Abstract
Fatty acid amide hydrolase (FAAH) is a pharmaceutical target whose inhibition may lead to valuable therapeutics. Sensitive substrates for high-throughput assays are crucial for the rapid-screening FAAH inhibitors. Here we describe the development of novel and highly sensitive fluorescent assays for FAAH based on substituted aminopyridines. Examining the relationship between the structure and the fluorescence of substituted aminopyridines suggested that a methoxy group in the para position relative to the amino group in aminopyridines greatly increased the fluorescence (i.e., quantum yields approach unity). These novel fluorescent reporters had a high Stokes’ shift of 94 nm, and their fluorescence in buffer systems increased with pH values from neutral to basic. Fluorescent substrates with these reporters displayed a very low fluorescent background and high aqueous solubility. Most importantly, fluorescent assays for FAAH based on these substrates were at least 25 times more sensitive than assays using related compounds with published colorimetric or fluorescent reporters. This property results in shorter assay times and decreased protein concentrations in the assays. Such sensitive assays will facilitate distinguishing the relative potency of powerful inhibitors of FAAH. When these fluorescent substrates were applied to human liver microsomes, results suggested that there was at least one amide hydrolase in addition to FAAH that could hydrolyze long-chain fatty acid amides. These results show that these fluorescent substrates are very valuable tools in FAAH activity assays including screening inhibitors by high-throughput assays instead of using the costly and labor-intensive radioactive ligands. Potential applications of novel fluorescent reporters are discussed.
Keywords: FAAH, Aminopyridines, Fluorescence, Sensitivity, Quantum yields, Substrates
Fatty acid amide hydrolase (FAAH)1 is a mammalian integral membrane enzyme that plays critical roles in regulating the levels of endogenous signaling lipids such as the cannabinoid anandamide, sleeping-inducing substance oleamide, anorexigenic compound N-oleoylethanolamide, and antiinflammatory agent N-palmitoylethanolamide [1–5]. Inhibition of FAAH is associated with treatments of multiple pathological disorders such as pain, anxiety, inflammation, and other disorders [6–15]. Therefore, compounds that inhibit this enzyme are of medical and therapeutic significance.
To meet increasing demands for identifying novel inhibitors from large libraries of chemical compounds, simple, highly sensitive, and specific assays, which are suitable for a high-throughput format such as colorimetric or fluorescent assays in 96- or 384-well plates or alternative formats, are crucial for screening. Most assays for FAAH activity are commonly associated with the use of radiolabeled substrates or chromatographic techniques, which are costly, labor-intensive, and incompatible with high-throughput format or have other limitations [16–24]. Recently, two assays that are suitable for high-throughput formats such as 96 or 384-well plate assays have been reported [25,26]. One is a coupled colorimetric assay, i.e., it utilizes the ability of FAAH to hydrolyze oleamide and measures the resultant production of ammonia by a NADH/NAD+-coupled enzyme [25]. Its application is limited by its sensitivity and the complexity of using a second enzyme (e.g., L-glutamate dehydrogenase). The other method is a fluorescent assay using the substrate arachidonyl 7-amino-4-methylcoumarin amide [26]. The application of this assay is also limited by the sensitivity of the assay, the poor aqueous solubility, and the instability of the substrate.
It is well known that 2-, 3-, and 4-aminopyridines are fluorescent compounds [27,28]. Among these aminopyridines, 2- and 3-substituted isomers show relatively stronger fluorescence than 4-substituted isomers. Therefore, both 2- and 3-isomers have been used as fluorescent reporters of hydrolytic activities of multiple enzymes such as nucleotide pyrophosphatase [29], transferases [30,31], and hyaluronidase [32]. However, the fluorescent strength (or quantum yield) of these aminopyridines is relatively low, compared with 7-aminocoumarin, and greatly varies with the change of pH values in media [27] due to protonization or deprotonization of both nitrogen atoms. Generally, strong fluorescence for aminopyridines is shown in acidic or neutral aqueous media, but weak fluorescence is shown in basic aqueous media. These properties severely limit the applications of simple aminopyridines in most enzymatic assays such as those for amide hydrolases and aminopeptidases, which show maximal hydrolytic activities in basic aqueous media. Extensive attention has been paid to investigating the fluorescence of more complicated compounds containing one or multiple pyridine rings [33,34]. However, there is little work on the use of simple aminopyridines as fluorescent reporters.
Here we describe an investigation of fluorescence of substituted 3- or 5-aminopyridines. Based on this investigation, a simple, novel, highly sensitive, and continuous fluorescent assay for FAAH activity has been developed. This sensitive assay is based on measuring the strong fluorescence of a substituted 3- or 5-aminopyridine after the amide hydrolysis. These reporters may be particularly valuable with enzymes where the size of the catalytic site is limited. The tools developed in this study will be useful for studying the hydrolytic activity of numerous amide hydrolases including FAAH and other enzymes.
Materials and methods
Chemicals and enzymes
Fluorescent reporters including 3-amino-4-methylpyridine (2), 3-amino-6-methylpyridine (3), 3-amino-2-fluoropyridine (4), 5-amino-2-fluoropyridine (5), 5-amino-2-methoxypyridine (6), 3-amino-2-methoxypyridine (7), and 3-amino-6-methoxy-2-picoline (8) were purchased from Lancaster Synthesis, Inc. (2; Windham, NH, USA), and AB Chem., Inc. (3; Quebec, Canada), Matrix Scientic (4; Columbia, SC, USA), Beta Pharma, Inc. (5; New Haven, CT, USA), Asychem (6, 7, 8; Durham, NC, USA), respectively. 3-Aminopyridine (1) and 7-amino-4-trifluoromethyl-coumarin (9) were purchased from Sigma–Aldrich (Saint Louis, MI, USA). Pooled human liver S9 and microsomes were purchased from BD Biosciences (San Jose, CA, USA). Rabbit anti-FAAH polyclonal antibody, peroxidase-conjugated anti-rabbit IgG, and ECL detection kit were purchased from Cayman Chemical (Ann Arbor, MI, USA), Sigma Chemical (Saint Louis, MI, USA), and GE Healthcare Bio-Sciences (Piscataway, NJ, USA), respectively. Structural identification and purity are based on 1H-NMR, 13C-NMR, GC/MS and LC/MS analysis. All substrates showed as single spots on TLC with no fluorescence at 254 or 280 nm. Data are presented in Supplemental information.
General synthetic procedures of substrate preparation
To an ice-cooled solution containing a fluorescent reporter (substituted aminopyridine or 7-amino-4-trifluo-romethylcoumarin, 100 mg) and dry CH2Cl2 (10 mL) was slowly added the appropriate acid chloride (1.05 eqv) and 4-methylmorpholine (1.05 eqv). The reaction was warmed slowly to room temperature and stirred overnight. The reaction mixture was washed with saturated sodium bicarbonate solution and extracted three times with ethyl acetate (3 × 20 mL). The combined organic phase was dried by magnesium sulfate. A crude solid product was obtained after filtration and evaporation under reduced pressure. The crude product was chromatographed on a gradient of hexane and ethyl acetate. Abbreviations of substrates (Fig. 1) are expressed by referring to the acid moiety followed by an abbreviation for the reporter such as N-(6-methoxypyridin-3-yl) acetamide (Acetyl-MP), N-(6-methoxy-2-methylpyridin-3-yl) acetamide (Acetyl-MMP), N-(4-(trifluoromethyl)-2-oxo-2H-chromen-7-yl) acetamide (Acetyl-TFMCoumarin), N-(6-methoxypyridin-3-yl) octanamide (Octanoyl-MP), N-(6-methoxy-2-methylpyridin-3-yl) octanamide (Octanoyl-MMP), N-(4-(trifluoromethyl)-2-oxo-2H-chromen-7-yl) octanamide (Octanoyl-TFMCoumarin), and N-(p-nitrophenyl) octanamide (Octanoyl-p-NP). Spectral characterization of these substrates is referred to the accompanying Supplemental information.
Fig. 1.

Structures of fluorescent reporters and substrates used in this study.
(5Z,8Z,11Z,14Z)-N-(6-methoxypyridin-3-yl) icosa-5,8,11,14-tetraenamide (Arachidonyl-MP)
Arachidonic acid (200 mg, 0.65 mmol) was added to an ice-cooled CH2Cl2 solution (10 mL) containing triethylamine (0.65 mmol) and 1-[3-(dimethylamino)propyl]-3-ethylcarbodiimide hydrochloride (139 mg, 0.73 mmol) under nitrogen protection. After 20 min reaction under an ice-cooled bath, a dichloromethane solution (2 mL) containing 5-amino-2-methoxypyridine (65 mg, 0.52 mmol) was added to the reaction. The reaction was stirred 3 h under the ice-cooled bath. The reaction mixture was washed twice with 1 M sodium bicarbonate solution (30 mL) and saturated NaCl solution (30 mL). The combined organic phase was dried under magnesium sulfate for 1 h. After filtration and evaporation, it gave rise to an oily product, which was chromatographed with a gradient of hexane and ethyl acetate. Slight yellowish oil (46 mg) was given (17% yield). Caution: For all procedures described here, aluminum foil was used to protect solution from being exposed to light. Spectral characterization of this substrate is referred to the accompanying Supplemental information.
Preparation of recombinant human FAAH
Transgenic expression of the human FAAH in a baculovirus expression system was performed by following a method described previously [35]. The cDNA encoding the human FAAH (GenBank Accession No. NM_001441) was amplified by PCR using a human liver cDNA library (Invitrogen, Carlsbad, CA, USA) as a template. The primer pair was 5′-AGATCTATGGTGCAGTACGAGCTGTGGGCC-3′ and 5′-GAATTCTCAGGATGACTGCTTTTCAGGGGT-3′. BglII and EcoRI endonuclease sites (underlines) were incorporated just upstream of the start codon and downstream of the stop codon of the coding sequence of FAAH, respectively. The PCR products were cloned into a pCR2.1 vector (Invitrogen) and the nucleotide sequence was verified by DNA sequencing (DBS DNA Sequencing Facility, University of California at Davis). The cDNA fragments were then excised and directionally ligated to the BglII and EcoRI sites of a baculovirus transfer vector pAcUW21 [36]. Recombinant baculoviruses harboring the human FAAH gene, Ac-hFAAH, were generated by cotransfection of cells derived from Spodoptera frugiperda (Sf21 cells) with the recombinant transfer vector plasmid and Bsu36I-cleaved BacPAK6 viral DNAs (Clontech Laboratories, Mountain view, CA, USA), as previously described [37]. Cells from Trichoplusia ni (High Five cells) (1 × 106 cells/mL) were inoculated with a high titer of Ac-hFAAH. At 72 h postinfection, the infected cells were harvested by centrifugation at 2000g for 20 min at 4°C and suspended in 50 mM Tris–HCl (pH 8.0) containing 150 mM NaCl, 1 mM EDTA, 1 μM pepstatin, 100 μM leupeptin, and 0.1 mg/mL aprotinin. The cell suspension was then homogenized using a Polytron homogenizer and centrifuged at 10,000g for 20 min at 4°C to collect the supernatant. The microsomal fraction was then isolated by ultracentrifugation of the supernatant at 100,000g for 60 min at 4°C. The pellet was resuspended in 20 mM Tris–HCl (pH 8.0) containing 10% (w/v) glycerol and 1% (w/v) Triton X-100 and stored at −80°C until use. Protein concentration was measured by BCA assay using BSA as a standard in this study. Partial characterization of FAAH is described in the accompanying Supplemental information.
Molar extinction coefficient
The molar extinction coefficients are determined according to Beer’s law, Aλ = ε * C * L. In this equation, Aλ is the absorbance at wavelength (λ nm), ε is the molar extinction coefficient, L is the width of the sample cuvette, and C is the concentration of the solute. In this study, a series of concentrations falling between 10 and 50 μM (final concentration) for each fluorescent reporter was used for determination of the molar extinction coefficient. Assays were performed in a quartz cuvette (1-cm optical passage). To 990 μL freshly distilled H2O was added 10 μL stock solution of a reporter in ethanol (1, 2, 3, 4, or 5 mM). Freshly distilled water was used to avoid a buildup of dissolved CO2, altering the pH. The resulting mixture was degassed by sonication with an ultrasonic cleaner Model 750 (VWR International, Wester, PA, USA) for 10 s at a power level of 9. Measurements were performed with a Spectra Max M2 spectrophotometer (Molecular Devices, Sunnyvale, CA, USA) at a wavelength 290 nm. A slope (or extinction coefficient) was obtained by using an average absorbance of triplicate samples for each concentration.
Maximal excitation and emission wavelengths
A stock solution in ethanol of each compound (10 μL, 5 mM) was added into 990 μL of sodium phosphate buffer (0.1 M, pH 8.0, final substrate concentration 50 μM) in a 1-cm quartz cuvette. The quartz cuvette was scanned from 200 to 500 nm to determine an absorption spectrum with a Spectra Max M2 spectrophotometer. Maximal excitation and emission wavelengths were obtained from scanning from 250 to 600 nm and from 360 to 600 nm by a 2-nm interval at 30°C, respectively.
Quantum yield
The fluorescence quantum yield, φf, was calculated according to the equation [38]
where ØF is the quantum yield of the tested fluorescent reporter, Østd is the fluorescence quantum yield of the standard, I and Istd are the integrated emission intensities of the sample and the standard, respectively, OD and ODstd are the absorbance of the sample and standard, respectively, at the desired wavelength λex, and n and nstd are the indexes of refraction of the sample and standard solutions, respectively.
In this study, 3-aminopyridine in distilled water at the maximal excitation wavelength 290 nm was used as a standard (Østd = 0.32, εmax = 2800 M −1 cm−1, [27]). To minimize inner filter effects, the optical densities of samples at the excitation wavelength (λ = 290 nm) were kept in the range of 0.095–0.13 (less than 0.150–0.165, [27]). The final concentration of each tested sample in a quartz cuvette was 10 μM with 1% ethanol in distilled water, same as that for the standard. The integrated area of fluorescent intensity for the standard or the samples was based on the area falling between 360 and 500 nm. The difference of the index of refraction between the standard and the sample was neglected because (1) all measurements were performed in the same quartz cuvette and spectrafluorimetry; (2) the final concentrations of the sample and the standard were the same, and (3) the structures of the sample and the standard were very similar—only one pyridine ring. The quantum yields presented in Table 1 were based on calculation of the average of OD and integrated fluorescent intensities of triplicates. The standard deviation for each quantum yield was less than ±5%.
Table 1.
Optical properties of different substituted aminopyridines
| Compound No. | Structure | ε max (M−1cm−1)a | Ex (nm)b | Em (nm)b | ØFc |
|---|---|---|---|---|---|
| 1 |

|
2890 (2800)d | 286 | 378 | 0.32d |
| 2 |
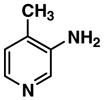
|
2580 | 280 | 376 | 0.53 |
| 3 |
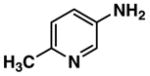
|
2700 | 290 | 384 | 0.62 |
| 4 |

|
3670 | 282 | 374 | 0.04 |
| 5 |

|
2360 | 292 | 384 | 0.72 |
| 6 |

|
2340 | 302 | 396 | 0.95 |
| 7 |

|
4670 | 284 | 378 | 0.27 |
| 8 |
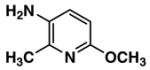
|
4130 | 304 | 392 | 0.97 |
Molar extinction coefficients were determined in H2O containing 1% ethanol at wavelength 290 nm and 30°C, and the final concentrations of the solute were 10, 20, 30, 40, and 50 μM.
Ex and Em were obtained from scanning from 250 to 600 nm and from 360 to 600 nm by a 2-nm interval at 30 °C, respectively, and 50 μM final concentration for each reporter in 0.1 M phosphate buffer. The standard deviation for each datum was ±2 nm.
ØF: standard deviations were under ±5%.
Data are from [27].
Dependence of relative fluorescent intensity on the pH values
To determine whether protonization/deprotonization will affect the relative fluorescent intensities of the reporters, we used 5-amino-2-methoxypyridine (6) as an example. A stock solution of 6 in ethanol (5 μL, 5 mM) was added into 995 μL 0.1 M sodium phosphate buffer (from pH 6.0 to pH 8.0) or 0.1 M Tris–HCl buffer (pH 9.0) in a quartz cuvette (final substrate concentration 25 μM). Relative fluorescent unit/intensity (RFU) for emission spectra was recorded by a 2-nm interval at room temperature using fixed excitation wavelength (λ = 302 nm). The RFU of each pH value was an average of triplicates and the standard deviation for each point was less than 1%. Similarly, relative fluorescent unit/intensity RFU for excitation spectra was recorded by a 2-nm interval at room temperature using fixed emission wavelength (λ = 396 nm) in 0.1 M, pH 8.0, phosphate buffer.
The quenching effect of proteins on the fluorescence is an important factor determining the assay sensitivity. This effect was determined by using 5-amino-2-methoxy-pyridine and BSA as an example. The detailed experimental procedure is available in the Supplemental information.
Comparison of aqueous solubility of substrates
The aqueous solubilities of the aminopyridine derivatives (e.g., Octanoyl-MP, Octanoyl-MMP) and the 7-amino-coumarin derivative (e.g., Octanoyl-TFMCoumarin) were determined in clear 96-well styrene flat-bottomed microtiter plates with a Spectra Max M2 spectrophotometer. Different concentrations of the substrates [(Octanoyl-MP, Octanoyl-MMP, and Octanoyl-TFMCoumarin); final concentration: 0.39, 0.78, 1.56, 3.13, 6.25, 12.5, 25, 50, 100 and 200 μM in 200 μL 0.1 M, pH 8.0, sodium phosphate buffer (1% ethanol)] were used and the absorbance was recorded at wavelength 800 nm at 37 °C. The measurements were performed in three replicates for each concentration. The wells were read at a wavelength where the substrate has little or no known absorbance. When a solution becomes saturated by the substrate and micelles or particulates form, the absorbance reading will increase dramatically due to light scattering. Based on this phenomenon, the estimated substrate solubility is determined by the point where the substrate concentration elicits the absorbance skewing from the horizontal line.
Comparison of the lowest detection limits of protein concentrations of FAAH toward colorimetric and fluorescent substrates
In general, all assays were performed in 96-well styrene flat-bottomed microtiter plates with Spectra Max M2 spectrophotometer at 37°C for 10 min. The total volume of each well was 201 μL consisting of 0.1 M, pH 8.0, sodium phosphate buffer (190 μL) containing 1% glycerol and 0.1% Triton X-100, stock or diluted protein solution (10 μL), and the substrate solution (1 μL) in ethanol (final concentration 200 μM for Octanoyl-MP and Octanoyl-MMP) or in DMSO (final concentration 25 μM for Octanoyl-TFMCoumarin and Octanoyl-p-NP, and 50 μM for Arachidonyl-MP). The lowest detection limits of microsomal fatty acid amide hydrolase by substrates were based on signals which were determined by the fact that the signals at 10 min after initiation of the kinetic measurements were equal to or over three times those of the noise or baseline. The colorimetric substrate (Octanoyl-p-NP) was evaluated in clear 96-well microtititer plates using an absorbance wavelength at 382 nm. The fluorescent substrates containing 5-amino-2-methoxypyridine (e.g., Octanoyl-MP, Arachidonyl-MP) were measured at excitation wavelength 302 nm, emission wavelength 396 nm, and auto cutoff wavelength 325 nm. Similarly, the fluorescent substrates containing 5-amino-2-methoxy-6-methyl pyridine (e.g., Octanoyl-MMP) were measured at an excitation wavelength 304 nm, emission wavelength 392 nm, and autocutoff wavelength 325 nm. In addition, excitation wavelength 366 nm, emission wavelength 496 nm, and autocutoff wavelength 495 nm were used for the substrate (e.g., Octanoyl-TFMCoumarin) containing 7-amino-4-tri-fluoromethyl coumarin.
Comparison of hydrolytic rates of different substrates by FAAH
To further compare the hydrolytic ratios of different substrates by FAAH, we incubated microsomal FAAH with different substrates at 37 °C for 90 min and recorded the hydrolysis using a 2-min interval. Assays were performed by the following procedures: a substrate solution (1 μL 10 mM in DMSO) was added into the protein-containing buffer (200 μL). This buffer consisted of 0.1 M, pH 7.4, phosphate buffer (or 0.1 M, pH 9.0, Tris–HCl buffer), 1% glycerol, 0.1% Triton X-100, 2.5% DMSO, and FAAH microsome (0.67 μg for Octanoyl-MP, Octanoyl-MMP, and Arachidonyl-MP; 100 μg for Octanoyl-TFM-Coumarin). The final concentration of the all examined substrates was 50 μM. The excitation and emission wavelengths of measurements were followed exactly as described above.
Kinetic study
The kinetic assays were also performed using the method described above for the sensitivity assay. Briefly, the total volume (201 μL) contained 0.1 M, pH 8.0, sodium phosphate buffer (190 μL) with 1% glycerol and 0.1% Triton X-100, protein solution (10 μL), and substrate solution (1 μL). The protein content was 0.67 μg per well (201 μL). The concentrations for Octanoyl-MP in DMSO were 200, 100, 50, 25, 12.5, 6.25, 3.13, 1.56, and 0.78 μM. Similarly, the concentrations of Arachidonyl-MP in DMSO were 100, 50, 25, 12.5, 6.25, 3.13, 1.56, 0.78, and 0.39 μM. Four replicates were used for each concentration. Reported kinetic data (Km and Vmax) were analyzed by the software Sigmaplot-Enzymatic Kinetics 2001 using the Michaelis–Menten equation.
Results and discussion
Optical properties
The molar extinction coefficient for absorption, fluorescence spectral profiles, and quantum yield of a fluorophore are the most important indexes determining its fluorescent intensity in the enzymatic reaction media. Due to deprotonization or protonization, aminopyridines (e.g., 2-aminopyridine) can display from over one (1.20) to almost zero (0.05) quantum yields in the different aqueous media [28]. Distilled water with 1% ethanol (dissolving the aminopyridines) was chosen as a medium for measuring the indexes described above. Results with distilled water as a medium (Table 1) indicated that these indexes varied greatly with the substituted group and position on the 3- or 5-aminopyridine. For example, the methoxy group located between the nitrogen on the pyridine ring and the carbon with an amino group of 3-aminopyridine (e.g., compound 7, Fig. 1) displayed a higher molar extinction coefficient but a much lower quantum yield than compound 6 with the same methoxy group and a different position (Fig. 1). The compounds 6 and 8 possessed quantum yields of almost unity (for efficient laser dyes it approaches unity) and redshifted excitation and emission wavelengths compared with those of other aminopyridines. However, the Stokes’ shift for all aminopyridines examined was almost the same. These interesting results provided us the impetus for further investigating their applications as fluorescent reporters of enzymatic activities.
Dependence of relative fluorescent intensity on the pH values
It is wellknown that hydroxide ions are fluorescent quenchers for aminopyridines and α- or β-naphthylamine [28,39]. This effect dramatically decreases in fluorescent intensity at maximal emission wavelength or quantum yields of aminopyridines from acid media to basic media. For example, the quantum yield of 3-aminopyridine drops from 1.07 in 0.1 N H2SO4 to 0.32 in water or to 0.03 in 1 N NaOH [28]. This may be one of the major limitations to the application of aminopyridines as fluorescent reporters of enzymatic activities.
To determine whether hydroxide ions are also fluorescent quenchers of simple substituted aminopyridines, we investigated the dependence on pH values using sodium phosphate buffer (pH 6.0–8.0) and Tris–HCl buffer (pH 9.0), in which the concentrations of hydroxide ions in buffer increase with the buffer pH values. Results with 5-amino-2-methoxypyridine (6) as an example (Fig. 2) indicated that (1) there was no redshift for either excitation or emission wavelength with an increase of pH from 6.0 to 9.0, (2) the relative fluorescent intensity of either excitation (only pH 8.0 is shown for clarity) or emission increased with an increase of pH value, (3) the relative fluorescent intensity of Octanoyl-MP emission at the maximal excitation wavelength of 6 was very low (less than 200 RFU), and (4) the profiles of excitation and emission wavelengths were completely separated. Similar properties were observed with compound 8 (data not shown here). It seems that hydroxide ions in these buffer systems do not strongly quench the fluorescence of 6 as it does on compound 1.
Fig. 2.

Fluorescent spectra of 5-amino-2-methoxypyridine (6) and its amide derivative (Octanoyl-MP). The dashed curve on the left side is the excitation spectrum of 5-amino-2-methoxypyridine (6) from 250 to 350 nm. This spectrum was determined by using 25 μM 6 in 0.1 M, pH 8.0, sodium phosphate buffer containing 1% ethanol at a fixed emission wavelength (396 nm) and room temperature. The five curves from top to bottom on the right side are curves of the emission spectra of the reporter 6 at pH 9, 8, 7, and 6 and N-(6-methoxypyridin-3-yl) octanamide (Octanoyl-MP) at pH 8, respectively. These five curves were determined at 25 μM final concentration of 6 or Octanoyl-MP in 0.1 M sodium phosphate buffer (pH 6–8) or Tris–HCl (pH 9.0) in 1-cm cuvette at a fixed excitation wavelength (302 nm) and room temperature.
Effects of protein amount on the relative fluorescent intensity
The quenching effect of proteins on the fluorescence of the reporters was studied. It is a common phenomenon that proteins quench the fluorescence of reporters. Data suggested that this effect is stronger for substituted aminopyridines than for coumarins. However, this quenching should not affect the sensitivity of fluorescent assays based on the substituted aminopyridines as fluorescent reporters. The detailed information on protein quenching is available in the Supplemental information.
Comparison of aqueous solubility of substrates
Substrate solubility in aqueous media is one of the important components when one considers a kinetic study. Although this may be easily solved by adding more cosolvent or detergents, some of the activities may be sacrificed or enzyme properties altered. Based on the catalytic properties displayed by FAAH for the panel of p-nitroaniline substrates, the length of acyl chain (C6 to C9) displayed a relatively strong enzyme–substrate interaction [40]. Thus, we chose compound C8 for comparison of substrates with p-nitroaniline, 7-amino-4-trifluoromethylcoumarin (9), and aminopyridines. With the same acid moiety, comparison of the aqueous solubility of pyridine substrates (e.g., Octanoyl-MP, Octanoyl-MMP) to a substrate with 7-amino-4-trifluoromethylcoumarin (9) suggested that pyridine substrates have much higher solubility in 0.1 M, pH 8.0, phosphate buffer (between 150 and 200 μM) than Octanoyl-TFMCoumarin (between 3.10 and 6.25 μM). This may partially contribute to the high substrate selectivity of FAAH described next. Partial characterization of FAAH suggested that expression of human FAAH was successful, and the FAAH protein was approximately 0.5 to 1% of the total microsome proteins (A more detailed discussion of the characterization of FAAH protein is in Supplemental information).
Comparison of the lowest detection limits of microsomal FAAH with different substrates
As described above, protein quenching and background noise resulting from some ingredients in proteins are two factors that contribute to the sensitivity of these assays. These aminopyridines have high molar extinction coefficients and quantum yields. However, another major factor determining sensitivity of assays of hydrolytic activities is the selectivity of hydrolases toward their substrates. For high-throughput assays, it is the ability to detect the lowest possible enzyme concentration with acceptable signal to noise ratio that determines practical sensitivity. The lowest detection limit of FAAH proteins for a particular substrate was determined from an assay with a signal after a 10 min kinetic measurement that was equal to three times that of the background noise. Results (Table 2) indicated that (1) the background noise or baselines varied with substrates, higher background for Octanoyl-MP and Octanoyl-MMP than for Octanoyl-TFMCoumarin and Arachidonyl-MP [may result from higher substrate concentration, i.e., 200 μM (Octanoyl-MP and Octanoyl-MMP) vs 50 μM (Octanoyl-TFMCoumarin and Arachidonyl-MP)]; (2) the lowest detection limits of FAAH proteins for Octanoyl-MP, Octanoyl-MM, and Arachidonyl-MP (0.55, 3.3, and 2.5 μg protein/mL, respectively) were determined but were not determinable for Octanoyl-TFMCoumarin and Octanoyl-p-NP even at 500 μg protein/mL in buffer system with 10% DMSO and 0.1% Triton X-100; and (3) the structure of reporters in substrates played a dramatic role in the sensitivity of assays for this hydrolase. For example, Octanoyl-TFMCoumarin with 7-amino-4-trifluoromethylcoumarin (9) was at least 900 times less sensitive than Octanoyl-MP with a 5-amino-2-methoxypyridine (6) reporter and 150 times less sensitive than Octanoyl-MMP with a 5-amino-2-methoxy-6-methylpyridine (8) reporter.
Table 2.
Comparison of the lowest detection limits of microsomal FAAH toward the colorimetric substrate (Octanoyl-p-NP) and fluorescent substrates (Octanoyl-MP, Octanoyl-MMP, Octanoyl-TFMCoumarin, and Arachidonyl-MP)a
| Substrate | Protein (μg/200 μL) | Signal (OD or RFU) | Average (OD or RFU) | Standard deviation | |
|---|---|---|---|---|---|
| Octanoyl-MP | 0.11 | Baseline | 310, 290, 296, 314 | 302 | 11 |
| Signal | 1083, 1126, 1069, 1086 | 1091* | 24 | ||
| Octanoyl-MMP | 0.67 | Baseline | 215, 161, 199, 201 | 194 | 23 |
| Signal | 760, 697, 694, 760 | 728* | 38 | ||
| Octanoyl-TFMCoumarin | 100 | Baseline | 134, 130, 124, 139 | 132 | 6 |
| Signal | 154, 150, 157, 182 | 161 | 15 | ||
| Octanoyl-p-NP | 100 | Baseline | 0.15, 0.16, 0.16, 0.16 | 0.16 | 0.01 |
| Signal | 0.16, 0.17, 0.18, 0.19 | 0.18 | 0.01 | ||
| Arachidonyl-MP | 0.5 | Baseline | 140, 140, 164, 140 | 148 | 11 |
| Signal | 566, 590, 534, 545 | 559* | 24 | ||
Assays were performed in a total volume (201 μL) containing 0.1 M, pH 8.0, sodium phosphate buffer with 1% glycerol and 0.1% Triton X-100 (190 μL), diluted protein solution (10 μL), and 1 μL substrate in ethanol (final concentration 200 μM for Octanoyl-MP and Octanoyl-MMP) or 1 μL substrate solution in DMSO (final concentration 25 μM for Octanoyl-TFMCoumarin, Octanoyl-p-NP, 50 μM for Arachidonyl-MP) at 37 °C. Absorbance wavelength for Octanoyl-p-NP was 382 nm. Excitation and emission wavelengths for Octanoyl-MP and Arachidonyl-MP were 302 and 396, for Octanoyl-MMP were 304 and 392, and for Octanoyl-MMP were 366 and 496, respectively.
Asterisks indicate that the datum point was equal to or over three times higher than the corresponding baseline.
Comparison of rates of hydrolysis of different substrates by FAAH
Four fluorescent substrates (Octanoyl-MP, Octanoyl-MMP, Octanoyl-TFMCoumarin, and Arachidonyl-MP) were chosen for comparison with 90-min incubation at 37 °C. Results (Figs. 3A and 3B) indicated that, under the same unit protein and same buffer system, the substrate hydrolysis by FAAH followed the same order: Octanoyl-MP ≫ Arachidonyl-MP > Octanoyl-MMP > Octanoyl-TFM-Coumarin. This order is in agreement with the results from the lowest detection limits of proteins described above. Although it seems that there was no hydrolysis for Octanoyl-TFMCoumarin at either pH 7.4 or 9.0 (Figs. 3A and 3B), the amount of the hydrolyzed Octanoyl-TFMCoumarin was very close to that of Arachidonyl-MP during the time course except for a 150-fold difference in amount of protein (0.67 μg proteins for Arachidonyl-MP and 100 μg proteins for Octanoyl-TFMCoumarin, respectively). The hydrolysis of Octanoyl-MP by FAAH did not follow a straight line during 90 min for either pH 7.4 (0.1 M phosphate buffer) or pH 9.0 (0.1 M Tris–HCl buffer), but it was a linear for the first 30 min. This assisted in the selection of the appropriate time for the following kinetic study. Although all substrates were hydrolyzed significantly faster at a higher pH value, the difference was very small (a 1.2-fold increase for Octanoyl-MP from pH 7.4 to pH 9.0).
Fig. 3.

Comparison of rates of hydrolysis of different substrates by FAAH. Assays were performed at 37 °C in 200 μL 0.1 M, pH 7.4, sodium phosphate buffer (A) or 0.1 M, pH 9.0, Tris–HCl buffer (B) containing 50 μM substrate, 1% glycerol, 0.1% Triton X-100, 3% DMSO, and microsomal protein (0.67 μg for Octanoyl-MP, Octanoyl-MMP, and Arachdonyl-MMP and 100 μg for Octanoyl-TFMCoumarin). The assays were recorded by a 2-min interval during 90 min. The excitation and emission wavelengths (nm) for Octanoyl-MP, Octanoyl-MMP, and Arachidonyl-MMP were 302 and 396, 304 and 392, and 302 and 396, respectively. Similarly, 366 and 496 were excitation and emission wavelengths (nm) for Octanoyl-TFMCoumarin.
Kinetic data
To further compare substrate selectivity with other substrates published in the literature, we conducted a kinetic study with Octanoyl-MP and Arachidonyl-MP. Results (Table 3) indicated that there was an eightfold difference between Octanoyl-MP and Arachidonyl-MP at Vmax under conditions of saturating substrate concentration, very close to the results (5 times) from the experiment determining the limit of detection of the proteins’ enzymatic activity (Table 2). Substrate selectivity of Octanoyl-MP was a little higher than that of Arachidonyl-MP (4.1 and 3.0 for Octanoyl-MP and Arachidonyl-MP, respectively). Although there is a difference in the percentage of FAAH protein in microsomes prepared from different laboratories and expression systems, this difference was negligible with respect to the substrate selectivity published in the literature (Table 3). Amazingly, the substrate selectivity of FAAH toward fluorescent substrates with a substituted aminopyridine as a reporter (e.g., Octanoyl-MP and Arachidonyl-MP) was over 50 times higher than that of the endogenous substrate (oleamide, Table 3) used in colorimetric assays or at least 25 times higher than that of fluorescent substrate with coumarin as a reporter (e.g., AAMCA, Table 3).
Table 3.
Comparison of kinetic data of FAAH toward different substrates
This amazing difference in sensitivity or substrate selectivity may result from a combination of multiple factors such as having higher aqueous solubility of the substrates and smaller reporters or forming electron donor or hydrogen bond interaction with the nitrogen of the pyridine ring. For example, the only difference between Octanoyl-MMP with a 5-amino-2-methoxy-6-methylpyridine (8) reporter and Octanoyl-MP with a 5-amino-2-methoxypyridine (6) reporter is one methyl group. The steric effects of the methyl group may partially block the nitrogen as electron donor or hydrogen bond, which may facilitate the hydrolysis by FAAH. This facilitation is possibly supported by the facts that (1) the sensitivity of FAAH toward Octanoyl-MMP is 6 times lower than that toward Octanoyl-MP and (2) the substrate selectivity of FAAH toward oleamide is over 50 times lower than that toward Octanoyl-MP or Arachidonyl-MP (Table 3). Of course, the effect of an increase in structural size from reporter 6 to 8 cannot be eliminated from these results. The steric effects of the reporters on the substrate selectivity are also supported by the difference between Octanoyl-MP and Octanoyl-TFMCoumarin (at least 900–fold difference, Table 2). A full explanation of the difference requires further investigation of structure–activity relationships.
Amidase activities of High Five Cells, Ac-hFAAH-infected cells, and human liver S9 microsomes were detected using these novel substrates. Results demonstrated that Arachidonyl-MP was the most selective substrate for Ac-hFAAH. They also indicated that, in addition to Ac-hFAAH, there must be another amide hydrolase that hydrolyzes long-chain fatty acid amides such as Octanoyl-MP and Octanoyl-MMP in human liver microsomes. Detailed evidence is placed in the accompanying Supplemental information.
Potential application of novel fluorescent reporters
Substituted aminopyridines display high fluorescence and represent a novel class of fluorescent reporters structurally different from aminocoumarins. They possess many advantages over aminocoumarins such as higher fluorescence, better aqueous solubility, and smaller size. In addition, as described above for FAAH, the nitrogen on the pyridine ring may work as an electron donor or may contribute a hydrogen bond and thus facilitate hydrolysis of the substrates by amide hydrolases. They are potentially useful for many enzymes that hydrolyze amide or carbamate bonds. These enzymes are not limited to FAAH but include nucleotide pyrophosphatase, transferases, hyaluronidase, numerous aminopeptidases, and other enzymes.
Conclusion
The relationship between structure of substituted aminopyridines and fluorescence was examined in this study. Results suggested that the substituted groups on the pyridine ring dramatically affected the fluorescence quantum yields of aminopyridines. A methoxy group para to the amino group in these aminopyridines greatly increased fluorescence quantum yields. Based on substrate selectivity, some novel fluorescent assays using substrates with aminopyridines as reporters were at least 50 times better than those assays (either fluorescent or colorimetric assays) published in the literature. Moreover, using novel fluorescent substrates, we found that there was at least one other amide hydrolase in addition to FAAH in human liver microsomes that could hydrolyze long chain fatty acid amides. In addition, the potential application of novel reporters was discussed.
Supplementary Material
Acknowledgments
We thank Drs. Paul D. Jones, Christophe Morisseau, and Mikeala Nichkova for some information during this study. This project was funded in part by NIEHS Grant R37 ES02710, NIEHS Superfund Grant P42 ES04699, and NIEHS Center grant P30 ES 05707.
Abbreviations used
- FAAH
fatty acid amide hydrolase
- BSA
bovine serum albumin
- RFU
relative fluorescent unit
- DMSO
dimethyl sulfoxide
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.ab.2006.10.041.
References
- 1.Mckinney MK, Cravett BF. Structure and function of fatty acid amide hydrolase. Annu Rev Biochem. 2005;74:411–432. doi: 10.1146/annurev.biochem.74.082803.133450. [DOI] [PubMed] [Google Scholar]
- 2.Maurelli S, Bisogno T, Petrocellis LD, Luccia AD, Marino G, Marzo VD. Two novel classes of neuroactive fatty acid amides are substrates for mouse neuroblastoma ‘anandamide amidohydrolase’. FEBS Lett. 1995;377:82–87. doi: 10.1016/0014-5793(95)01311-3. [DOI] [PubMed] [Google Scholar]
- 3.Boger DL, Fecik RA, Patterson JE, Miyauchi H, Patricelli MP, Cravett BF. Fatty acid amide hydrolase substrate specificity. Bioorg Med Chem Lett. 2000;10:2613–2616. doi: 10.1016/s0960-894x(00)00528-x. [DOI] [PubMed] [Google Scholar]
- 4.Rodríguez de Fonseca F, Navarro M, Gómez R, Escuredo L, Nava F, Fu J, Murillo-Rodríguez E, Giuffrida A, LoVerme J, Gaetani S, Kathuria S, Gall C, Piomelli D. An anorexic lipid mediator regulated by feeding. Nature. 2001;414:209–212. doi: 10.1038/35102582. [DOI] [PubMed] [Google Scholar]
- 5.Goparaju SK, Ueda N, Taniguchi K, Yamamoto S. Enzymes of porcine brain hydrolyzing 2-arachidonoylglycerol, an endogenous ligand of cannabinoid receptors. Biochem Pharmacol. 1999;57:417–423. doi: 10.1016/s0006-2952(98)00314-1. [DOI] [PubMed] [Google Scholar]
- 6.Cravatt BF, Demarest K, Patricelli MP, Bracey MH, Giang DK, Martin BR, Lichtman AH. Supersensitivity to anandamide and enhanced endogenous cannabinoid signaling in mice lacking fatty acid amide hydrolase. Proc Natl Acad Sci USA. 2001;98:9371–9376. doi: 10.1073/pnas.161191698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Massa F, Marsicano G, Hermann H, Cannich A, Monory K, Cravatt BF, Ferri G, Sibaev A, Storr M, Lutz B. The endogenous cannabinoid system protects against colonic inflammation. J Clin Invest. 2004;113:1202–1209. doi: 10.1172/JCI19465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cravatt BF, Saghatelian A, Hawkins EG, Clement AB, Bracey MH, Lichtman AH. Functional disassociation of the central and peripheral fatty acid amide signaling systems. Proc Natl Acad Sci USA. 2004;101:10821–10826. doi: 10.1073/pnas.0401292101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kathuria S, Gaetani S, Fegley D, Valino F, Duranti A, Tontini A, Mor M, Tarzia G, La Rana G, Calignano A, Giustino A, Tattoli M, Palmery M, Cuomo V, Piomelli D. Modulation of anxiety through blockade of anandamide hydrolysis. Nat Med. 2003;9:76–81. doi: 10.1038/nm803. [DOI] [PubMed] [Google Scholar]
- 10.Lictman AH, Shelton CC, Advani T, Cravatt BF. Mice lacking fatty acid amide hydrolase exhibit a cannabinoid receptor-mediated phenotypic hypoalgesia. Pain. 2004;109:319–327. doi: 10.1016/j.pain.2004.01.022. [DOI] [PubMed] [Google Scholar]
- 11.Walker JM, Huang SM, Strangman NM, Tsou K, Sanudo-Pena MC. Pain modulation by release of the endogenous cannabinoid anandamide. Proc Natl Acad Sci USA. 1999;96:12198–12203. doi: 10.1073/pnas.96.21.12198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cravatt BF, Lichtman AH. Fatty acid amide hydrolase: an emerging therapeutic target in the endocannabinoid system. Curr Opin Chem Biol. 2003;7:469–475. doi: 10.1016/s1367-5931(03)00079-6. [DOI] [PubMed] [Google Scholar]
- 13.Bisogno T, De Petrocellis L, Di Marzo V. Fatty acid amide hydrolase, an enzyme with many bioactive substrates: possible therapeutic implications. Curr Pharm Des. 2002;8:533–547. doi: 10.2174/1381612023395655. [DOI] [PubMed] [Google Scholar]
- 14.Cravatt BF, Demarest K, Patricelli MP, Bracey MH, Giang DK, Martin BR, Lichtman AH. Supersensitivity to anandamide and enhanced endogenous cannabinoid signaling in mice lacking fatty acid amide hydrolase. Proc Natl Acad Sci USA. 2001;98:9371–9376. doi: 10.1073/pnas.161191698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hohmann AG, Suplita RL, Bolton NM, Neely MH, Fegley D, Mangieri R, Krey JF, Walker JM, Holmes PV, Crystal JD, Duranti A, Tontini A, Mor M, Tarzia G, Piomelli D. An endocannabinoid mechanism for stress-induced analgesia. Nature. 2005;435:1108–1112. doi: 10.1038/nature03658. [DOI] [PubMed] [Google Scholar]
- 16.Deutsch DG, Chin SA. Enzymatic synthesis and degradation of anandamide, a cannabinoid receptor agonist. Biochem Pharmacol. 1993;46:791–796. doi: 10.1016/0006-2952(93)90486-g. [DOI] [PubMed] [Google Scholar]
- 17.Maccarrone M, Bari M, Finazzi-AgrÒ A. A sensitive and specific radiochromatographic assay of fatty acid amide hydrolase activity. Anal Biochem. 1999;267:314–318. doi: 10.1006/abio.1998.2964. [DOI] [PubMed] [Google Scholar]
- 18.Thumser AEA, Voysey J, Wilton DC. A fluorescence displacement assay for the measurement of arachidonoyl ethanolamide (anandamide) and oleoyl amide (octadecenoamide) hydrolysis. Biochem Pharmacol. 1997;53:433–435. doi: 10.1016/s0006-2952(96)00720-4. [DOI] [PubMed] [Google Scholar]
- 19.Wilson SJ, Lovenberg TW, Barbier AJ. A high-throughput-compatible assay for determining the activity of fatty acid amide hydrolase. Anal Biochem. 2003;318:270–275. doi: 10.1016/s0003-2697(03)00217-3. [DOI] [PubMed] [Google Scholar]
- 20.Qin C, Lin SY, Lang WS, Goutopoulos A, Pavlopoulos S, Mauri F, et al. Determination of anandamide amidase activity using ultraviolet-active amine derivatives and reverse-phase high-performance liquid chromatography. Anal Biochem. 1998;261:8–15. doi: 10.1006/abio.1998.2713. [DOI] [PubMed] [Google Scholar]
- 21.Patterson JE, Ollmann IR, Cravatt BF, Boger DL, Wong CH, Lerner RA. Inhibition of oleamide hydrolase catalyzed hydrolysis of the endogenous sleep-inducing lipid cis-9-octadecenamide. J Am Chem Soc. 1996;118:5938–5945. [Google Scholar]
- 22.Desarnaud F, Cadas H, Piomelli D. Anandamide amidohydrolase activity in rat brain microsomes—identification and partial characterization. J Biol Chem. 1995;270:6030–6035. doi: 10.1074/jbc.270.11.6030. [DOI] [PubMed] [Google Scholar]
- 23.Koutek B, Prestwich GD, Howlett AC, Chin SA, Salehani D, Akhavan N, Deutsch DG. Inhibitors of arachidonoyl ethanolamide hydrolysis. J Biol Chem. 1994;269:22937–22940. [PubMed] [Google Scholar]
- 24.Omeir RL, Chin S, Hong Y, Ahern DG, Deutsch DG. Arachidonoyl ethanolamide-[1, 2-14C] as a substrate for anandamide amidase. Life Sci. 1995;56:1999–2005. doi: 10.1016/0024-3205(95)00181-5. [DOI] [PubMed] [Google Scholar]
- 25.De Bank PA, Kendall DA, Alexander SPH. A spectrophotometric assay for fatty acid amide hydrolase suitable for high-throughput screening. Anal Biochem. 2005;69:1187–1193. doi: 10.1016/j.bcp.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 26.Ramarao MK, Murphy EA, Shen MWH, Wang YR, Bushell KN, Huang N, Williams C, Clark JD. A fluorescence-based assay for fatty acid amide hydrolase compatible with high-throughput screening. Anal Biochem. 2005;343:143–151. doi: 10.1016/j.ab.2005.04.032. [DOI] [PubMed] [Google Scholar]
- 27.Weisstuch A, Testa AC. A fluorescent study of aminopyridines. J Phys Chem. 1968;72:1982–1987. [Google Scholar]
- 28.Rusakowicz R, Testa AC. 2-Aminopyridine as a standard for low-wavelength spectrofluorimetry. J Phys Chem. 1968;72:2680–2681. [Google Scholar]
- 29.Anderson BM, Yuan JH, Vercellotti SV. Studies of 3-aminopyridine adenine dinucleotide phosphate. Mol Cell Biochem. 1975;8:89–96. doi: 10.1007/BF02116237. [DOI] [PubMed] [Google Scholar]
- 30.Kato H, Satomura S. Preparation of glycosyl amino acid derivatives as substrates for sugar transferases. 1994. JP 06065300. [Google Scholar]
- 31.Uozumi N, Teshima T, Yamamoto T, Nishikawa A, Gao YE, Miyoshi EJ, Gao CX, Noda K. A fluorescent assay method for GDP-L-Fuc: N-acetyl-β-D-glucosaminide α 1-6 fucosyltransferase activity, involving high performance liquid chromatography. J Biochem. 1996;120:385–392. doi: 10.1093/oxfordjournals.jbchem.a021424. [DOI] [PubMed] [Google Scholar]
- 32.Nakamura T, Majima M, Kubo K, Takagaki K, Tamura S, Endo M. Hyaluronidase assay using fluorogenic hyaluronate as a substrate. Anal Biochem. 1990;191:21–24. doi: 10.1016/0003-2697(90)90380-r. [DOI] [PubMed] [Google Scholar]
- 33.Nishikawa S, Kurono M, Shibayama K, Okuno S, Inagaki M, Kashimura N. Synthesis and cytokinin activity of fluorescent 7-phenylethynylimidazo[4,5-b]pyridine and its riboside. J Agric Food Chem. 2000;48:2559–2564. doi: 10.1021/jf0000225. [DOI] [PubMed] [Google Scholar]
- 34.Groziak MP, Wilson SR, Clauson GL, Leonard NJ. Fluorescent heterocyclic systems: syntheses, structures, and physicochemical properties of dipyrido-substituted 1, 3, 4, 6-tetraazapentalenes. J Am Chem Soc. 1986;108:8002–8006. [Google Scholar]
- 35.Nishi K, Huang H, Kamita SG, Kim I, Morisseau C, Hammock BD. Characterization of pyrethroid hydrolysis by human liver carboxylesterases hCE-1 and hCE-2. Arch Biochem Biophys. 2006;445:115–123. doi: 10.1016/j.abb.2005.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.López-Ferber M, Sisk WP, Possee RD. Baculovirus transfer vectors. In: Richardson CD, editor. Baculovirus Expression Protocols. Humana Press; Totowa, NJ, USA: pp. 25–63. [Google Scholar]
- 37.O’Reilly DR, Miller LK, Luckow VA. Baculovirus Expression Vectors: A Laboratory Manual. Oxford University Press; New York: 1992. [Google Scholar]
- 38.Horspool WH, Song PS, editors. CRC Handbook of Organic Photochemistry I. CRC Press; Boca Raton, FL: 1995. pp. 234–235. [Google Scholar]
- 39.Boaz H, Rollefson GK. The quenching of fluorescence derivations from the Stern-Volmer Law. J Am Chem Soc. 1950;72:3435–3443. [Google Scholar]
- 40.Patricelli MP, Cravatt BF. Characterization of manipulation of the acyl chain selectivity of fatty acid amide hydrolase. Biochemistry. 2001;40:6107–6115. doi: 10.1021/bi002578r. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


