. [DOI] [PubMed] [Google Scholar]
- Zhou H, Li SH, Li XJ. Chaperone suppression of cellular toxicity of huntingtin is independent of polyglutamine aggregation. J Biol Chem. 2001;276:48417–48424. doi: 10.1074/jbc.M104140200. [DOI] [PubMed] [Google Scholar]