Abstract
Site-specific integrative recombination of bacteriophage λ involves unequal partners. The minimal phage att site is composed of approximately 240-base pairs and four distinct binding sites for Int protein, at least three of which are crucial for function. This ‘donor site’ recombines efficiently with a smaller ‘recipient site’ that lacks the extensive interactions with Int protein.
The integration/excision pathway of bacteriophage λ is a particularly favourable system for studying site-specific recombination at the molecular level (for reviews see refs 1, 2). Integration of the phage genome into the chromosome of the host occurs by a reciprocal recombination between the phage att site (POP’) and the bacterial att site (BOB’). The products are two prophage att sites which comprise the left (BOP’) and right (POB’) junctures between bacterial DNA and the integrated prophage (the formal equivalent of this recombination is shown in Fig. 1b). Both the integration and excision reactions require the phage-encoded protein Int as well as the products of several host Escherichia coli genes3. An additional requirement for the excision reaction is the phage-encoded protein Xis. The Int and Xis proteins are 356 and 72 amino acids respectively, as determined from their encoding DNA sequences4. Recent in vivo experiments suggest that in some situations, Xis can partially replace, or be replaced by, the host factors5. Highly purified Int protein forms specific stable complexes with att DNA, has an enzymatic activity for cutting and reseating DNA strands, and is functional in an in vitro intermolecular recombination system6–11.
Fig. 1.
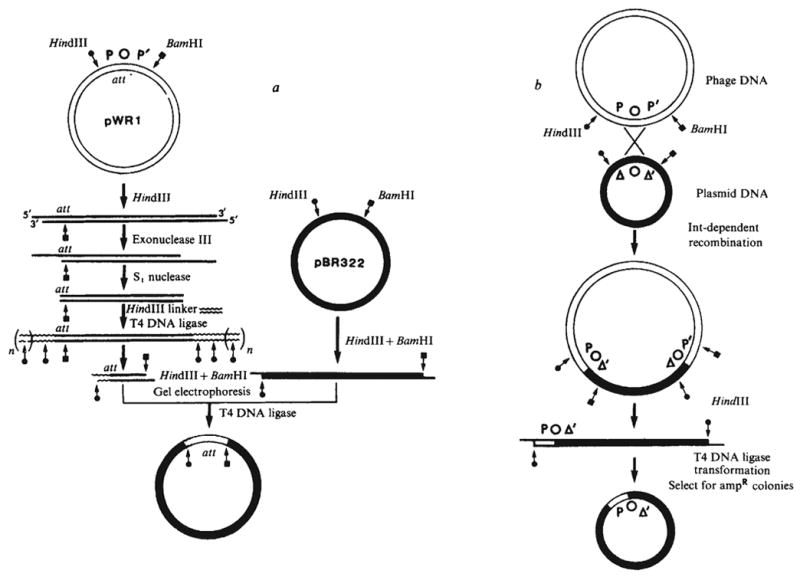
Construction of plasmids carrying resected att sites. a, Plasmid pWR1 was first linearised, for example with HindIII for shortening the P arm. Digestion of the linear DNA with a combination of exonuclease III from E. coli (New England Biolabs) and nuclease S1 (Miles) generates a series of shortened plasmids carrying flush base-paired termini. A chemically synthesised decamer HindIII linker (Collaborative Research) was ligated to the blunt end by T4 DNA ligase. Digestion with HindIII and BamHI generates a family of fragments with unaltered P′ arms and P arms shortened to different extents. These were fractionated by gel electrophoresis and regions corresponding to the desired size were excised and eluted. Finally, the size selected fragments were recloned between the HindIII and BamHI sites of pBR322 and the plasmid carried in each clone characterised by restriction fragment analysis. In an analogous fashion, shortening of the P′ arm would be carried out by using BamHI in the place of HindIII to linearise the plasmid and using a BamHI linker in place of the HindIII linker. b, Cells harbouring a plasmid with a shortened att site arm were infected with λ phage carrying a wild-type att site. As a result, some of the infecting phage underwent Int-dependent recombination with the att site on the plasmid and thus acquired the ability to transduce ampR (D. Kamp and K. Mizuuchi, personal communication). The att site at each phage-plasmid junction has one arm derived from the plasmid and the other arm derived from the phage. Recovery of the recombinant att site on a self-replicating plasmid was accomplished by digesting the plasmid-phage hybrid DNA with HindIII, recircularising the DNA (by means of the HindIII ends) and selecting ampicillin-resistant transformants. These procedures will be described in detail elsewhere (P.-L. H. and A.L., in preparation).
A structural feature of integrative recombination, both in vivo and in vitro, is that the phage att site must be on a negatively supercoiled DNA molecule8,11,12. The cross-over event of Int-dependent recombination must take place within, or at the boundaries of, a 15-base pair core sequence that is common to all four att sites and is designated ‘O’ in the att site nomenclature13. DNA sequences of the four arms of the att sites, designated P, P′, B and B′, are all different from each other. The four att sites can also be distinguished from each other genetically by their distinctive behaviour in pairwise crosses14–16.
The results reported here define the functional limits of the phage att site. We have also extended our studies of the interaction between Int protein and att sites9. The minimal phage att site is quite large (approximately 240 base pairs) and has several widely spaced Int binding sites that differ in size and in response to heparin challenge. In contrast, the minimal sequence required for a phage att site partner (such as the bacterial att site) may not be much larger than the 15-base pair common core. We suggest a model in which integrative recombination involves two unequal partners; accordingly the phage att site is denoted the ‘donor’ and the bacterial att site or its analogue the ‘recipient’.
Generating resected att sites
The λ phage att site is in the centre of a 492-base pair HindIII-BamHI restriction fragment that extends from positions −251 to +242. (For numbering DNA sequences in this region of the λ chromosome, the centre of the common core region is taken as 0; positive numbers extend to the right, through the int and xis genes, and negative numbers extend to the left13.) This att-containing restriction fragment was cloned into pBR322 in place of the HindIII-BamHI fragment from the tet gene. The resulting plasmid, pWR1, carries all of the information necessary to specify a functional phage att site as determined by its competence for Int-dependent recombination with a bacterial att site in vitro (see below). This plasmid was used as the starting point for defining the functional limits of the phage att site. The question is then, how much DNA can be removed from around the phage att site without impairing its function?
When comparing different plasmids for relative amounts of phage att site function it is desirable that only one of the two phage att site arms should be altered at a time, and that non-att sequences (that is, plasmid DNA) adjacent to the different shortened termini should all be the same. We therefore generated phage att sites with incremental lengths of one arm as summarised in Fig. 1. The first procedure involved exonucleolytic digestion of the terminal nucleotides and cloning of the shortened att sites. An additional procedure involved reassortment of shortened arms between two att sites by Int-dependent recombination and recovery of the products.
Protocols were designed so that the final att-containing restriction fragments always have one HindIII and one BamHI terminus, thus facilitating subsequent cloning, restriction analysis and sequencing. The details for the construction of each plasmid will be described elsewhere.
Function of resected phage att sites
Competence for phage att site function was tested in an in vitro Int-dependent recombination system10,11. In the assay conditions used the phage att site must be on a supercoiled molecule11 and the bacterial att site functions best when it is on a linear molecule (K. Mizuuchi, personal communication). Control experiments established that this assay registers only phage att site function on the supercoiled parent, and not prophage att site function (Fig. 3, right panel).
Fig. 3.
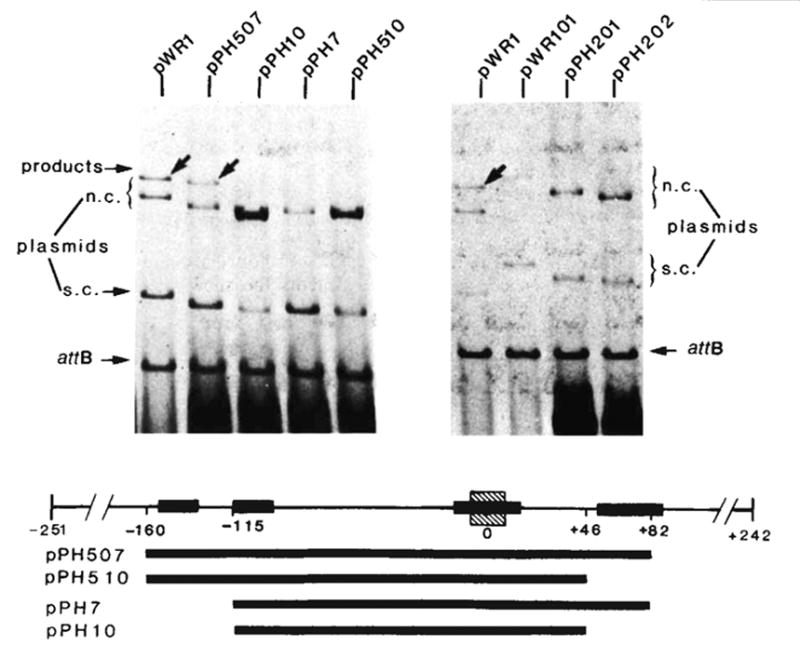
Integrative recombination of the minimal phage att site. The in vitro recombination reaction and gel electrophoresis were carried out as described in Fig. 2 legend. The structure and boundaries of the cloned fragments used in the left panel were confirmed by restriction fragment analysis and by DNA sequencing. The extent of each cloned fragment is indicated by a long bar beneath the map of the att site region. The att sites carried by plasmids in the right panel are: pWR1-phage att site (HindIII-BamHI-492), pWR101-bacterial att site (EcoRI-BamHI-1,600), pPH201-left prophage att site (EcoRI-BamHI-1,300) and pPH202-right prophage att site (HindIII-BamHI-800). Positions on the electropherogram are indicated for the ‘recipient’ linear DNA containing the bacterial att site (attB), the ‘donor’ circular plasmid DNA containing truncated or intact phage att site and migrating as supercoils (s.c.) or nicked circles (n.c.) and the linear products of recombination (arrows). Binding sites for Int protein (■) (see legends to Figs 4 and 5), and the 15-base pair common core sequence (hatched) are indicated on the linear map of the att site region.
All plasmids carrying a P arm equal to or longer than that of pPH54 (−160) recombined efficiently with the bacterial att site, but none of those carrying a shorter arm (−115 or less) did so (Fig. 2 and Table 1). Thus some DNA between positions −160 and −115 is essential for the proper functioning of the P arm in a phage att site.
Fig. 2.
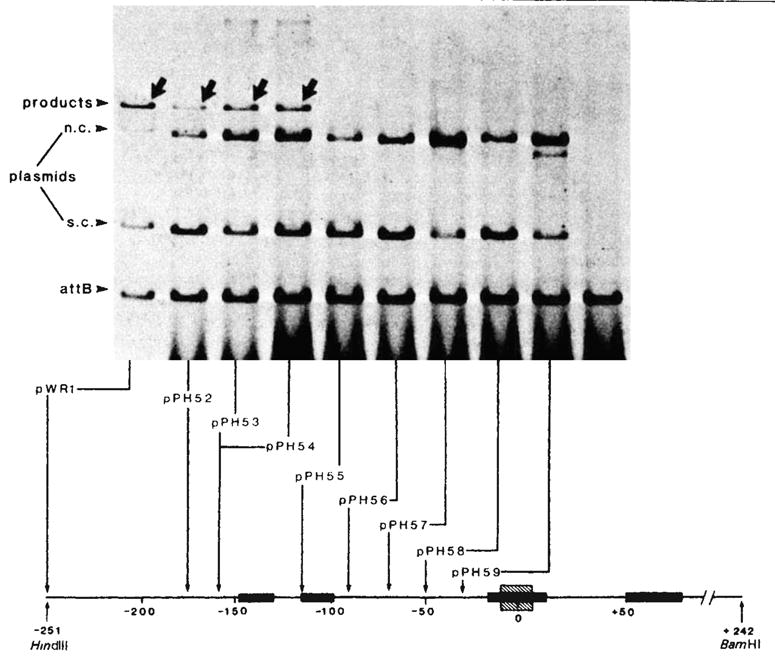
Integrative recombination of plasmids with resected P arms. Each reaction mixture contained ~1 μg supercoiled plasmid DNA with an intact, or P arm-resected, phage att site and 1μg of linear DNA with a bacterial att site (EcoRI-BamHI fragment; 1,600-base pairs) at a molar ratio of 1:3. The recombination was carried out in a 20-μl mixture containing 50 mM Tris-HCl (pH 7.4), 5 mM EDTA, 40 mM KCl, 12.5 mM spermidine, 4 μl of sonicated bacterial extract and 1.5 μl of Int traction 1 (refs 8, 10) After incubation at 25 °C for 40 min, 30 μl of a buffer containing 0.3 M NaCl, 10 mM Tris-HCl (pH 7.4) was added and the samples were extracted with phenol-CHCl3 before being loaded onto a 1 % agarose slab gel and electrophoresed at 35 V for 16 h in E buffer (40 mM Tris, 20 mM Na-acetate, 2 mM EDTA, 18 mM NaCl, adjusted to pH 7.9 with acetic acid). The gel was strained with EtBr(1 μg ml−1) for 15 min and then photographed under UV light. For quantitation the fragment containing the bacterial att site was first end-labelled with 32P by T4 polynucleotide kinase9. After recombination and gel electrophoresis, portions of the gels corresponding to the unreacted linear DNA and the recombination product were excised and counted individually for 32P radioactivities. The percentage of phage att site molecules that recombined with labelled bacterial att site DNA (present in threefold molar excess) was calculated as follows:
The P arm end point of each resected plasmid, as determined by restriction enzyme analysis or by DNA sequencing is shown at the bottom (also see Table 1). All plasmids contain a P′ arm extending to the BamHI site at position + 242. Positions on the electropherogram are indicated for the 32P-labelled linear DNA containing the bacterial att site (attB), the unlabelled circular plasmid DNA containing the intact or resected phage att sites and migrating as supercoils (s.c.) or nicked circles (n.c.), and the linear 32P-labelled products of recombination (arrows). Binding sites for Int protein (■) (see legends to Figs 4 and 5), and the 15-base pair common core sequence (hatched) are indicated on the linear map of the att site region.
Table 1.
Integrative recombination of plasmids with resected P arms or resected P′ arms
| End point
|
% Relative recombination† |
|||
|---|---|---|---|---|
| Plasmid | P arm | P′ arm | Expt 1 | Expt 2 |
| pWR1 | −251 | +242 | 100(41) | 100(30) |
| pPH52* | −174 | +242 | 48 | 100 |
| pPH53 | −160 | +242 | 95 | 75 |
| pPH54 | −160 | +242 | 99 | 106 |
| pPH55 | −115 | +242 | <0.5 | <0.5 |
| pPH56 | −90 | +242 | <0.5 | <0.5 |
| pPH57* | −70 | +242 | <0.5 | <0.5 |
| pPH58* | −50 | +242 | <0.5 | <0.5 |
| pPH59* | −30 | +242 | <0.5 | <0.5 |
| pWR1 | −251 | +242 | 100 (32) | 100 (20) |
| pPH303 | −251 | + 124 | 113 | 80 |
| pPH304* | −251 | +100 | 95 | 75 |
| pPH307 | −251 | +82 | 69 | 90 |
| pPH308 | −251 | +64 | <0.5 | <0.5 |
| pPH310 | −251 | +46 | <0.5 | <0.5 |
The end points of these plasmids were determined by restriction fragment analysis. Ail other end points were determined by DNA sequencing.
See Fig. 2 for quantitation. The results are normalised to 100% for pWR1. The absolute value for pWR1 in each experiment is shown in parentheses.
A family of P′ arm-shortened plasmids generated as outlined in Fig. 1a, had a constant left terminus at −115. Because this did not include all the information required for a functional P arm (see above and Fig. 2), a longer P arm was restored to each plasmid as outlined in Fig. 1b. All att sites with P′ arms equal to or greater than that of pPH307 (+82) were efficient recombination partners, whereas those with shorter P′ arms (+64 or less) failed to recombine efficiently (Table 1). The shortest P′ arm that functions as a phage att site contains essentially all the DNA identified as forming heparin-resistant complexes with purified Int protein9. The longest P′ arm which fails to function as a phage att site contains only half of this sequence.
To test whether the combination of a minimal P arm and a minimal P′ arm is sufficient for full function, a phage att site carrying both minimal arms was constructed. A HindIII-HinfI fragment from −160 to −115 (from pPH54) and a HinfI-BamHI fragment from −115 to +82 (from pPH307) were cloned together into the HindIII-BamHI sites of pBR322. The resultant plasmid, pPH507 (−160 to +82), thus contains all the elements of a minimal P arm and a minimal P′ arm, as defined by the experiments described above.
The function of this plasmid was compared with that of pPH7 (−115 to +82), pPH10 (−115 to +46) and pPH510 (−160 to +46). In recombination with the bacterial att site pPH507 recombined as well (60–100%) as the control plasmid pWR1, whereas the other three gave no detectable recombination (Fig. 3). In these assay conditions, supercoiled plasmid DNAs carrying the bacterial att site or the left or right prophage att sites all failed to recombine efficiently with the linear bacterial att site (Fig. 3). Therefore, the competence of pPH507 must reflect phage att site function (and not prophage or bacterial att site function). We conclude that the sequences delimited by the minimal P and P′ arms (−160 to +82) are both necessary and sufficient for phage att site function.
Sequence and Int binding of minimal phage att site
Most of the DNA comprising the minimal region required for phage att site function was included in our reported sequence extending from −114 to +203 (ref. 13), within which two specific sites of Int interaction were detected9 (see below). One of these 30–35-base pair binding sites has at its centre the 15-base pair common core sequence where the cross-over occurs in recombination. The second site is in the P′ arm (+50 to +86) and the sequence protected by Int extends to the right boundary of (3 to 4 bases beyond) the minimal length P′ arm. Clones lacking part or all of this P′ arm Int binding site failed to function as phage att sites (Fig. 3 and Table 1), indicating a requirement for this Int interaction for phage att site function (see below).
We now report the sequence for the remaining DNA in the P arm that is required for phage att site function as well as sequence extending further to the left (Fig. 4). The sequence from −250 to −200 is 47% A+T, in contrast to a relatively uniform distribution of 62% A+T from −200 to −115. When combined with the previously determined sequence13 this yields an average base composition of 71% A + T for the minimal phage att site.
Fig. 4.
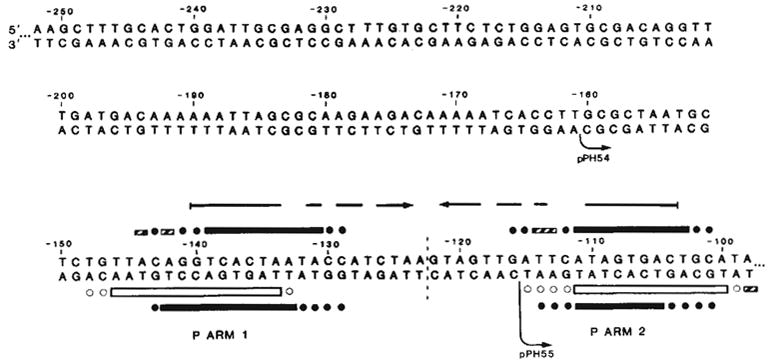
Sequence of the left portion of the P arm and location of Int binding sites P arm 1 and P arm 2. P arm sequence is given negative numbers proceeding leftward from the centre of the common core region (0). We have previously reported the sequence from −114 to +203 (ref. 13). DNA sequence was determined for both strands (with the exception of −252 to −237) by the method of Maxam and Gilbert27 using 5′ termini labelled with 32P by polynucleotide kinase9: top strand sequence from −241 to −90 was obtained from the HindIII end of the HindIII (−251)-Alu(−6) fragment (see ref. 13); top strand sequence from −252 to −241 was determined using the EcoRI site of pBR322 in the plasmid carrying the HindIII(−251)-BamHI(+242) att fragment (pWR1. see text). Complementary sequence from −116 to −237, was obtained from the HinfI end of fragment HinfI(−115)-Mnl 1(recognition site at —231). The left boundaries of the shortest functional P arm (−160, pPH54) and the longest nonfunctional P arm (−115, pPH55) obtained in these experiments are indicated (↪) The locations of Int-binding sites P arm 1 and P arm 2 were determined by the method of DNase I footprinting17 with modifications described previously9 (see also Fig. 5). Int protection of sequences in the top strand from neocarzinostatin28,29 digestion (■) was carried out using two different fragments, each labelled at the HindIII end: HindIII(−251)-Alu(−6), carrying only P arm sequences, and HindIII(−251)-HpaII(+305), carrying the entire minimal phage att site sequence (shown in Fig. 5a). Int protection of the bottom strand from neocarzinostatin (■) or DNase I (□) digestion was carried out on a fragment from plasmid pPH310 containing att sequence from +46 to −251 (HindIII), labelled at the BamHI linker adjacent to position +46. Partially protected bases are designated (
 ). Ambiguities in the precise boundaries of the protected regions are due to the failure of neocarzinostatin (●) or DNase I (○) to cut certain bases in the control digests. An inverted repeat structure (→ ←) which includes sequence from each of the two Int-binding sites plus additional sequence between the two sites is indicated.
). Ambiguities in the precise boundaries of the protected regions are due to the failure of neocarzinostatin (●) or DNase I (○) to cut certain bases in the control digests. An inverted repeat structure (→ ←) which includes sequence from each of the two Int-binding sites plus additional sequence between the two sites is indicated.
The newly sequenced P arm region was examined for additional Int binding sites using the technique of DNase footprinting17. In the presence or absence of Int protein a singly end-labelled restriction fragment was digested with a nonspecific nuclease in conditions where each molecule was cut approximately once. On a DNA sequencing gel the end-labelled fragments form a ladder, with gaps at those positions protected by Int. Using a restriction fragment that extends in both directions beyond the minimal phage att site, we observed that Int protects four regions from digestion by the AT-specific antitumour agent neocarzinostatin (Fig. 5a). Two of these regions correspond to the common core and P′ arm binding sites9. The two others are located in the P arm, within the minimal phage att site; P arm site 1 extends from −148 to −129, and P arm site 2 from −116 to −98 (see also Fig. 4). Good agreement with these results was obtained by monitoring DNase I digestion of the complementary (bottom) strand (Figs 4 and 5b). These two binding sites are most probably the basis for nitrocellulose filter retention of Int complexed with P arm-containing restriction fragments18.
Fig. 5.
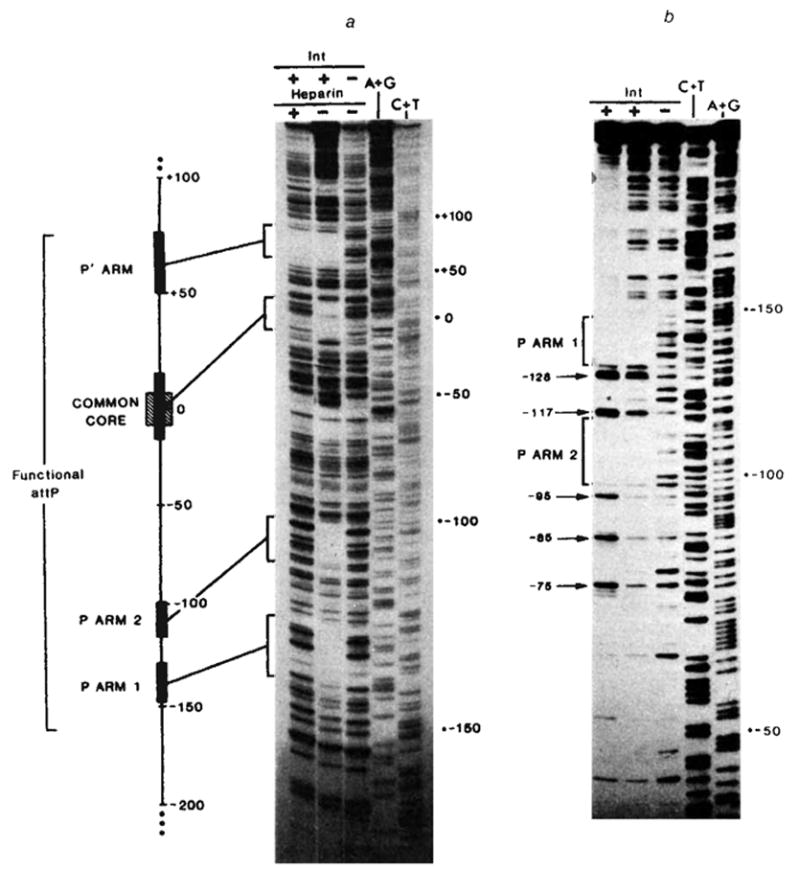
Footprints of Int binding on restriction fragments containing: a, the entire minimal phage att site (using neocarzinostatin); or b, the P arm and common core regions (using DNase I), a, In lanes 1–3 the restriction fragment HindIII(−251)-HpaII(+305), 32P-labeIled at the 5′ end of the top strand (HindIII), was partially digested with neocarzinostatin28,29 in the absence (lane 3) or presence of purified Int protein at 10 μg ml−1 (lane 2) or 20 μgml−1 (lane 1). The Int-DNA complex in lane 1 was challenged with heparin before neocarzinostatin digestion. Sequence markers for this restriction fragment, A+G (lane 4), and C+T (lane 5), were prepared according to the method of Maxam and Gilbert27. Details of these methods and electrophoresis conditions have been described previously9. A linear map of the minimal phage att site region depicts the relative sizes and positions of the four Int protected sequences (■) seen in the neocarzinostatin footprint. Also indicated are the boundaries (−160 and +82) of the smallest functional attP region obtained in these experiments (see Fig. 3) and the 15-base pair common core sequence (hatched), b, In lanes 1–3 a restriction fragment from plasmid pPH310, containing att sequence from +46 to −251(HindIII) (labelled at the 5′ end of the bottom strand at the BamHI linker adjacent to position +46), was partially digested with DNase I in the absence (lane 3) or presence of purified Int protein at 2.5 μg ml−1 (lane 2) or 5 μg ml−1 (lane 1). The footprinting and electrophoresis conditions were as described previously9. The common core region has been run off this gel. Positions of enhanced cutting by DNase I (→) at ~ 10-base pair intervals (−75, −85, −95, −117, −128) are indicated in lane 1.
The importance of P arm site 1 is indicated by the fact that its removal abolished phage att site function (Figs 2,3). However, it cannot be determined from exonuclease resection experiments whether P arm site 2 is also necessary. Base changes or deletions within P arm site 2 will be required to clarify this point.
An additional feature of Int-DNA interaction was visualised by DNase I, but not neocarzinostatin digestion. With Int concentrations higher than the minimum required to observe specific protection there was a series of strongly enhanced bands at 10-base pair intervals (Fig. 5b, lane 1, positions −75, −85, −95, −117 and −128). Similar patterns of enhanced DNase I cutting were also observed to varying extents on other restriction fragments in regions surrounding the common core and P′ arm binding sites (data not shown), but not to the left of P arm site 1 (Fig. 5b). These results are to be distinguished from the possibly related observation that exonuclease digestion produced a 10-base pair pattern within the Int-protected sequence of the P′ arm18.
Structural relations between Int binding sites
The P arm Int binding sites (15–20 base pairs) are each shorter than either the common core or the P′ arm sites, but are comparable in size to the 15-base pair protected region in the attB common core9 (Fig. 6). However, the P arm sequences do not share homology with the common core sequence or with the core-arm juncture sequences9.
Fig. 6.
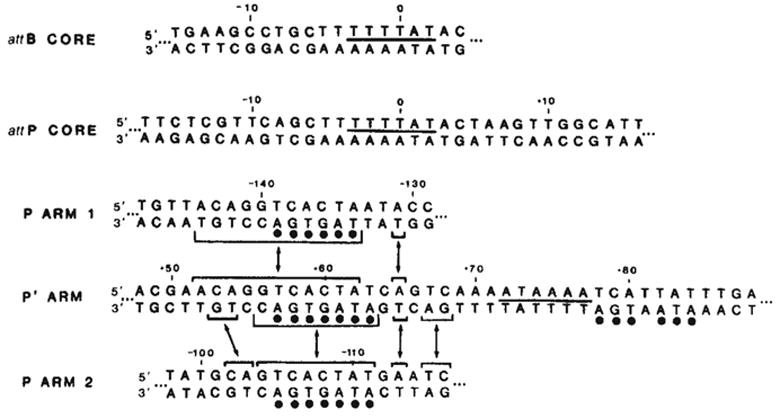
Comparison of Int-protected sequences in the bacterial and phage att sites. The sequence shown for each binding site is the maximum length of DNA protected by Int from either neocarzinostatin or DNase I digestion (see Figs 4 and 5 and ref. 9). The sequence of the P arm 2 site has been inverted (compare Fig. 4) to facilitate comparison of sequence homologies. A 6-base pair homology between the two common core sites and the P′ arm site is indicated by a line between the two strands of sequence (—). Homologies between the left portion of the P′ arm site and each of the two P arm sites are indicated(–) (see text). A 7-base pair sequence in the left half of the P′ arm protected region, found within the sequence shared with the two P arm sites, is also found, with one mismatch, in the right half of the P′ arm site (●).
The common core and P′ arm Int binding sites show very little sequence homology with each other, possibly suggesting their interactions with different domains of the Int protein. In contrast, however, each of the P arm sites shares a considerable uninterrupted homology (11-base pairs in P arm 1 and 8-base pairs in P arm 2) with the left portion of the P′ arm site. The two P arm sites also share this homologous sequence but with an inverted orientation, such that P arm 1 is a direct repeat and P arm 2 is an inverted repeat of the P′ homology. A number of bases in the sequence shared among the P′ and P arm protected regions have been identified as ‘contact’ sites in the Int-DNA interaction, as defined by methylation-protection experiments with the P′ site (data not shown). Included within the left half of the P′ site is the 7-base pair sequence, TCACTAT, which, in addition to being part of the homology with the two P arm sites, is found (with one mismatch) in the right half of the P′ site.
Int binding in the P′ arm has the unique property of resistance to challenge by the polyanion heparin (Fig. 5a and ref. 9). Furthermore, in studies with DNA carrying only the P′ arm site (data not shown) we have established that the heparin-resistant binding is not dependent on the presence of the other binding sites. Thus, Int binding and resistance to heparin challenge are intrinsic features of the P′ arm sequence. As Int binding at sequences homologous to the left half of the P′ arm site (P arm sites 1 and 2) did not show the property of heparin resistance (Fig. 5a), the right half of this site, which is essential for recombination (Table 1), might be responsible for heparin resistance.
We have also used the footprinting assay to examine restriction fragments carrying other combinations of one to three Int binding sites (data not shown). The P arm 1 site was capable of binding Int when present singly. The common core region, when tested using a fragment containing att sequence from −115 to +46, bound Int independently of the P arm 1 and P′ sites. Quantitative measurements to determine whether there are subtle differences in binding affinities as a function of the simultaneous presence of two or more binding sites have not yet been carried out.
Efficient partners for phage att site
From in vivo Int-dependent crosses it was found that the phage att site recombines most efficiently with the bacterial att site and less efficiently with itself or either of the prophage att sites14–16. Similarly, in the in vitro conditions used here the phage att site recombined most efficiently with the bacterial att site. (Both in vivo and in vitro relative recombination values are strongly affected by reaction conditions and it is therefore difficult to make quantitative comparisons between them16,19–21).
In our in vitro experiments the supercoiled phage att site recombined with the linear bacterial att site at a level of approximately 50% and with the linear right prophage att site (attR) at a level of approximately 5–10% (Fig. 7). With the linear left prophage att site (attL) the level of recombination was just above background in this experiment (not visible in the reproduced photograph), and recombination with the linear phage att site was not detectable. The relative recombination efficiencies were affected significantly by factors such as Int concentration, and this aspect of the reaction is being investigated. Nevertheless, we always found that a linear bacterial att site gave very high values and a linear phage att site gave very low values in their respective recombination patterns with supercoiled phage att site. Is this difference due to the presence of unique features that confer competence on the linear bacterial att site, or is it due to certain features on the linear phage att site (and linear attL) that inhibit recombination with a supercoiled phage att site?
Fig. 7.
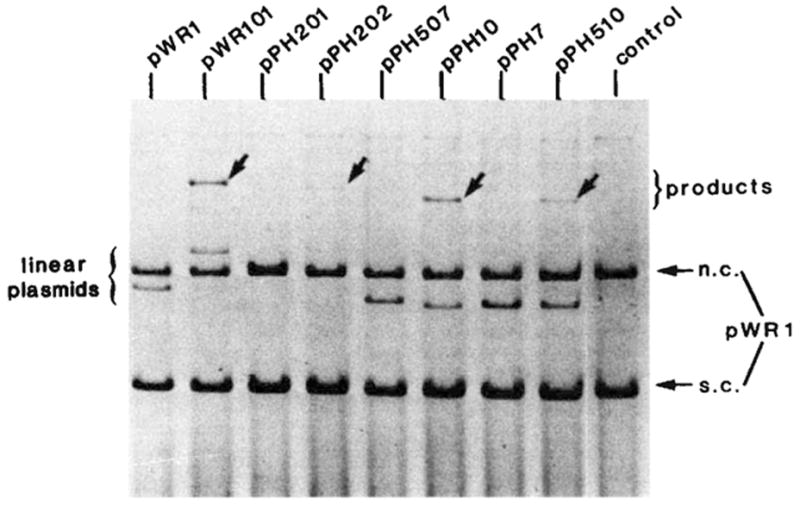
The efficiency of different att sites as linear recombination partners. The in vitro recombination reactions and gel electrophoresis were performed as described in Fig. 2 legend. Each reaction mixture contained supercoiled pWR1 (1.5μg) and linear recipient DNA in a three to one molar ratio. The linear plasmids were generated by PstI digestion of pPH7, 10, 507, 510 or by EcoRI digestion of pWR1, pWR101, pPH201 and pPH202. The origin and boundaries of the restriction fragments carried by each plasmid are described in Fig. 3 legend. Positions are indicated on the electropherogram for DNA containing different ‘recipient’ att sites (linear plasmid), the ‘donor’ circular plasmid DNA (pWR1) containing the phage att site and migrating as supercoils (s.c.) or nicked circles (n.c.) and the linear product of recombination (arrows).
Four plasmids containing the common core and different combinations of P or P′ arm Int binding sites (as described in Fig. 3) were used as linear partners for a supercoiled phage att site (Fig. 7). It is clear from the fact that linear pPH10 recombines efficiently with supercoiled attP that an efficient partner for the phage att site requires neither the P arm 1 nor the P′ binding sites. In this test of integrative recombination the bacterial (art site does not possess any unique features that cannot be provided or mimicked by a phage att site lacking the P arm 1 and P′ Int binding sites (compare lanes 2 and 6, Fig. 7).
The only difference between the two att sites that recombined efficiently with attP and the two that failed to recombine significantly was the presence of the P′ arm Int binding site. Thus, the presence of the P′ arm site on the linear partner was unnecessary, and actually interfered with its ability to recombine with a supercoiled phage att site. The P arm 1 binding site did not contribute as much interference (Fig. 7, compare pPH10 and pPH510). The interfering effect did not seem to be due to titration by the P′ arm binding site as it was not abolished by higher concentrations of Int.
Discussion
Our results reveal the involvement in recombination function of approximately 240-base pairs of DNA adjoining the cross-over locus. Similar experiments, using a different method for obtaining shortened att sites, have been carried out by Mizuuchi and Mizuuchi22. In addition, they found function for an att site in which bacterial DNA is substituted for phage DNA up to position −152 (8-base pairs less than our shortest functional P arm, pPH54 and pPH507). Therefore, it is probable that a clone carrying phage att site DNA from positions −152 to +82 (235-base pairs) would also be functional.
The diversity of the Int binding regions, both in sequence and in size (Fig. 6) is surprising. Considerable sequence homology is found among the three sites in the P and P′ arms, but the sequence in the common core binding region does not share this homology. Sequence differences are almost certainly responsible for the unique heparin-resistant character of binding in the P′ arm site. The lack of homology between the core and the other sites could reflect more than one DNA-binding domain within the Int protein, or a single domain responding differently to the respective sequences. It is clear, however, that Int has at least two distinct capabilities. In addition to the ‘structural’ or DNA-binding properties involving sequences quite distant from the cross-over site of recombination, Int also has a nicking-closing (topoisomerase) activity10. Although no specificity for the phage att site has yet been demonstrated in the topoisomerase assays, it is clear from our results that Int does interact specifically with the normal substrate for this enzymatic activity9.
The nature of the role of the distant Int binding sites in the P and P′ arms is much less apparent. Int binding seems to be an intrinsic feature of the individual DNA sequences since protection was seen for the P arm 1 and P′ sites when present singly and for the common core in the absence of the P arm 1 and P′ sites. The kinetics has not yet been studied, however, and it is possible that there is facilitation of binding at one site by Int bound at another site.
The two size classes of Int binding sites, approximately 30–35-base pairs and 15-base pairs, could reflect differences in the number of bound Int monomers. Similarities between the two P arm sites and each half of the larger P′ site also suggest the possibility of higher order Int complexes. The linear distance between the P arm 1 and P′ Int binding sites, or between the common core and either of these sites, is not incompatible with a three-dimensional structure in which a loop of DNA would bring any of these sites together in the same Int complex. For example, the distance between non-contiguous sequences which are in close proximity in the three-dimensional structure of the nucleosome is estimated to be approximately 80-base pairs23. By analogy with studies on nudeosomes24,25 and DNA gyrase complexes26 the 10-base pair pattern of enhanced cutting around the binding sites (Fig. 5b) is suggestive of DNA wrapping around, or lying on, the surface of a higher order oligomer of Int. This idea is consistent with the electron microscope observation of Int complexes (8–10 monomers) interacting with the phage att site (Bob Yuan, personal communication).
Although their specific functions have not yet been defined, it is clear that the P arm 1 and P′ Int binding sites have critical roles in recombination. Further experiments will be necessary to ascertain whether the P arm 2 binding site is also critical and whether the observed Int-DNA interactions are influenced by host factor proteins or by negatively supercoiled DNA substrate.
Our results suggest a model for integrative recombination that involves two unequal partners. The large size and complexity of the phage att site contrasts with the smaller size and simplicity of its partner. We refer to the phage att site, which must be supercoiled and which provides most of the structural information and specificity for the reaction, as the ‘donor’ molecule. A ‘donor’ att site recombines efficiently with an appropriate ‘recipient’ att site, such as the bacterial att site. Although we have yet to define the minimal requirements for ‘recipient’ att sites, several properties are known. We have shown previously that Int binding to the bacterial att site results in the protection of a short (approximately 15-base pairs) DNA sequence at the left core-arm juncture9. The present results (and additional data, not shown) establish that a ‘recipient’ att site does not require the presence of the P arm 1 or P′ arm binding sites. Furthermore, the absence of the P′ site is necessary for a good ‘recipient’ att site. The sequence homology between the shortened phage att site (pPH10) and the bacterial att site, together with their respective Int protection patterns, suggests that an efficient ‘recipient’ site may not be much larger than the common core and partially homologous core-arm junctures13. Additional experiments will be necessary to confirm, and possibly extend, the applicability of this ‘donor-recipient’ model for integrative recombination.
Acknowledgments
We thank Howard Nash and Yoshiko Kikuchi for providing purified Int protein, Kiyoshi Mizuuchi and Howard Nash for discussions, Julie Smith for help in preparing the manuscript, Eric Johnson for technical assistance and Carl Foeller and Monika Buraczynska for sequencing some of the att plasmids. This work was supported by grants AI13544 from the NIH and 1-543 from the National Foundation, March of Dimes. P.-L.H. is the recipient of an NIH Postdoctoral Research Fellowship (GM07046) and W.R. is a recipient of a Brown University Graduate Fellowship. A.L. is a Faculty Research Associate of the American Cancer Society.
References
- 1.Gottesman ME, Weisberg RA. In: The Bacteriophage Lambda. Hershey AD, editor. Cold Spring Harbor Laboratory; New York: 1971. pp. 113–138. [Google Scholar]
- 2.Nash HA. Curr Topics Microbiol Immun. 1978;78:171–199. doi: 10.1007/978-3-642-66800-5_6. [DOI] [PubMed] [Google Scholar]
- 3.Miller HI, Kikuchi A, Nash HA, Weisberg RA, Friedman DI. Cold Spring Harb Symp quant Biol. 1979;43:1121–1126. doi: 10.1101/sqb.1979.043.01.125. [DOI] [PubMed] [Google Scholar]
- 4.Hoess R, Foeller C, Bidwell K, Landy A. Proc natn Acad Sci USA. doi: 10.1073/pnas.77.5.2482. (in the press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gottesman S, Abremski K. J molec Biol. 138 (in the press) [Google Scholar]
- 6.Kotewicz M, Chung S, Takeda Y, Echols H. Proc natn Acad Sci USA. 1977;74:1511–1515. doi: 10.1073/pnas.74.4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kikuchi Y, Nash HAJ. biol Chem. 1978;253:7149–7157. [PubMed] [Google Scholar]
- 8.Kikuchi Y, Nash HA. Cold Spring Harb Symp quant Biol. 1979;43:1099–1109. doi: 10.1101/sqb.1979.043.01.122. [DOI] [PubMed] [Google Scholar]
- 9.Ross W, Landy A, Kikuchi Y, Nash HA. Cell. 1979;18:297–307. doi: 10.1016/0092-8674(79)90049-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kikuchi A, Nash HA. Proc natn Acad Sci USA. 1979;76:3760–3764. doi: 10.1073/pnas.76.8.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mizuuchi K, Mizuuchi M. Cold Spring Harb Symp quant Biol. 1979;43:1111–1114. doi: 10.1101/sqb.1979.043.01.123. [DOI] [PubMed] [Google Scholar]
- 12.Mizuuchi K, Gellert M, Nash HA. J molec Biol. 1978;121:375–392. doi: 10.1016/0022-2836(78)90370-4. [DOI] [PubMed] [Google Scholar]
- 13.Landy A, Ross W. Science. 1977;197:1147–1160. doi: 10.1126/science.331474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guerrini F. J molec Biol. 1969;46:523–542. doi: 10.1016/0022-2836(69)90194-6. [DOI] [PubMed] [Google Scholar]
- 15.Echols H. J molec Biol. 1970;47:575–583. doi: 10.1016/0022-2836(70)90324-4. [DOI] [PubMed] [Google Scholar]
- 16.Guarneros G, Echols H. Virology. 1973;52:30–38. doi: 10.1016/0042-6822(73)90395-4. [DOI] [PubMed] [Google Scholar]
- 17.Galas DJ, Schmitz A. Nucleic Acids Res. 1978;5:3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davies RW, Schreier PH, Kotewicz ML, Echols H. Nucleic Acids Res. 1979;7:2255–2273. doi: 10.1093/nar/7.8.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pollock TJ, Abremski K. J molec Biol. 1979;131:651–654. doi: 10.1016/0022-2836(79)90013-5. [DOI] [PubMed] [Google Scholar]
- 20.Nash HA. Proc natn Acad Sci USA. 1975;72:1072–1076. doi: 10.1073/pnas.72.3.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gottesman S, Gottesman ME. J molec Biol. 1975;91:489–499. doi: 10.1016/0022-2836(75)90275-2. [DOI] [PubMed] [Google Scholar]
- 22.Mizuuchi M, Mizuuchi K. Proc natn Acad Sci USA. in the press. [Google Scholar]
- 23.Finch JT, et al. Nature. 1977;269:29–36. doi: 10.1038/269029a0. [DOI] [PubMed] [Google Scholar]
- 24.Noll M. Nucleic Acids Res. 1974;1:1573–1578. doi: 10.1093/nar/1.11.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lutter LC. Nucleic Acids Res. 1979;6:41–56. doi: 10.1093/nar/6.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu LF, Wang JC. Cell. 1978;15:979–984. doi: 10.1016/0092-8674(78)90281-7. [DOI] [PubMed] [Google Scholar]
- 27.Maxam A, Gilbert W. Proc natn Acad Sci USA. 1977;74:560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D’Andrea AD, Haseltine W. Proc natn Acad Sci USA. 1978;75:3608–3612. doi: 10.1073/pnas.75.8.3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hatayama T, Goldberg I, Takeshita M, Grollman A. Proc natn Acad Sci USA. 1978;75:3603–3607. doi: 10.1073/pnas.75.8.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]


