Summary
Prions are self-propagating, infectious aggregates of misfolded proteins. The mammalian prion, PrPSc, causes fatal neurodegenerative disorders. Fungi also have prions. While yeast prions depend upon glutamine/asparagine(Q/N)-rich regions, the Podospora anserina HET-s and PrP prion proteins, lack such sequences. Nonetheless, we show that the HET-s prion domain fused to GFP propagates as a prion in yeast. Analogously to native yeast prions: transient overexpression of the HET-s fusion induces ring-like aggregates that propagate in daughter cells as cytoplasmically-inherited, detergent-resistant dot aggregates. Efficient dot propagation, but not ring formation, is dependent upon the Hsp104 chaperone. The yeast prion [PIN+] enhances HET-s ring formation, suggesting that prions with and without Q/N-rich regions interact. Finally, HET-s aggregates propagated in yeast are infectious when introduced into Podospora. To our knowledge, this is the first report of prion propagation in a truly foreign host. Since yeast can host non Q/N-rich prions, such native yeast prions may exist.
Introduction
Prions are misfolded, self-propagating, infectious proteins. Several neurodegenerative disorders such as “mad cow disease”, Scrapie and Creutzfeldt-Jakob disease are caused by the conversion of a normal cellular protein, PrPc, into a β-sheet rich infectious prion isoform, PrPsc (Prusiner, 1998). The discovery that prions also exist in fungi has greatly facilitated the unraveling of the prion mystery (Wickner et al., 2004)
In Saccharomyces cerevisiae, three non-Mendelian elements have been shown to be prions: [URE3], the prion isoform of Ure2p which is a nitrogen catabolite repression regulator (Wickner, 1994); [PSI+], the prion isoform of Sup35p, a translational termination factor (Wickner et al., 1995) and [PIN+], the prion form of Rnq1p, a protein of unknown function (Sondheimer and Lindquist, 2000; Derkatch et al., 2001).
In the filamentous fungus Podospora anserina, the het-s locus has two antaganostic alleles: het-s and het-S (Rizet, 1952). HET-s, the protein product of the het-s allele can exist either as the non-prion isoform, [Het-s*] or as the infectious prion form, [Het-s] (Coustou et al., 1997). In contrast, the protein encoded by the het-S allele, HET-S, never folds in a prion form. Fusion of [Het-s] and het-S strains results in cell death, i.e. heterokaryon incompatibility for somatic cells (Rizet, 1952; Saupe, 2000) and spore killing for the sexual cycle (Dalstra et al., 2005). In contrast, interactions between [Het-s*] and het-S strains are neutral.
Like the mammalian prion protein PrP, fungal prions form insoluble amyloid-like aggregates in vivo and in vitro (Speransky et al., 2001; Kimura et al., 2003; see Wickner et al., 2004 for review). Regions of prion proteins: Sup35p (Ter-Avanesyan et al., 1994; Derkatch et al. 1996; King et al., 1997), Ure2p (Masison and Wickner, 1995; Masison et al., 1997; Taylor et al., 1999); Rnq1p (Sondheimer and Lindquist, 2000; Vitrenko et al., 2007) and HET-s (residues 218–289) (Balguerie et al., 2003), defined as prion domains (PrD), are essential and sufficient for prion propagation in vivo, and the formation of fibers in vitro. In vivo infectivity caused by in vitro formed fibers of purified full length or prion domain fragments of HET-s (Maddelein et al., 2002), Sup35p (King and Diaz-Avalos, 2004; Tanaka et al., 2004), Ure2p (Brachmann et al., 2005) and Rnq1p (Patel and Liebman, 2007) has definitively proven the ‘protein-only’ hypothesis for prion propagation.
Prion domain sequences facilitate both self-aggregation and breakage of aggregates into smaller infective “seeds” (Borchsenius et al., 2001; Osherovich et al., 2004). The PrD’s of all three known yeast prions have (Q/N)-rich regions, which are apparently essential for prion protein aggregation (DePace et al., 1998; Osherovich et al., 2004; Ross et al., 2005). The PrD’s of HET-s and PrP aggregate and propagate via another mechanism since they are not Q/N rich. The PrD’s of native Sup35p (Serio et al., 2000), Ure2p (Thual et al., 2001; Pierce et al., 2005), HET-s (Balguerie et al., 2003) and PrP (Viles et al., 2001) are flexible and poorly structured. Structural data of all prions suggest a cross-β conformation of the prion isoforms (Baxa et al., 2006). So far, the positions of the β-strand structural elements have only been precisely defined for the HET-s PrD. The four HET-s β-strands are proposed to fold into a β-roll composed of two stacked β-strand-turn-β-strand motifs (Ritter et al., 2005). Recent STEM and electron diffraction data further support this cross-β, β-roll model (Sen et al., 2006). This is unlike data for the yeast prion fibrils of Sup35p, which support an in-register parallel β-sheet structure (Shewmaker et al., 2006). The HET-s PrD differs markedly from yeast PrDs not only because it is not Q/N-rich but also because it is rich in charged residues which are sparse in the Sup35p and Ure2p PrDs.
Prion aggregation and propagation in vivo requires additional cellular factors. Yeast lacking the chaperone Hsp104 are unable to propagate any of the known yeast prions (Chernoff et al., 1995; Derkatch et al., 1997; Moriyama et al., 2000; Sondheimer and Lindquist, 2000). Hsp104 appears to disaggregate and shear high molecular weight aggregates into propagons or seeds, which are required for efficient transmission to daughter cells (Paushkin et al. 1996; Ferreira et al., 2001; Jung and Masison, 2001; Wegrzyn et al., 2001; Kryndushkin et al., 2003; Shorter and Lindquist, 2006). Interestingly, Hsp104 is needed only for prion propagation, but not for the initial aggregation of yeast prion proteins (Osherovich and Weissman, 2001).
While the mechanisms responsible for the spontaneous in vivo appearance of prions in the absence of infection are unknown, it is clear that in yeast and fungi the de novo appearance of prions is greatly enhanced by overproduction of the corresponding normal protein or prion domain (Chernoff et al., 1993; Wickner, 1994; Masison and Wickner, 1995; Derkatch et al., 1996; Coustou et al., 1997). Presumably the increased number of molecules improves the chance that some will fold in the infectious form and/or their higher concentration allows them to interact with each other more frequently, promoting the formation of a prion seed. In yeast, this process is facilitated by the presence of heterologous pre-existing prions (Derkatch and Liebman, 2007).
Interestingly, both positive and negative interactions have been identified amongst the Q/N-rich yeast prions (Derkatch et al., 1997; Bradley et al., 2002; Schwimmer and Masison, 2002; Bradley and Liebman, 2003). Also, overexpression of other cellular proteins with Q/N-rich domains, and non-prion polyQ amyloid aggregates associated with Huntington’s Diseases, facilitates the de novo appearance of yeast prions (Osherovich and Weissman, 2001; Derkatch et al., 2001, 2004). In contrast, two non-polyQ amyloid proteins, transthyretin and α-synuclein, did not promote [PSI+] induction (Derkatch et al., 2004). These results suggest that amyloid aggregates with similar domains can cross-seed each other in vivo.
It is not known how relevant information on Q/N-rich yeast prions will be for non Q/N-rich prion proteins such as PrPSc. While PrP forms aggregates in yeast (Ma and Lindquist, 1999), these aggregates have not been shown to be infectious. Here, we investigated the non Q/N-rich HET-s prion protein of Podospora anserina to gain insight into the biology of non Q/N-rich prions in yeast. The HET-s prion domain fused to GFP, HET-s(PrD)-GFP, has been previously shown to form infectious aggregates in Podospora anserina (Balguerie et al., 2003). Here we show that in yeast this same fusion protein can exist either in a non-aggregated, non-prion form, [het-s]y or an infectious aggregated prion form, [Het-s]y. Extracts of [Het-s]y but not [het-s]y yeast efficiently infect Podospora anserina with the [Het-s] prion. We also show that Hsp104 is required for efficient [Het-s]y propagation and that the Q/N-rich yeast prion, [PIN+], enhances the de novo appearance of [Het-s]y.
RESULTS
HET-s forms heritable aggregates in yeast
To ask if the PrD of HET-s could form aggregates in yeast, a [pin−][psi−] strain was transformed with a plasmid bearing a fusion of the HET-s(PrD) and GFP under a GAL1 promoter. Initially, by growing the transformants on 0.05% galactose (gal) media selective for the plasmid we induced a low level of expression of the HET-s(PrD)-GFP fusion which was maintained in exponential or stationary phase (Figure 1A). When analyzed under a fluorescent microscope, most of the cells showed dull diffuse fluorescence (Figure 1B) and very rare cells (less than 0.5%) contained ring, dot or line shaped aggregates. Since overproduction of the normal form of a prion protein is known to dramatically increase the de novo of appearance of that prion, we overexpressed HET-s(PrD)-GFP to see if that would promote it to form a prion. Expression of the fusion increased considerably when the transformants were grown on 2% gal compared to 0.05% gal in both exponential and stationary phase (Figure 1A). Also, we observed frequent huge ring-like aggregates of HET-s(PrD)-GFP like those seen when Sup35(PrD)-GFP was overexpressed in a [PIN+] strain (Zhou et al., 2001). Similar to Sup35(PrD)-GFP rings, HET-s(PrD)-GFP formed smooth, branched, twisted, theta-like and internal rings (Figure 1C-H). We also observed very few cells with dots (Figure 1I). In contrast, a T266P mutant of HET-s(PrD)-GFP that was shown earlier to cause loss of the prion in Podospora anserina due to a proline insertion in β-strand β3 (Coustou et al., 1999; Ritter et al., 2005) did not show any aggregation in yeast (Figure 1J) although the expression level of the HET-s(PrD)-GFP was not affected by the mutation (Figure S1A).
Figure 1. Aggregates of HET-s (PrD)-GFP in yeast.
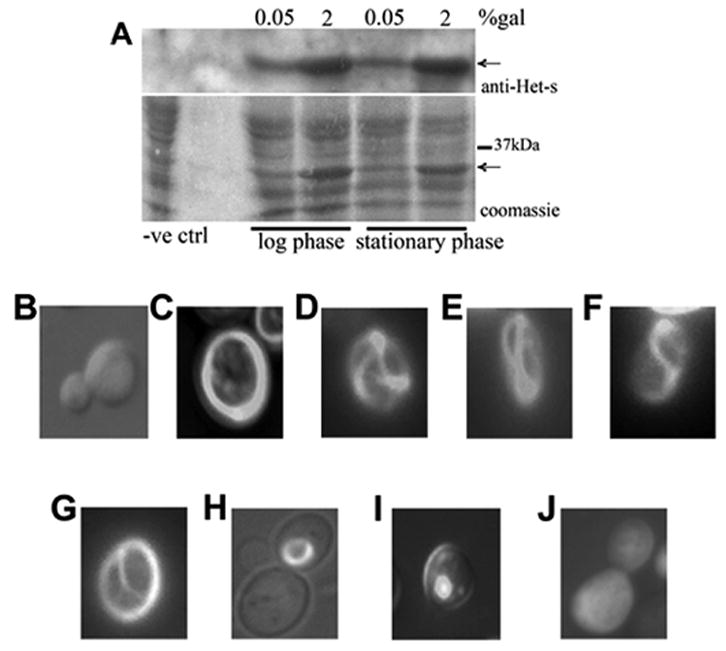
The HET-s(PrD)-GFP fusion construct was expressed in a [pin−][psi−] strain. (A) The expression levels of Het-s(PrD)-GFP fusion on 0.05% gal and 2% gal media in log (OD600nm ~0.3) and stationary phase (OD600nm ~1.2). Arrows indicate the Het-s(PrD)-GFP fusion. (B-J) Cells were examined under a fluorescent microscope after 48 hours of growth at 30°C. Diffuse fluorescence (B) was observed when cells were grown on 0.05% gal where the fusion is expressed at a ‘low level’. Aggregates of different shapes were formed when cells were grown on 2% gal, which induced ‘high expression’ of the fusion. Rings were (C) smooth, (D) branched, (E-F) twisted, (G) theta-like, or (H) internal. In addition, there were dots (I), and the T266P mutant of PrD-GFP showed no aggregates (J).
Since the presence of the Q/N-rich yeast prion [PIN+] enhances the de novo induction of the heterologous Q/N-rich yeast prions [PSI+] (Derkatch et al., 2001) and [URE3] (Bradley et al., 2002) as well as polyQ aggregation in yeast (Osherovich and Weissman, 2001), we asked if the Q/N-rich [PIN+] prion could promote aggregation of the non Q/N-rich HET-s prion protein. Indeed, the presence of [PIN+] caused a more than two fold increase in the percentage of cells with HET-s rings and dots (Figure 2A) although the expression level of HET-s(PrD)-GFP was not affected (Figure S1A).
Figure 2. Quantitative measure of HET-s(PrD)-GFP aggregation in yeast.
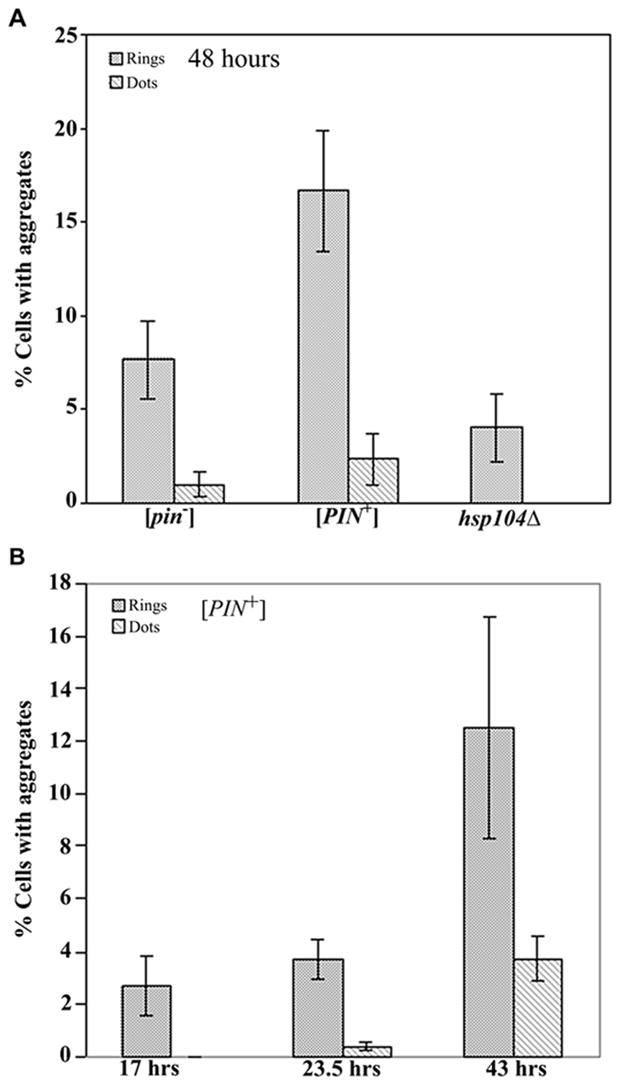
Yeast [psi−] strains containing the HET-s(PrD)-GFP plasmid were induced to form aggregates on 2% gal. The percentage of cells with aggregates in three (A), and four (B), independent transformants was determined by examining 300–600 cells for each. Error bars indicate standard deviations. (A) The induction of Het-s(PrD)-GFP rings and dots after 48 hours of growth in 2% gal in strains with the indicated genotypes. (B) The appearance of rings and dots when a [PIN+] strain is grown in 2% gal for the indicated periods of time.
Rings first appear after 3–4 generations (10–12 hours) of growth in 2% gal. The dot-like HET-s aggregates appear 3–5 generations after the appearance of the rings (Figure 2B). When cells with HET-s rings were diluted and re-grown in 2% gal, the number of cells with dots increased in addition to rings (Figure 3A). This suggests, by analogy with [PSI+] (Zhou et al., 2001), that rings are indicative of newly appearing prion intermediates and that cells with rings bud off daughter cells with dots, some of which can stably propagate the prion.
Figure 3. HET-s(PrD)-GFP ring-like aggregates give rise to dots.
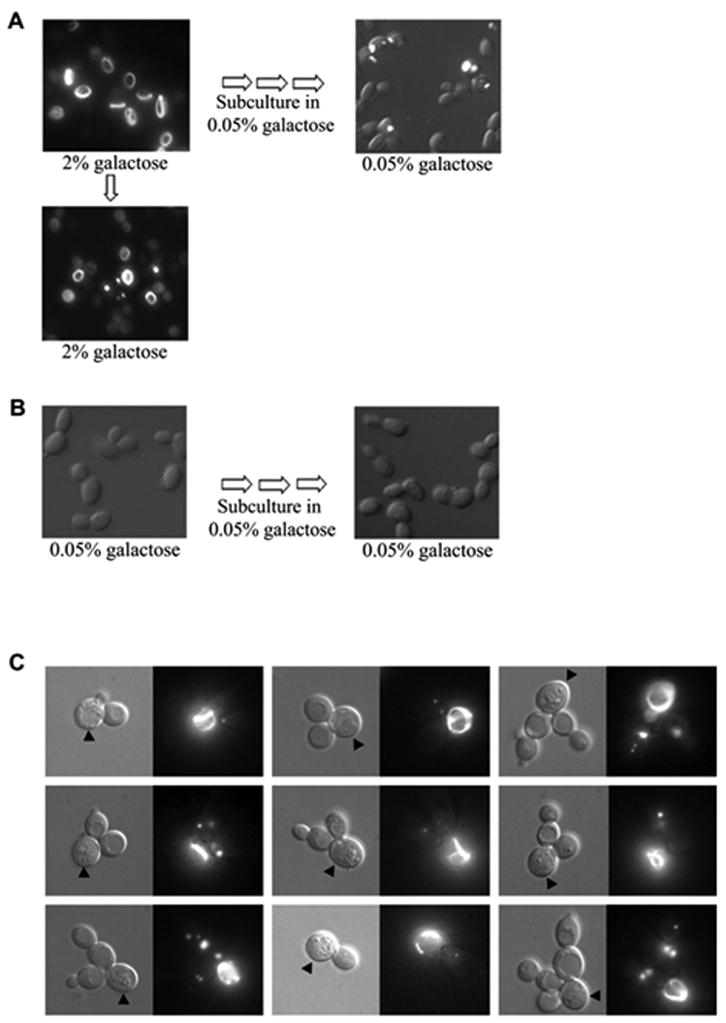
A. [PIN+][psi−] yeast with HET-s(PrD)-GFP was induced to form rings on 2% gal (left panel) for 48 hours. The cells were then passed again to 2% gal (bottom panel) or 3-times on 0.05% gal (right panel). B. As a control, the cells were grown only on 0.05% gal. C. A liquid culture in 2% gal was briefly sonicated and plated as isolated cells on solid 2% gal. Microcolonies of 2 to 8-cells were observed after 12 hours. Cells with dots mainly correspond to daughters or granddaughters of cells with rings. Since rings can sometimes only be visualized in their entirety by varying the focal plane, cells with rings are marked with an arrowhead for clarity.
To ask if HET-s(PrD)-GFP aggregates can be propagated in the absence of overexpression, cells with ring-like aggregates formed by induction on 2% gal were serially passed on low expression medium (0.05% gal). Ring aggregates were no longer present. Instead, about 15% of the cells had dot aggregates while the remainder had diffuse fluorescence (Figure 3A, see also Figure 5A). In contrast, there was little or no aggregation in control cells serially passed on 0.05% gal without any prior induction (Figure 3B). Consistent with the idea that cells with rings give rise to cells with aggregates that propagate as dots, mother-cells containing rings and daughter cells containing dots were observed in microcolonies (Figure 3C).
Figure 5. Hsp104 is required for the maintenance of HET-s(PrD)-GFP aggregates.
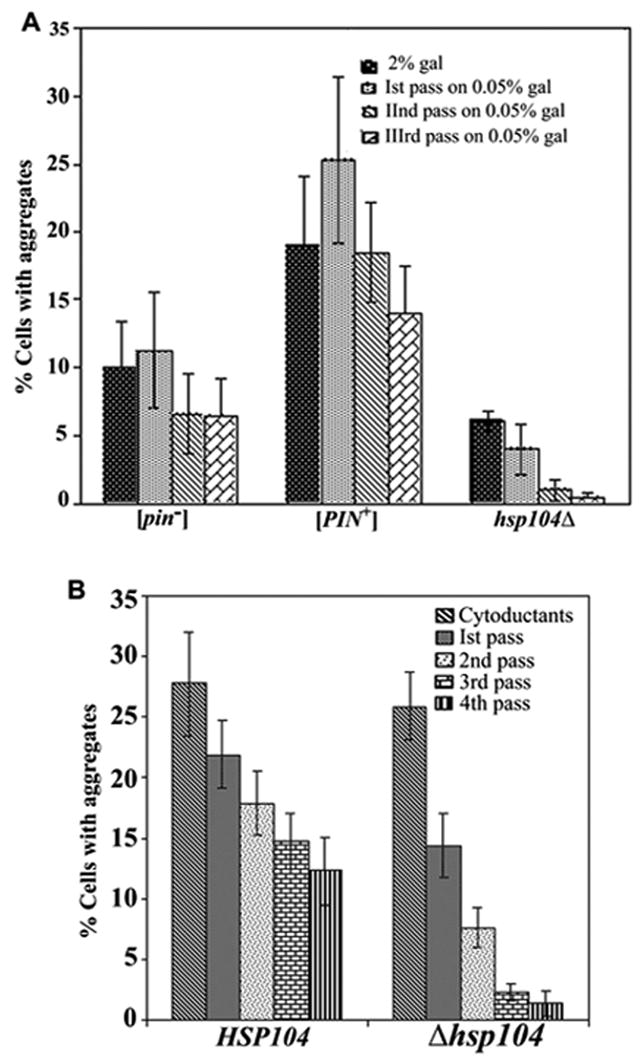
A. The maintenance of HET-s aggregates in the indicated strains was monitored on three serial passes of the cells on 0.05% gal after induction of HET-s(PrD) on 2% gal. The percentage of cells with aggregates was determined after 36–40 hours of growth on inducing media and subsequent passes on 0.05% gal in four transformants (300–500 cells each) for each strain. B. HSP104 donor strain with Het-s dots was mated with both HSP104 and hsp104Δ strains. The cytoductants obtained on SG-Trp+0.05% gal+Cyh were passed serially four times on SR-Trp+0.05% gal. The percentage of Het-s dots in eight independent cytoductants and subsequent passes were determined after 36 hours of growth on the respective media.
In order to obtain separate cultures with HET-s(PrD)-GFP propagated in the aggregated vs. non-aggregated state, we plated a liquid 0.05% gal culture (previously grown in 2% gal) containing 7% of cells with dots on solid 0.05% gal medium and after 5 days screened 205 individual colonies for the presence of dot-containing cells. We identified 9 colonies that contained over 95% of the cells with dots (the remaining colonies had less than 0.5% cells with dots) (Figure S2). When streaked or dispersed in water and re-plated on solid or liquid 0.05% gal medium, these cultures lost dots at rates per generation of less than 1% and 4%, respectively (Tables S1). Colonies without dots never regained dots unless induced on 2% gal. We refer to colonies containing HET-s(PrD)-GFP in the propagating aggregated state as [Het-s]y and colonies with the non-aggregated HET-s(PrD)-GFP as [het-s]y. The difference between these cultures was not caused by a difference in the expression level of HET-s(PrD)-GFP since no increase in expression was found in a [Het-s]y culture propagated on 0.05% gal vs. a [het-s]y culture (Figure S1B).
When 2% gal cultures with newly induced rings were plated on 0.05% gal and individual colonies were screened for dots, most had no or only a few aggregates, but 12 out of 150 had an average of 30% of cells showing HET-s dots. Two such colonies were again plated on 0.05% gal and we obtained 9 out of 150 colonies with an average of 60% of the cells showing HET-s dots.
HET-s aggregates are cytoducible in yeast
One of the distinguishing characteristics of yeast prions is that they are transferred with cytoplasmic material. Cytoplasmic transfer can be accomplished by cytoduction, which involves mating donor and recipient strains in the presence of a mutation that inhibits nuclear fusion. Daughter cells with the recipient haploid nucleus and a mixture of the parental cytoplasms (cytoductants) can then be obtained from this dikaryon (Conde and Fink, 1976).
To test if HET-s(PrD)-GFP aggregates are cytoducible, donors showing HET-s dots vs. diffuse fluorescence were used. We chose four colony purified [Het-s]y donors for cytoduction (~60% of cells in these colonies showed HET-s dots). As a control, we took three [het-s]y colonies from the same 0.05% plates showing essentially diffuse fluorescence (0.2% of cells showed HET-s dots). The recipient strains also expressed HET-s(PrD)-GFP but were grown only on 0.05% gal and lacked any HET-s dots. Generally, we characterized cytoductants obtained in a patch when the mating mixtures were replicaplated to 0.05% gal Cyh media. Dots present in the donors were efficiently cytoduced into and propagated in the recipients as expected for a prion (Figure 4). However, when [Het-s]y was cytoduced into recipients carrying the pT266P-GFP plasmid, [Het-s]y dots propagated with a very low efficiency (<2%) (Figure 4). This confirmed that the T266P mutation prevents the efficient propagation of [Het-s]y. When [het-s]y donor cultures with diffuse fluorescence were used, only rare cytoductants (less than 0.2%) had dots (Figure 4). We also crossed donor colonies with 90% and 0% dots to the recipient and instead of examining a patch of cytoductants we spread the mating mixtures for single colonies on media selective for cytoductants. While none of the 100 cytoductants examined from the 0% donor had dots, 70% (72/101) of the cytoductant colonies obtained from the 90% donor contained cells with dots.
Figure 4. HET-s(PrD)-GFP aggregates formed in yeast are cytoducible.
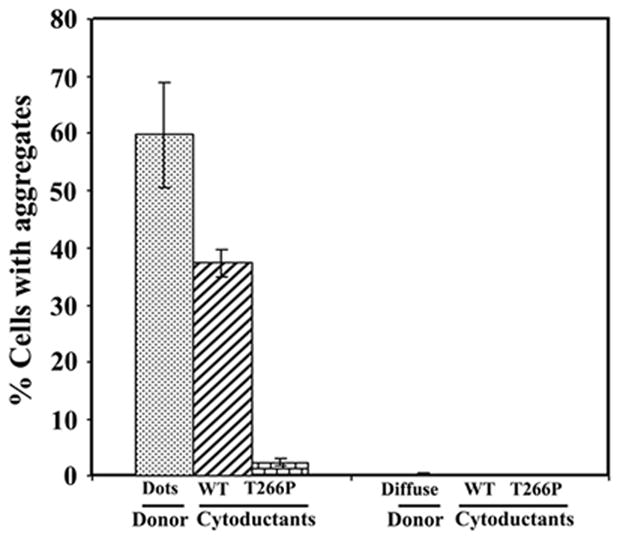
[Het-s]y or [het-s]y [PIN+] HSP104 donor strains grown on 0.05% gal after induction on 2% gal and with, respectively Het-s aggregates (dots) or diffuse fluorescence, were cytoduced into a [rho−] [pin−] [het-s]y HSP104 cyh2R recipient carrying either pHET-s(PrD)-GFP (WT) or mutant pT266P-GFP plasmids. Recipients and cytoductants were grown only on 0.05% gal. Data is the average of five and three independent cytoductions (300–500 cells for each) respectively for [Het-s]y and [het-s]y donor strains.
Effect of an HSP104 deletion on [Het-s]y propagation
High expression of the HET-s(PrD)-GFP fusion in a hsp104Δ strain caused the appearance of ring-like aggregates that were indistinguishable from the rings formed in wild type HSP104 strains (data not shown). However, there was an approximately 4- and 2-fold reduction in the frequency of ring appearance in the HSP104 deletion strain relative to, respectively, isogenic [PIN+] and [pin−] HSP104 strains (Figure 2A). Clearly, the HSP104 deletion has an additional effect beyond the loss of the [PIN+] prion (Derkatch et al., 1997; Sondheimer and Lindquist, 2000). Furthermore, unlike either [PIN+] or [pin−] HSP104 strains, hardly any hsp104Δ cells showed dots after 48 hours of growth on inducing media (Figure 2A). Furthermore, when hsp104Δ transformants with ring aggregates previously induced on 2% gal were serially passed on 0.05% gal, cells with rings gave rise to cells with dots which were lost after two or three passes on 0.05% gal (Figure 5A). In contrast, [Het-s]y dots were more faithfully maintained in both [PIN+] and [pin−] HSP104 strains (Figures 3A and 5A). Similar results were obtained when Hsp104 was inactivated by expressing a dominant negative allele of HSP104 (data not shown). Thus while overexpression of HET-s(PrD)-GFP in a hsp104Δ strain caused the de novo aggregation of HET-s(PrD)-GFP, Hsp104 is needed to efficiently propagate the [Het-s]y prion.
Dots from four colony purified [Het-s]y HSP104 donors (containing ~ 60% of cells with dots) were successfully cytoduced into a hsp104Δ recipient strain. However with each pass on 0.05% gal, the fraction of cells with dots decreased considerably in the hsp104Δ, compared to the HSP104, strain (Figure 5B). Interestingly, the low percentage of HET-s dots obtained in a hsp104Δ strain following the first pass of cells (Figure 5A) with rings to low galactose could be cytoduced into a [PIN+] HSP104 recipient where they propagated as [Het-s]y (data not shown).
HET-s aggregates are resistant to detergent
The [PSI+] and [PIN+] prions are not dissolved into monomers when treated with SDS in the absence of boiling, but break into SDS-stable subparticles that can be resolved on agarose gels (Kryndushkin et al., 2003; Bagriantsev and Liebman, 2004). While unheated 2% SDS did break HET-s(PrD)-GFP aggregates into monomers (data not shown), unheated 0.5% SDS, 1% SDS (data not shown) or 2% sarkosyl (an anionic detergent like SDS) (Figure 6) gave rise to subparticles, and the sarkosyl subparticles were even stable at 37°C or 60°C (Figure S3). Interestingly, the HET-s rings obtained in the hsp104Δ strain, produced much larger sarkosyl resistant subparticles than those in either [PIN+] or [pin−] HSP104 strains (Figure 6A). Also, the sarkosyl resistant subparticles formed upon induction in 2% gal in [PIN+] and [pin−] (Figure 6A) yeast differ in size although the level of HET-s(PrD)-GFP is not affected by [PIN+] (Figure S1A). The fluorescent dots obtained after two passes in low galactose medium, also produced sarkosyl resistant subparticles, however, the sizes of the subparticles made in [PIN+] and [pin−] cells could no longer be reliably distinguished (Figure 6B).
Figure 6. HET-s(PrD)-GFP aggregates formed in yeast are sarkosyl resistant.
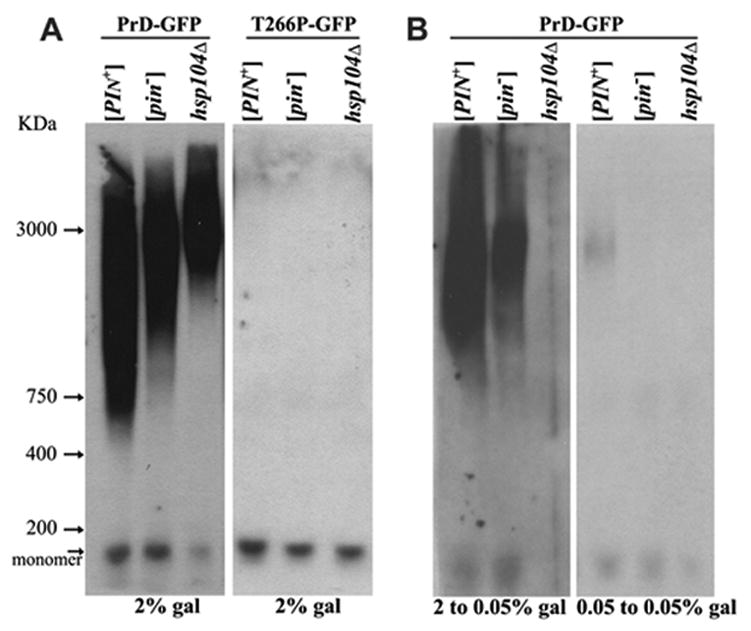
Precleared yeast lysates normalized for total protein and treated with 2% sarkosyl at room temperature for 10 min were resolved on 1.5% agarose gels and probed with anti-GFP antibody. Stained chicken pectoralis muscle extract (Kim and Keller, 2002) provided molecular weight markers. The position of Het-s(PrD)-GFP monomer is shown, although the monomer does not reliably transfer on agarose gels. Indeed, we established that neither [PIN+], hsp104Δ or T266P altered the total level of Het-s(PrD)-GFP in cells by probing blots of acrylamide gels with anti-GFP antibody (Figure S1A) A. Left panel: The HET-s aggregates formed in the indicated strains following growth in 2% gal were broken into sarkosyl resistant subparticles. Right panel represents strains expressing the T266P-GFP fusion in 2% gal. B. Left panel: HSP104 cells with HET-s(PrD)-GFP aggregates obtained in 2% gal even after two serial passes in 0.05% gal gave rise to sarkosyl resistant subparticles. Subparticles were lost in hsp104Δ. Right panel represents strains after two serial passes in 0.05% gal without prior growth in 2% gal.
No subparticles were found in cells expressing the same levels of a control non-aggregating T266P mutant of HET-s(PrD)-GFP (Figures 6A and S1A). Also, in agreement with our in vivo observations (Figure 3B), we obtained very little or no sarkosyl resistant particles from HET-s(PrD)-GFP cells that were grown only on 0.05% gal without any prior induction on 2% gal (Figure 6B) or when hsp104Δ cells from inducing media were passed twice on 0.05% gal (Figure 6B).
[Het-s]y is infectious in Podospora anserina
In order to determine whether the [Het-s]y yeast aggregates are infectious for Podospora anserina, we performed protein transfection experiments with crude cell extracts of colony purified [Het-s]y and [het-s]y cells. We selected one [HET-s]y colony with 95% of the cells having dots and one [het-s]y colony with diffuse fluorescence from the same 0.05% gal plate. Protein transfection of P. anserina [Het-s*] mycelium was then carried out with the mechanical shearing method (Benkemoun et al., 2006). Crude extracts from the [Het-s]y culture induced the appearance of the [Het-s] prion while no significant infectivity was detected with extracts from the [het-s]y culture (Table 1). Thus the [Het-s]y aggregates propagating in yeast display [Het-s]-prion infectivity when introduced into Podospora anserina cells. Likewise, cell extracts from cultures with ring aggregates are also infectious (data not shown).
Table 1.
Ability of S. cerevisiae protein extracts to infect Podospora anserina
| Yeast Strain | Exp. 1 | Exp. 2 | Exp. 3 | Total | Mean±SD |
|---|---|---|---|---|---|
| [Het-s]y | 25/36 | 19/48 | 25/36 | 69/120 | 59,4%±17,8% |
| [het-s]y | 0/36 | 1/48 | 0/36 | 1/120 | 0.6%±1.2% |
| no extract | 0/12 | 0/48 | 0/36 | 0/96 | 0 |
Extracts from the indicated yeast strains were used to transfect [Het-s*] Podospora anserina with the [Het-s] prion. The number of successful infections (numerators) out of the total transfection attempts (denominators) is shown.
Discussion
[Het-s] is a prion from Podospora anserina. While the Q/N rich PrD’s of native yeast prions are indispensable for prion propagation, [Het-s] lacks a Q/N-rich region and in contrast to yeast PrDs is rich in charged residues. Here, we show that, despite these differences, the prion domain of HET-s fused to GFP propagates in yeast as the [Het-s]y prion. As the [Het-s] phenotype of heterokaryon incompatibility cannot be detected in yeast, we used the aggregation state of the HET-s fusion to score for the [Het-s]y prion state. We established two distinct HET-s(PrD)-GFP states in yeast grown under identical conditions: propagating aggregates, [Het-s]y; and non-aggregated, [het-s]y. Furthermore, we showed that cytoplasm from [Het-s]y, but not [het-s]y, cells are infectious in yeast as well as in Podospora. Since the aggregates are resistant to 1% SDS and 2% sarkosyl treatment and the T266P mutation which inhibits amyloid formation by acting as a β-strand breaker (Ritter et al., 2005) abolishes aggregate formation in both yeast and Podospora, it appears that the aggregates are amyloid-like. In contrast to [PIN+] and [PSI+] aggregates, but like recombinant HET-s(PrD) prion amyloids formed in vitro (R. S. and S.J.S., unpublished data), HET-s(PrD)-GFP aggregates formed in yeast are not resistant to 2% SDS.
As the known yeast prions are rich in Q/N sequences, approaches to search for new prions have focused on exploring candidates with similar domains. Our work demonstrates that yeast cells can form and propagate non Q/N-rich prions, so it now makes sense to search for such native prions in yeast.
The inheritance of prions requires both aggregate formation and propagation. While many proteins aggregate, these aggregates are not infectious and thus are not prions. We have shown that in yeast the non Q/N-rich HET-s fusion, which does not aggregate when expressed at low levels, forms ring-like aggregates when highly expressed. Upon mitotic growth, cells with rings were replaced with cells with dots, which continued to propagate even after they were serially passed many times at a low expression level. The finding that when cells with newly induced ring-aggregates are grown on 0.05% gal only a small fraction of the cells stably propagate dots in mitotic progeny is reminiscent of the findings that newly appearing amyloid-like aggregates of yeast prion proteins are often unstable and only a fraction of the cells with such aggregates can propagate a stable prion (Chernoff et al., 2000; Derkatch et al., 2000; Kushnirov et al., 2000; Li and Lindquist, 2000; Santoso et al., 2000; Zhou et al., 2001; Salnikova et al., 2005). The prion nature of the dot-like HET-s fusion aggregates was confirmed by the ability of [Het-s]y, but not [het-s]y, yeast lysates to infect Podospora anserina.
Yeast can now be used to identify interactions with other cellular factors involved in the propagation of [Het-s]y. Similar to Sup35 (Osherovich and Weissman, 2001), HET-s aggregates can be formed in the absence of Hsp104, but appear to need Hsp104 to efficiently shear the aggregates into smaller fragments that can be transmitted to the daughter cells. However, in the absence of Hsp104, HET-s ring aggregates do give rise to dots. Furthermore, dots obtained in HSP104, and cytoduced into hsp104Δ strains are maintained for sometime before being lost. It appears that HET-s(PrD)-GFP aggregates can break in yeast, albeit inefficiently, even in the absence of Hsp104.
Our observation that deletion of HSP104 causes a slight decrease, relative to HSP104 [pin−] strains, in the induction of [Het-s]y (Figure 2A) is consistent with the finding that Hsp104 catalyzes the formation of Sup35 fibril formation in vitro (Krzewska and Melki, 2006; Shorter and Lindquist, 2006). Like the detergent resistant subparticles of [PSI+] aggregates, detergent resistant subparticles of [Het-s]y grow bigger in size (Figure 6A) in the absence of Hsp104 before the prion is lost. Our data suggest that Hsp104 has a role in both the de novo appearance of HET-s aggregates, and the propagation of the prion. Interestingly, in Podospora anserina the Hsp104 homologue is not strictly required for [Het-s] maintenance. [Het-s] can be maintained in Podospora without Hsp104, but propagon numbers, propagation rates, meiotic stability and spontaneous emergence are greatly reduced in the absence of Hsp104 (L.M and S.J.S, unpublished data).
Heterologous Q/N-rich yeast prions have been shown to facilitate the initial formation of other Q/N-rich yeast prions. In vivo, the de novo appearance of [PSI+] was dramatically enhanced by the presence of [PIN+] or other Q/N-rich amyloids but not by non Q/N-rich amyloids (Derkatch et al., 2001; Osherovich and Weissman, 2001; Derkatch et al., 2004). [PIN+] also enhances the de novo formation of [URE3] but generally only a 10-fold effect was seen with this N-rich prion (Bradley et al., 2002). Since HET-s is not Q/N-rich, we did not expect [PIN+] to influence the appearance of [Het-s]y. Surprisingly, [PIN+] doubled the frequency of the de novo formation of [Het-s]y. This suggests an in vivo cross-talk between Q/N-rich and non Q/N-rich amyloid proteins.
A number of models have been proposed to explain the ability of heterologous prions to enhance the de novo appearance of other prions in yeast: titration of inhibitory factors by heterologous prions (Derkatch et al., 2001; Osherovich and Weissman, 2001; Uptain et al., 2001); activation of stimulatory factors by heterologous prions (Schwimmer and Masison, 2002); ability of heterologous prions to directly cross-seed the de novo formation of another prion. Each of these models can also explain the interactions between Q/N-rich and non Q/N-rich amyloids and indeed, more than one mechanism could be involved.
In vitro evidence supports the “cross-seeding” model because both Q/N rich and non Q/N-rich amyloids were shown to stimulate the aggregation of Sup35 (Derkatch et al., 2004). Indeed, all amyloid aggregates share common structural features and can interact to affect the fibrillization of non-related amyloid proteins (Chiti and Dobson, 2006). Several studies point to Q and N residues as being particularly important in the initial steps of the amyloid assembly process (Perutz et al., 1994). Interstrand N-N stacking is proposed to serve as a primer for in-register parallel β-strand assembly. Such N ladders were described in amyloid fibrils formed by a Sup35-derived peptide (Nelson et al., 2005). Interestingly, while overall the HET-s PrD is not N-rich, it contains an N residue in each of the 4 β-strands (Ritter et al., 2005). It is possible, that the effect of the Q/N-rich [PIN+] prion on HET-s PrD aggregation only requires this minimal similarity to promote the formation of N ladders. Such prion-prion interactions are not only important to understand the de novo appearance of prions but have broad implications in amyloid-associated neurodegenerative diseases.
Interestingly, prion proteins can exist in different heritable states defined as prion “strains” or “variants” (Derkatch et al., 1996; Schlumpberger et al., 2000; Bradley et al., 2002; Bruce, 2003). Evidence suggests that heritable structural differences are the basis of prion variants (King and Diaz-Avalos, 2000; Tanaka et al., 2004; Krishnan and Lindquist, 2005). Variants can be distinguished by differences in the size of detergent-resistant subparticles of the prion aggregates (Kryndushkin et al., 2003; Bagriantsev and Liebman, 2004). Since the subparticles of HET-s fusion rings obtained in the presence vs. the absence of [PIN+] are distinctly different (Figure 6A), [Het-s]y may adopt a different conformation if templated by [PIN+] than if formed in a [pin−] cell. This supports the idea that cross-seeding can give rise to “variants”. However, the difference in the subparticle size was not clearly distinguishable after the rings became dots. Thus, we cannot define these as “true variants” of [Het-s]y. Instead, HET-s might form different intermediate products in the presence and absence of [PIN+] which lead to the same final prion variant.
Heterologous prion propagation has previously been achieved for foreign proteins that are orthologous to a native prion from the host species. Transgenic mice expressing PrP sequences from a variety of different mammalian species were able to propagate PrPSc (Scott et al., 1989; Baron, 2002; Tamguney et al., 2006). Similarly, Sup35p and Ure2p orthologs from different yeast species have been expressed and propagated as prions in S. cerevisiae (Edskes et al., 1999; Chernoff et al., 2000; Kushnirov et al., 2000; Nakayashiki et al., 2001; Resende et al., 2002; Baudin-Baillieu et al., 2003). In contrast to these examples, there are no HET-s orthologs in S. cerevisiae, the foreign host genome used here. Nevertheless, [Het-s]y is able to propagate in this completely alien cellular context. This suggests that, in contrast to most host-infectious agent relations, prion propagation in yeast is promiscuous, and furthermore, that an evolutionary fine-tuning between the prion protein and the cellular machinery is not required.
Experimental Procedures
Strains, media and plasmids
Analysis of HET-s(PrD)-GFP aggregates in vivo
Transformants of 74-D694 and its derivatives bearing pHET-s(PrD)-GFP and pT266P-GFP were induced to express HET-s fusion constructs on galactose media. For serial passes, cells were diluted in water and re-streaked or were harvested, washed and reinoculated to an OD of 0.02. HET-s aggregates were visualized under a fluorescent microscope (Zeiss Axioskop 2) and photographed with a digital camera (Zeiss, AxioCam). The percentage of cells with HET-s aggregates were calculated as the number of cells with aggregates divided by total number of cells with and without aggregates. Cells were counted in 2–3 random fields with 150–200 cells per field.
Analysis of HET-s-GFP subparticles on semi-denaturing detergent agarose gel electrophoresis (SDD-AGE)
Transformants carrying pHET-s(PrD)-GFP and pT266P-GFP were grown in low (0.05% gal) or high expression (2% gal) media. Cells were passed twice in 0.05% gal and harvested each time after 24 hours of incubation. Cell lysates (prepared as described in Supplementary Experimental Procedures) were subjected to agarose electrophoresis to resolve the HET-s(PrD)-GFP subparticles and transferred to PVDF membrane using a widened mini-gel cassette as described previously (Bagriantsev and Liebman, 2004; Liebman et al., 2006). The HET-s (PrD)-GFP subparticles were probed with anti-GFP antibody.
Cytoduction
[Het-s]y or [het-s]y donor strains were mated with L2598 and L2736, [rho−], [het-s]y, cyh2R recipient strains defective for nuclear fusion (kar1). The donors carried pHET-s(PrD)-GFP. The recipient strains carried either pHET-s(PrD)-GFP or pT266P-GFP. Recipients were grown on SR-Trp+0.05% gal and had no aggregates. Mating was on SR-Trp+0.05% gal to maintain HET-s fusion expression. Cytoductants were selected on SG-Trp+0.05% gal+Cyh which also maintains low level expression of HET-s(PrD)-GFP and selects against diploids and donors, which cannot grow on cycloheximide (the cyh2R marker being recessive) and against recipients, which cannot grow on glycerol. Only recipients that carry cytoplasm from the donor (cytoductants) can grow.
Protein transfection of [Het-s*] mycelium with crude yeast extracts
The protein transfection of [Het-s*] mycelium was performed using the mechanical shearing method (Benkemoun et al., 2006) outlined in the Supplementary Experimental Procedures.
Supplementary Material
Acknowledgments
We thank Namitha Vishveshwara, Sapan Shan, Axelle Balguerie and Vidhu Mathur for help in completing some of the experiments and Anita Manogaran, Namitha Vishveshwara and Joo Yun Hong for helpful comments on the manuscript. We also thank Christophe Cullin, in whose laboratory N. Talarek performed some of the experiments and Michel Aigle for helpful advice. This work was supported by grants from the National Institutes of Health (R01 GM056350-10 to S.W.L.), the French Ministry of Research (to S. J. S.) and GIS "Infections à Prions" (to C. Cullin).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bagriantsev S, Liebman SW. Specificity of prion assembly in vivo. [PSI+] and [PIN+] form separate structures in yeast. J Biol Chem. 2004;279:51042–51048. doi: 10.1074/jbc.M410611200. [DOI] [PubMed] [Google Scholar]
- Balguerie A, Dos Reis S, Ritter C, Chaignepain S, Coulary-Salin B, Forge V, Bathany K, Lascu I, Schmitter JM, Riek R, Saupe SJ. Domain organization and structure-function relationship of the HET-s prion protein of Podospora anserina. EMBO J. 2003;22:2071–2081. doi: 10.1093/emboj/cdg213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron T. Mouse models of prion disease transmission. Trends Mol Med. 2002;8:495–500. doi: 10.1016/s1471-4914(02)02416-4. [DOI] [PubMed] [Google Scholar]
- Baudin-Baillieu A, Fernandez-Bellot E, Reine F, Coissac E, Cullin C. Conservation of the prion properties of Ure2p through evolution. Mol Biol Cell. 2003;14:3449–3458. doi: 10.1091/mbc.E03-01-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxa U, Cassese T, Kajava AV, Steven AC. Structure, function, and amyloidogenesis of fungal prions: filament polymorphism and prion variants. Adv Protein Chem. 2006;73:125–180. doi: 10.1016/S0065-3233(06)73005-4. [DOI] [PubMed] [Google Scholar]
- Benkemoun L, Sabate R, Malato L, Dos Reis S, Dalstra H, Saupe SJ, Maddelein ML. Methods for the in vivo and in vitro analysis of [Het-s] prion infectivity. Methods. 2006;39:61–67. doi: 10.1016/j.ymeth.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Borchsenius AS, Wegrzyn RD, Newnam GP, Inge-Vechtomov SG, Chernoff YO. Yeast prion protein derivative defective in aggregate shearing and production of new ‘seeds’. EMBO J. 2001;20:6683–6691. doi: 10.1093/emboj/20.23.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann A, Baxa U, Wickner RB. Prion generation in vitro: amyloid of Ure2p is infectious. EMBO J. 2005;24:3082–3092. doi: 10.1038/sj.emboj.7600772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley ME, Edskes HK, Hong JY, Wickner RB, Liebman SW. Interactions among prions and prion “strains” in yeast. Proc Natl Acad Sci USA . 2002;99(Suppl 4):16392–16399. doi: 10.1073/pnas.152330699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley ME, Liebman SW. Destabilizing interactions among [PSI+] and [PIN+] yeast prion variants. Genetics. 2003;165:1675–1685. doi: 10.1093/genetics/165.4.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce ME. TSE strain variation. Br Med Bull. 2003;66:99–108. doi: 10.1093/bmb/66.1.99. [DOI] [PubMed] [Google Scholar]
- Chernoff YO, Derkach IL, Inge-Vechtomov SG. Multicopy SUP35 gene induces de-novo appearance of psi-like factors in the yeast Saccharomyces cerevisiae. Curr Genet. 1993;24:268–270. doi: 10.1007/BF00351802. [DOI] [PubMed] [Google Scholar]
- Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [PSI+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- Chernoff YO, Galkin AP, Lewitin E, Chernova TA, Newnam GP, Belenkiy SM. Evolutionary conservation of prion-forming abilities of the yeast Sup35 protein. Mol Microbiol. 2000;35:865–876. doi: 10.1046/j.1365-2958.2000.01761.x. [DOI] [PubMed] [Google Scholar]
- Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- Conde J, Fink GR. A mutant of Saccharomyces cerevisiae defective for nuclear fusion. Proc Natl Acad Sci USA. 1976;73:3651–3655. doi: 10.1073/pnas.73.10.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coustou V, Deleu C, Saupe S, Begueret J. The protein product of the het-s heterokaryon incompatibility gene of the fungus Podospora anserina behaves as a prion analog. Proc Natl Acad Sci USA. 1997;94:9773–9778. doi: 10.1073/pnas.94.18.9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coustou V, Deleu C, Saupe SJ, Begueret J. Mutational analysis of the [Het-s] prion analog of Podospora anserina. A short N-terminal peptide allows prion propagation. Genetics. 1999;153:1629–1640. doi: 10.1093/genetics/153.4.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalstra HJ, van der Zee R, Swart K, Hoekstra RF, Saupe SJ, Debets AJ. Non-mendelian inheritance of the HET-s prion or HET-s prion domains determines the het-S spore killing system in Podospora anserina. Fungal Genet Biol. 2005;42:836–847. doi: 10.1016/j.fgb.2005.05.004. [DOI] [PubMed] [Google Scholar]
- DePace AH, Santoso A, Hillner P, Weissman JS. A critical role for amino-terminal glutamine/asparagine repeats in the formation and propagation of a yeast prion. Cell. 1998;93:1241–1252. doi: 10.1016/s0092-8674(00)81467-1. [DOI] [PubMed] [Google Scholar]
- Derkatch IL, Liebman SW. Prion-prion interactions. In: Chernoff YO, editor. Protein-based inheritance. Austin: Landes Bioscience; 2007. in press. [Google Scholar]
- Derkatch IL, Chernoff YO, Kushnirov VV, Inge-Vechtomov SG, Liebman SW. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics. 1996;144:1375–1386. doi: 10.1093/genetics/144.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch IL, Bradley ME, Zhou P, Chernoff YO, Liebman SW. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics. 1997;147:507–519. doi: 10.1093/genetics/147.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch IL, Bradley ME, Masse SV, Zadorsky SP, Polozkov GV, Inge-Vechtomov SG, Liebman SW. Dependence and independence of [PSI+] and [PIN+]: a two-prion system in yeast? EMBO J. 2000;19:1942–1952. doi: 10.1093/emboj/19.9.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: the story of [PIN+] Cell. 2001;106:171–182. doi: 10.1016/s0092-8674(01)00427-5. [DOI] [PubMed] [Google Scholar]
- Derkatch IL, Uptain SM, Outeiro TF, Krishnan R, Lindquist SL, Liebman SW. Effects of Q/N-rich, polyQ, and non-polyQ amyloids on the de novo formation of the [PSI+] prion in yeast and aggregation of Sup35 in vitro. Proc Natl Acad Sci USA. 2004;101:12934–12939. doi: 10.1073/pnas.0404968101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edskes HK, Gray VT, Wickner RB. The [URE3] prion is an aggregated form of Ure2p that can be cured by overexpression of Ure2p fragments. Proc Natl Acad Sci USA. 1999;96:1498–1503. doi: 10.1073/pnas.96.4.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira PC, Ness F, Edwards SR, Cox BS, Tuite MF. The elimination of the yeast [PSI+] prion by guanidine hydrochloride is the result of Hsp104 inactivation. Mol Microbiol. 2001;40:1357–1369. doi: 10.1046/j.1365-2958.2001.02478.x. [DOI] [PubMed] [Google Scholar]
- Jung G, Masison DC. Guanidine hydrochloride inhibits Hsp104 activity in vivo: a possible explanation for its effect in curing yeast prions. Curr Microbiol. 2001;43:7–10. doi: 10.1007/s002840010251. [DOI] [PubMed] [Google Scholar]
- Kim K, Keller TC., 3rd Smitin, a novel smooth muscle titin-like protein, interacts with myosin filaments in vivo and in vitro. J Cell Biol. 2002;156:101–111. doi: 10.1083/jcb.200107037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y, Koitabashi S, Fujita T. Analysis of yeast prion aggregates with amyloid-staining compound in vivo. Cell Struct Funct. 2003;28:187–193. doi: 10.1247/csf.28.187. [DOI] [PubMed] [Google Scholar]
- King CY, Tittmann P, Gross H, Gebert R, Aebi M, Wuthrich K. Prion-inducing domain 2–114 of yeast Sup35 protein transforms in vitro into amyloid-like filaments. Proc Natl Acad Sci USA. 1997;94:6618–6622. doi: 10.1073/pnas.94.13.6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King CY, Diaz-Avalos R. Protein-only transmission of three yeast prion strains. Nature. 2004;428:319–323. doi: 10.1038/nature02391. [DOI] [PubMed] [Google Scholar]
- Krishnan R, Lindquist SL. Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature. 2005;435:765–772. doi: 10.1038/nature03679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryndushkin DS, Alexandrov IM, Ter-Avanesyan MD, Kushnirov VV. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J Biol Chem. 2003;278:49636–49643. doi: 10.1074/jbc.M307996200. [DOI] [PubMed] [Google Scholar]
- Krzewska J, Melki R. Molecular chaperones and the assembly of the prion Sup35p, an in vitro study. EMBO J. 2006;25:822–833. doi: 10.1038/sj.emboj.7600985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnirov VV, Kochneva-Pervukhova NV, Chechenova MB, Frolova NS, Ter-Avanesyan MD. Prion properties of the Sup35 protein of yeast Pichia methanolica. EMBO J. 2000;19:324–331. doi: 10.1093/emboj/19.3.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Lindquist S. Creating a protein-based element of inheritance. Science. 2000;287:661–664. doi: 10.1126/science.287.5453.661. [DOI] [PubMed] [Google Scholar]
- Liebman SW, Bagriantsev SN, Derkatch IL. Biochemical and genetic methods for characterization of [PIN+] prions in yeast. Methods. 2006;39:23–34. doi: 10.1016/j.ymeth.2006.04.010. [DOI] [PubMed] [Google Scholar]
- Ma J, Lindquist S. De novo generation of a PrPSc-like conformation in living cells. Nat Cell Biol. 1999;1:358–361. doi: 10.1038/14053. [DOI] [PubMed] [Google Scholar]
- Maddelein ML, Dos Reis S, Duvezin-Caubet S, Coulary-Salin B, Saupe SJ. Amyloid aggregates of the HET-s prion protein are infectious. Proc Natl Acad Sci USA. 2002;99:7402–7407. doi: 10.1073/pnas.072199199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masison DC, Wickner RB. Prion-inducing domain of yeast Ure2p and protease resistance of Ure2p in prion-containing cells. Science. 1995;270:93–95. doi: 10.1126/science.270.5233.93. [DOI] [PubMed] [Google Scholar]
- Masison DC, Maddelein ML, Wickner RB. The prion model for [URE3] of yeast: spontaneous generation and requirements for propagation. Proc Natl Acad Sci USA. 1997;94:12503–12508. doi: 10.1073/pnas.94.23.12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama H, Edskes HK, Wickner RB. [URE3] prion propagation in Saccharomyces cerevisiae: requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Mol Cell Biol. 2000;20:8916–8922. doi: 10.1128/mcb.20.23.8916-8922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayashiki T, Ebihara K, Bannai H, Nakamura Y. Yeast [PSI+] “prions” that are crosstransmissible and susceptible beyond a species barrier through a quasi-prion state. Mol Cell. 2001;7:1121–1130. doi: 10.1016/s1097-2765(01)00259-3. [DOI] [PubMed] [Google Scholar]
- Nelson R, Sawaya MR, Balbirnie M, Madsen AO, Riekel C, Grothe R, Eisenberg D. Structure of the cross-beta spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osherovich LZ, Weissman JS. Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI+] prion. Cell. 2001;106:183–194. doi: 10.1016/s0092-8674(01)00440-8. [DOI] [PubMed] [Google Scholar]
- Osherovich LZ, Cox BS, Tuite MF, Weissman JS. Dissection and design of yeast prions. PLoS Biol. 2004;2:442–451. doi: 10.1371/journal.pbio.0020086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel BK, Liebman SW. Prion-proof” for [PIN+]: Infection with in vitro-made amyloid aggregates of Rnq1p-(132-405) induces [PIN+] J Mol Biol. 2007;365:773–782. doi: 10.1016/j.jmb.2006.10.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paushkin SV, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD. Propagation of the yeast prion-like [PSI+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 1996;15:3127–3134. [PMC free article] [PubMed] [Google Scholar]
- Perutz MF, Johnson T, Suzuki M, Finch JT. Glutamine repeats as polar zippers: their possible role in inherited neurodegenerative diseases. Proc Natl Acad Sci USA. 1994;91:5355–5358. doi: 10.1073/pnas.91.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce MM, Baxa U, Steven AC, Bax A, Wickner RB. Is the prion domain of soluble Ure2p unstructured? Biochemistry. 2005;44:321–328. doi: 10.1021/bi047964d. [DOI] [PubMed] [Google Scholar]
- Prusiner SB. Prions. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resende C, Parham SN, Tinsley C, Ferreira P, Duarte JA, Tuite MF. The Candida albicans Sup35p protein (CaSup35p): function, prion-like behaviour and an associated polyglutamine length polymorphism. Microbiology. 2002;148:1049–1060. doi: 10.1099/00221287-148-4-1049. [DOI] [PubMed] [Google Scholar]
- Ritter C, Maddelein ML, Siemer AB, Luhrs T, Ernst M, Meier BH, Saupe SJ, Riek R. Correlation of structural elements and infectivity of the HET-s prion. Nature. 2005;435:844–848. doi: 10.1038/nature03793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizet G. Les phénomènes de barrage chez Podospora anserina. I Analyse de barrage entre les souches s et S. Rev Cytol Biol Veg. 1952;13:51–92. [Google Scholar]
- Ross ED, Minton A, Wickner RB. Prion domains: sequences, structures and interactions. Nat Cell Biol. 2005;7:1039–1044. doi: 10.1038/ncb1105-1039. [DOI] [PubMed] [Google Scholar]
- Salnikova AB, Kryndushkin DS, Smirnov VN, Kushnirov VV, Ter-Avanesyan MD. Nonsense suppression in yeast cells overproducing Sup35 (eRF3) is caused by its non-heritable amyloids. J Biol Chem. 2005;280:8808–8812. doi: 10.1074/jbc.M410150200. [DOI] [PubMed] [Google Scholar]
- Santoso A, Chien P, Osherovich LZ, Weissman JS. Molecular basis of a yeast prion species barrier. Cell. 2000;100:277–288. doi: 10.1016/s0092-8674(00)81565-2. [DOI] [PubMed] [Google Scholar]
- Saupe SJ. Molecular genetics of heterokaryon incompatibility in filamentous ascomycetes. Microbiol Mol Biol Rev. 2000;64:489–502. doi: 10.1128/mmbr.64.3.489-502.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlumpberger M, Wille H, Baldwin MA, Butler DA, Herskowitz I, Prusiner SB. The prion domain of yeast Ure2p induces autocatalytic formation of amyloid fibers by a recombinant fusion protein. Protein Sci. 2000;9:440–451. doi: 10.1110/ps.9.3.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwimmer C, Masison DC. Antagonistic interactions between yeast [PSI+] and [URE3] prions and curing of [URE3] by Hsp70 protein chaperone Ssa1p but not by Ssa2p. Mol Cell Biol. 2002;22:3590–3598. doi: 10.1128/MCB.22.11.3590-3598.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott M, Foster D, Mirenda C, Serban D, Coufal F, Walchli M, Torchia M, Groth D, Carlson G, DeArmond SJ, et al. Transgenic mice expressing hamster prion protein produce species-specific scrapie infectivity and amyloid plaques. Cell. 1989;59:847–857. doi: 10.1016/0092-8674(89)90608-9. [DOI] [PubMed] [Google Scholar]
- Sen A, Baxa U, Simon MN, Wall JS, Sabate R, Saupe SJ, Steven AC. Mass analysis by scanning transmission electron microscopy and electron diffraction validate predictions of the stacked beta -solenoid model of HET-s prion fibrils. J Biol Chem. 2006;282:5545–5550. doi: 10.1074/jbc.M611464200. [DOI] [PubMed] [Google Scholar]
- Serio TR, Cashikar AG, Kowal AS, Sawicki GJ, Moslehi JJ, Serpell L, Arnsdorf MF, Lindquist SL. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science. 2000;289:1317–1321. doi: 10.1126/science.289.5483.1317. [DOI] [PubMed] [Google Scholar]
- Shewmaker F, Wickner RB, Tycko R. Amyloid of the prion domain of Sup35p has an in-register parallel {beta}-sheet structure. Proc Natl Acad Sci USA. 2006;103:19754–19759. doi: 10.1073/pnas.0609638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J, Lindquist S. Destruction or potentiation of different prions catalyzed by similar Hsp104 remodeling activities. Mol Cell. 2006;23:425–438. doi: 10.1016/j.molcel.2006.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondheimer N, Lindquist S. Rnq1: an epigenetic modifier of protein function in yeast. Mol Cell. 2000;5:163–172. doi: 10.1016/s1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- Speransky VV, Taylor KL, Edskes HK, Wickner RB, Steven AC. Prion filament networks in [URE3] cells of Saccharomyces cerevisiae. J Cell Biol. 2001;153:1327–1336. doi: 10.1083/jcb.153.6.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamguney G, Giles K, Bouzamondo-Bernstein E, Bosque PJ, Miller MW, Safar J, Dearmond SJ, Prusiner SB. Transmission of elk and deer prions to transgenic mice. J Virol. 2006;80:9104–9114. doi: 10.1128/JVI.00098-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Conformational variations in an infectious protein determine prion strain differences. Nature. 2004;428:323–328. doi: 10.1038/nature02392. [DOI] [PubMed] [Google Scholar]
- Taylor KL, Cheng N, Williams RW, Steven AC, Wickner RB. Prion domain initiation of amyloid formation in vitro from native Ure2p. Science. 1999;283:1339–1343. doi: 10.1126/science.283.5406.1339. [DOI] [PubMed] [Google Scholar]
- Ter-Avanesyan MD, Dagkesamanskaya AR, Kushnirov VV, Smirnov VN. The SUP35 omnipotent suppressor gene is involved in the maintenance of the non-Mendelian determinant [PSI+] in the yeast Saccharomyces cerevisiae. Genetics. 1994;137:671–676. doi: 10.1093/genetics/137.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thual C, Bousset L, Komar AA, Walter S, Buchner J, Cullin C, Melki R. Stability, folding, dimerization, and assembly properties of yeast prion Ure2p. Biochemistry. 2001;40:1764–1773. doi: 10.1021/bi001916l. [DOI] [PubMed] [Google Scholar]
- Uptain SM, Sawicki GJ, Caughey B, Lindquist S. Strains of [PSI+] are distinguished by their efficiencies of prion-mediated conformational conversion. EMBO J. 2001;20:6236–6245. doi: 10.1093/emboj/20.22.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viles JH, Donne D, Kroon G, Prusiner SB, Cohen FE, Dyson HJ, Wright PE. Local structural plasticity of the prion protein. Analysis of NMR relaxation dynamics. Biochemistry. 2001;40:2743–2753. doi: 10.1021/bi002898a. [DOI] [PubMed] [Google Scholar]
- Vitrenko YA, Pavon ME, Stone SI, Liebman SW. Propagation of the [PIN+] prion by fragments of Rnq1 fused to GFP. Curr Genet. 2007;51:309–319. doi: 10.1007/s00294-007-0127-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegrzyn RD, Bapat K, Newnam GP, Zink AD, Chernoff YO. Mechanism of prion loss after Hsp104 inactivation in yeast. Mol Cell Biol. 2001;21:4656–4669. doi: 10.1128/MCB.21.14.4656-4669.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner RB. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- Wickner RB, Masison DC, Edskes HK. [PSI] and [URE3] as yeast prions. Yeast. 1995;11:1671–1685. doi: 10.1002/yea.320111609. [DOI] [PubMed] [Google Scholar]
- Wickner RB, Liebman SW, Saupe SJ. Chapter 7 “Prions of Yeast and Filamentous Fungi: [URE3+], [PSI+], [PIN+], and [Het-s]”. In: Prusiner SB, editor. Prion Biology and Diseases. Cold Spring Harbor Laboratory Press; 2004. pp. 305–372. [Google Scholar]
- Zhou P, Derkatch IL, Liebman SW. The relationship between visible intracellular aggregates that appear after overexpression of Sup35 and the yeast prion-like elements [PSI+] and [PIN+] Mol Microbiol. 2001;39:37–46. doi: 10.1046/j.1365-2958.2001.02224.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


