Abstract
The human antibody b12 recognizes a discontinuous epitope on gp120 and is one of the rare monoclonal antibodies that neutralize a broad range of primary HIV-1 isolates. We previously reported the isolation of B2.1, a dimeric peptide that binds with high specificity to b12 and competes with gp120 for b12 antibody binding. Here, we show that the affinity of B2.1 was improved 60-fold over its synthetic-peptide counterpart by fusing it to the N-terminus of a soluble protein. This affinity, which is within an order of magnitude of that of gp120, probably more closely reflects the affinity of the phage-borne peptide. The crystal structure of a complex between Fab of b12 and B2.1 was determined at 1.8 Å resolution. The structural data allowed the differentiation of residues that form critical contacts with b12 from those required for maintenance of the antigenic structure of the peptide, and revealed that three contiguous residues mediate B2.1's critical contacts with b12. This single region of critical contact between the B2.1 peptide and the b12 paratope is unlikely to mimic the discontinuous key binding residues involved in the full b12 epitope for gp120, as previously identified by alanine scanning substitutions on the gp120 surface. These structural observations are supported by experiments that demonstrate that B2.1 is an ineffective immunogenic mimic of the b12 epitope on gp120. Indeed, an extensive series of immunizations with B2.1 in various forms failed to produce gp120 cross-reactive sera. The functional and structural data presented here, however, suggest that the mechanism by which b12 recognizes the two antigens is very different. Here, we present the first crystal structure of peptide bound to an antibody that was originally raised against a discontinuous protein epitope. Our results highlight the challenge of producing immunogens that mimic discontinuous protein epitopes, and the necessity of combining complementary experimental approaches in analyzing the antigenic and immunogenic properties of putative molecular mimics.
Keywords: HIV-1, neutralizing, antibody, b12, discontinuous epitope, peptide, B2.1, mimotope, structure, immunogenicity
Introduction
A major obstacle to HIV-1 vaccine design is the difficulty in generating a protective humoral immune response against the viral envelope proteins (Env), gp120 and gp41. One strategy to develop such a vaccine is to design immunogens that elicit antibodies similar to the rare human monoclonal antibodies (MAbs) that are effective at neutralizing a broad range of primary HIV-1 isolates (e.g., b12, 447-52D, 2G12, 2F5 and 4E10). However, the design of such immunogens is proving a significant challenge. The main barrier is the limitation in designing immunogens that faithfully reproduce the complex structural features of the viral epitopes that originally induced such neutralizing antibodies.
Antibody b12 was isolated from a phage-displayed Fab library that was produced from the bone marrow of an HIV-1 infected individual.1,2 Antibody b12 neutralizes a wide range of primary HIV-1 isolates from diverse geographic origins in vitro,1,2,3 and also protects against HIV-1 infection in passive immunization experiments in animals,4,5 whether injected intravenously,4,6,7 or applied in a topical gel.7 Antibody b12 recognizes an epitope overlapping the CD4-binding site of gp120. Ala substitution of surface-exposed residues on gp120 has demonstrated that b12 binds to a discontinuous epitope,8 which indicates that residues widely-separated in sequence, but not in space on the protein surface, are required for b12 binding. The crystal structure of intact IgG1 b12 was previously determined at 2.7 Å resolution,9 revealing a 15 Å vertical projection of the 18-residue complementarity determining region (CDR)-H3 ringed by two canyons, one of which is formed between CDRs L1, L3 and H3 and the other between CDRs H1, H2 and H3.10 Ala substitution studies on b12 have implicated CDRs H3 and L1 as regions for its interaction with gp120.11
Phage-displayed peptide libraries can be highly effective tools in selecting peptide ligands for antibodies against linear as well as discontinuous protein epitopes.12 Moreover, peptides selected from phage-display libraries with antibodies have acted as “immunogenic mimics” in eliciting cross-reactive13,14,15 antibody responses to several pathogens in various animal models; in some cases, these antibody responses have conferred neutralization in-vitro16,17 and protection18,19,20. By screening a panel of phage-displayed peptide libraries, we previously isolated and characterized a putative “peptide mimic”, termed B2.1, that is specific for b1221 and which effectively blocks the interaction of b12 and gp120. B2.1 is a homodimeric peptide, comprising two identical 18-residue peptide chains joined by a disulfide bridge. It does not react with non-neutralizing antibodies from sera derived from HIV-positive patients (unpublished data, X. Wang, C. Wang, and J.K.S.), suggesting that this peptide recognizes unique features of the neutralizing b12 paratope. In addition, Ala substitution studies on b12 revealed that a number of the mutations that affect binding to gp120 also influence binding to B2.1.11 Considering these findings, we explored the possibility that B2.1 mimics the b12 epitope on gp120 and might serve as an immunogenic mimic of the gp120 epitope.
Here, we elucidate the mechanism by which b12 recognizes the B2.1 peptide. Our previous study showed that the affinity of b12 for phage-borne B2.1 is probably much greater than its affinity for synthetic B2.1 peptide.21 In this study, we show that the dissociation constant at equilibrium (Kd) of b12 for B2.1 fused to the E. coli maltose binding protein (MBP) is between 20 and 60 nM, which is significantly lower than that of b12 for the synthetic peptide, with Kds between 2.5 and 6.9 μM. Thus, the affinity of B2.1-MBP fusion most likely reflects that observed for the phage-displayed B2.1 peptide. Alanine substitution studies revealed that 12 out of the 15 residues in the B2.1 sequence contribute to binding to b12, but only six of them are critical. The crystal structure of B2.1 in complex with the b12 Fab revealed that three of these critical binding residues (CBRs) form side-chain contacts with the b12 paratope, whereas the remaining three CBRs are required for the structural integrity of the peptide. Immunization studies with B2.1 in various forms failed to produce cross-reactive antibodies with gp120. Moreover, the fine specificity of the immune sera differed significantly from that of b12, since the sera produced different patterns of reactivity with Ala-substituted B2.1 peptides. Taken together, these results indicate that while B2.1 is bound by b12 with high affinity, it is a poor immunogenic mimic of the b12 epitope, most likely because it does not structurally mimic the full discontinuous epitope on gp120. Our results reveal caveats encountered in defining mechanisms of peptide mimicry of discontinuous protein epitopes, and their relevance to epitope and antibody-targeted vaccines.
Results
b12 affinity for B2.1 synthetic peptide and B2.1-MBP fusion protein
In previous work, we showed that the B2.1 sequence is functional only as a dimeric synthetic peptide or a phage-displayed recombinant protein dimer. Fab b12 binds tightly to B2.1 in ELISA if B2.1 is displayed in the context of a recombinant protein (i.e., displayed by fusion to the pVIII phage coat protein) or if B2.1 is adsorbed onto plastic as a biotinylated synthetic peptide; however, b12 Fab binds relatively poorly to biotinylated B2.1 synthetic peptide captured by streptavidin.21 We also reported a relatively weak in-solution affinity of b12 for B2.1 synthetic peptide as measured by a KinExA instrument.21
To probe the difference in binding of Fab b12 to B2.1 as a synthetic peptide versus B2.1 as a recombinant protein, we tested the affinity of the B2.1 sequence in a context that would be more akin to the phage-displayed, recombinant form. As B2.1 is displayed on phage as a disulfide-bridged fusion to the N-terminus of two pVIII molecules, we fused the B2.1 sequence to the N-terminus of MBP, and gel-purified it as a dimeric protein (B2.1-MBP) that is ostensibly formed via B2.1's disulfide bridge. The Kd of b12 IgG for B2.1-MBP were in agreement, being 20 nM and 60 nM as determined by surface plasmon resonance (SPR) and KinExA, respectively (Table 1). Agreement between the Kd values derived by different approaches was also observed for the B2.1 synthetic peptide; equilibrium and steady-state SPR analyses using peptide in solution produced Kds of 5.0 and 6.9 μM, respectively, compared to and in-solution Kd of 2.5 μM previously obtained with the KinExA instrument.21 Thus, the difference in b12's affinity B2.1 as an MBP fusion and a synthetic peptide is significant, being between 42 and 345 fold. Taken together, these results confirm our hypothesis that b12 binds with higher affinity to the B2.1 sequence displayed at the N-terminus of a protein, compared to its synthetic-peptide counterpart, even though both forms are dimers comprising two identical disulfide-bridged chains. Thus, we suppose that phage-borne B2.1, which was selected and optimized in the context of the phage,21 has an affinity closer to that of the B2.1-MBP fusion than to the B2.1 synthetic peptide.
Table 1.
Affinity constants (Kds) obtained for the B2.1 sequence as a synthetic peptide and as a fusion at the N-terminus of the maltose binding protein of E. coli (B2.1-MBP) as determined by a kinetic exclusion assay (KinExA) and surface plasmon resonance (SPR).
| Reactants1 | Method | Sequence | Kd (M) |
|---|---|---|---|
| B2.1 peptide- IgG2 | KinExA | NH3-HERSYMFSDLENRCIAAE-Orn(biotin)-KK-NH2 | 2.5 × 10−6 |
| B2.1 peptide- Fab | SPR, Equil/soln | NH3-HERSYMFSDLENRCIAAE-Orn(biotin)-KK-NH2 | 5.0 × 10−6 |
| Fab b12-B2.1 | SPR, Steady-state | NH3-HERSYMFSDLENRCIAAE-Orn(biotin)-KK-NH2 | 6.9 × 10−6 |
| IgG b12-B2.1/MBP | KinExA | NH3-HERSYMFSDLENRCIAAEE-MBP | 6.0 × 10−8 |
| IgG b12-B2.1/MBP | SPR, Kinetic | NH3-HERSYMFSDLENRCIAAEE-MBP | 2.0 × 10−8 |
For the kinetic and the steady-state methods in SPR, the first of the reactants in each pair listed is the one immobilized on the sensor chip, the second is the in-solution analyte that is passed over the immobilized ligand.
Previously published result (21).
Ala scanning identifies discontinuous CBRs on the B2.1 peptide
Single Ala substitutions were introduced into the phage-displayed B2.1 peptide sequence to identify residues that are critical or important for b12 binding. DNA sequencing confirmed the mutations, and SDS-PAGE showed that phage-associated levels of recombinant pVIII were roughly equivalent for all the Ala-substituted clones (data not shown). CBRs are defined here as those that, if substituted with Ala, decrease binding in phage-based ELISA to both IgG and Fab b12 of >90%. Important binding residues are defined as those whose substitution with Ala causes an >80% drop in binding to Fab without significantly affecting IgG binding.
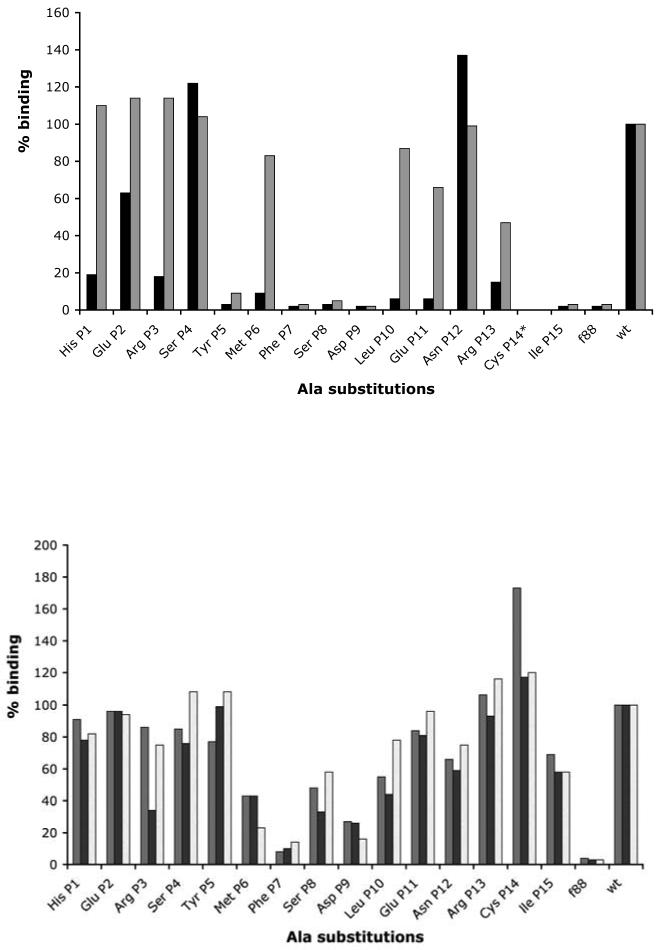
Ala substitutions within B2.1 (Figure 1(a)) revealed two sets of contiguous CBRs (Phe P7, Ser P8, Asp P9, and Cys P14, Ile P15), as well as Tyr P5. Other residues (Met P6, Leu P10, Glu P11 and Arg P13) had a significant effect on Fab binding, and, hence, are important but not critical. It is unlikely that all of the CBRs make Fab contacts. For example, Cys P1421 and other residues may play a unique role in providing a stable framework for the B2.1 structure. To understand the molecular basis of the B2.1-b12 interaction, we determined the crystal structure of the antibody-antigen complex.
Figure 1.
Alanine-substitution scanning of the B2.1 sequence. (a) Binding of IgG b12 (grey bars) and Fab b12 (black bars) to phage displaying Ala-substituted B2.1 sequences. (b) Binding of sera from each of three mice (grey, black, and white bars) immunized with B2.1 synthetic peptide-OVA conjugate. Results are expressed as % binding of each mutant phage with respect to wild-type (wt) B2.1 phage; f88 is a negative control for phage bearing only wild-type pVIII protein. *Ala replacement of the Cys residue was not included in this study, as previous work showed substitution of this Cys by Ser abrogates b12 binding.21
Antibody crystal structures indicate conservation of free and bound b12 Fab conformations
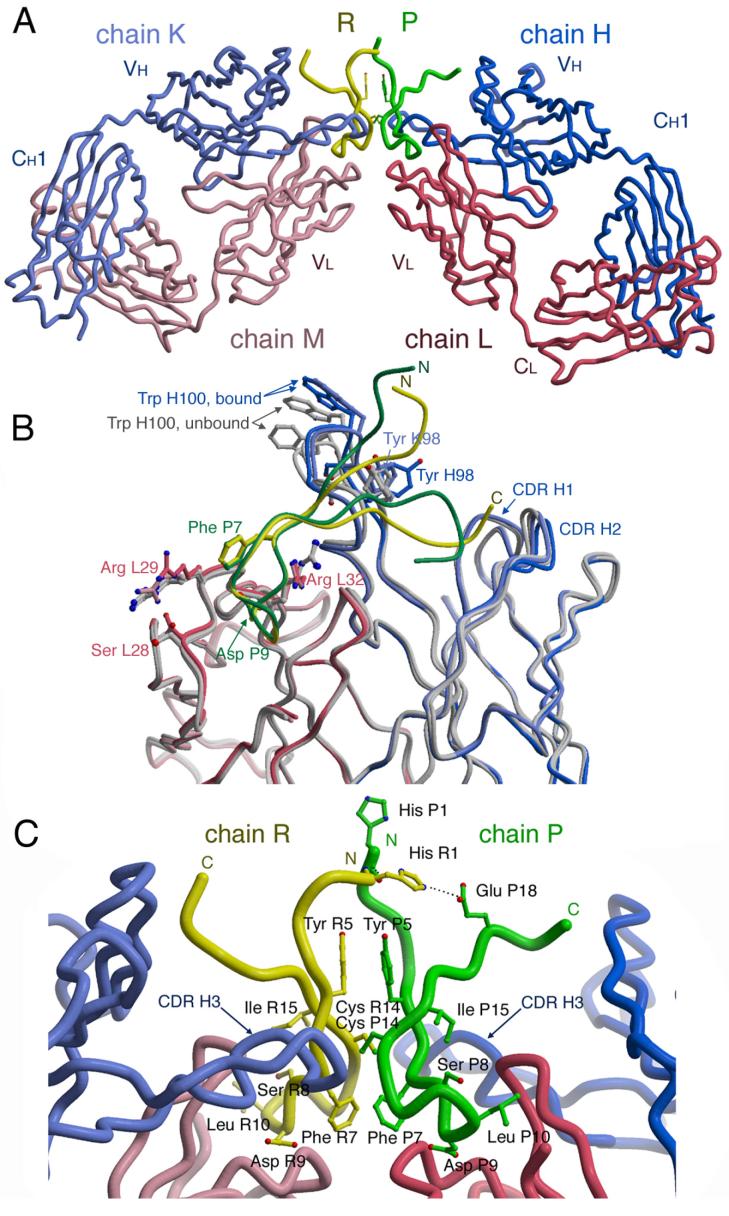
The b12-B2.1 structure was determined to 1.8 Å by molecular replacement using Fab regions of the IgG1 b12 structure10 as search models and refined to an Rcryst of 22.0 and an Rfree of 25.2 (Table 2). The asymmetric unit of the crystals consists of a single B2.1 molecule, comprising a disulfide-bridged homodimer (chains P and R), in simultaneous contact with two opposing Fab (chains L, H, and M, K) (Figure 2(a)). The two Fabs in the asymmetric unit are essentially identical in structure. In addition, the structures of the two Fab b12 regions in complex with B2.1 peptide are nearly identical to the unliganded b12 Fabs contained within the intact b12 IgG1.9,10 The only noticeable difference is for the CDR-H3s of the peptide-bound b12 structures, which have a slightly taller vertical projection than the equivalent CDR-H3s of the unbound b12 structures (0.6-3 Å root-mean-square deviation (rmsd) for the main chain, and 3-8 Å rmsd for all atoms, including the distal tip of the Trp H100 side chain; Figure 2(b)). Arg L29, which contacts Phe P7 of the peptide, and Arg L32, which contacts Asp P9, also adopt slightly different conformations in the bound and unbound structures, due to ligand interaction. In addition, Ser L28 adopts a different rotamer from the unbound structures, although it does not contact peptide (Figure 2(b)). Otherwise, the overall similarity in structures between the bound and free b12 Fabs (average rmsd. of 0.74 Å for all main-chain atoms in all possible combinations of free and bound forms) implies that no gross conformational changes accompany peptide recognition, and that the extended CDR-H3 maintains its rigidity upon B2.1 binding.
Table 2.
Summary of crystallographic data and refinement statistics.
| Data collection | |||
| Wavelength (Å) | 0.965 Å | ||
| Space group | P21 | ||
| Unit cell dimensions | a=51.6Å, b=184.4Å, c=56.2Å, β=103.1° | ||
| Resolution (Å)1 | 45-1.78 (1.81-1.78) | ||
| # reflections | 215,495 observed; 94,383 (4468) unique | ||
| Completeness1 (%) | 92.0 (87.1) | ||
| Rsym1,2 (%) | 6.0 (38.5) | ||
| I/σ(I)1 | 17.6 (1.5) | ||
| Refinement statistics | |||
| Number of reflections | 84,895 | ||
| Number in test set3 | 9419, 1967 | ||
| Rcryst4 (%) | 22.0 | ||
| Rfree5 (%) | 25.2 | ||
| Number of residues | 876 antibody, 41 peptide | ||
| Number of other atoms6 | 83 ligand atoms, 725 waters | ||
| Average B values (Å2) | |||
| Fab fragments | 31.3 | ||
| Peptide | 47.0 (35.7 in contact hairpin7) | ||
| Ramachandran plot (%) | Root mean square deviations (rmsd) | ||
| Most favored | 89.0 | Bond lengths (Å) | 0.005 |
| Additional allowed | 10.5 | Angles (°) | 1.4 |
| Generously allowed | 0.2 | Dihedral (°) | 26.7 |
| Disallowed8 | 0.2 | Improper (°) | 0.80 |
Numbers in parentheses refer to highest resolution shell.
Rsym = Σ| I - <I> | / |<I>|, where <I> is the mean intensity of a set of equivalent reflections
A 10% test set was used for initial rounds of refinement, and a 2.3% test set was used for the final round of refinement 56. Reflections in the final, smaller test set were also in the original, larger test set.
Rcryst = Σhkl |Fo-Fc|/ Σhkl |Fo|
Rfree was calculated as for Rcryst, but on the test set portion of the data excluded from the refinement 57.
14 atoms from a CAPS molecule, 48 atoms from glycerols, 20 atoms from sulfate ions, and one potassium atom.
The contact hairpin includes residues Ser P8 – Leu P10 and Ile P15.
Includes Val L51 of both Fab light chains. This residue exists in a well-defined γ turn in almost all antibody structures, but still continues to be designated by PROCHECK 58 as an outlier despite its inclusion in a well-defined and well-documented secondary structure.
Figure 2.
Crystal structure of the b12-B2.1 complex. (a) the B2.1 peptide comprises a homodimer formed by chains P (green) and R (yellow). Chain P (green) is bound to Fab LH, which is designated in the deposited PBD coordinates by heavy chain H (blue) and light chain L (red). Chain R (yellow) is bound to Fab MK, which is designated by heavy chain K (light blue) and light chain M (light pink). (b) Each of the two chains of the B2.1 peptide independently contacts one of the two b12 Fabs in the asymmetric unit. Key side chains of B2.1 are illustrated in ball-and-stick. The extended CDR H3s of the b12 antibody reach across each peptide monomer and do not contact the opposing chain. (c) Bound and unbound structures of Fab b12 are superimposed. The two unliganded Fabs from the IgG structure 9 are colored grey, whereas the two B2.1-bound Fabs are colored as in panels a and b. Only those side chains that adopt conformers different between free and bound Fab structures are represented. Trp H100 adopts similar conformers, but varies up to 8Å in position because of the 1-3 Å adjustment of the CDRH3 main chain.
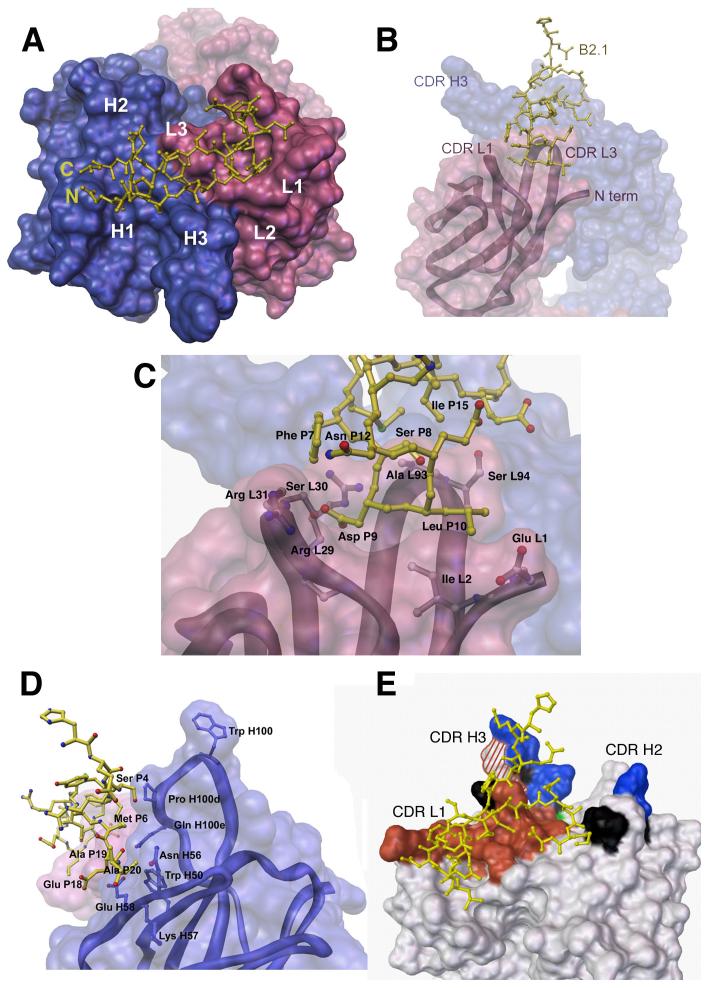
Approximately 610 Å2 of B2.1 peptide and 650 Å2 of b12 antibody molecular surface are buried by the interaction (as calculated with ms25 ).This interaction surface is larger than of that observed for most antibody-peptide complexes (on average, 480 Å2 of peptide and 550 Å of antibody surface),26,27,28 owing to the larger B2.1 peptide epitope (21 ordered residues), as compared to an average of 9-10 ordered residues in typical antibody-peptide structures in the Protein Data Bank. This value likely corresponds to about half of the putative b12 paratope as defined by other antibody-protein complexes, with no indication of conformational changes induced by binding to B2.1 peptide (i.e., no major “induced fit”).
Only a few contiguous peptide residues are critical for Fab12 interaction
Each polypeptide chain of the B2.1 peptide forms a hairpin structure, with the N-terminal arms of the two hairpins of the dimer extending roughly in parallel and the C-terminal arms extending away from each other (Figure 2c). The hairpin turn of each chain in the peptide consists of Phe 7, Ser P8, Asp P9, and Leu P10 and kinks away from the extended strands on either side at Phe P7. This region is identical in both peptide chains, P and R, with each turn region reaching into one the antigen-binding sites of adjacent Fabs. The interface between the two chains of B2.1 comprises its two Tyr P5 residues, which form aromatic face-to-face π-stacking interactions, and its two Cys P14 residues, which form an interchain disulfide bridge (Figure 2c and Supplementary Table 1). The B2.1 hairpin structure appears to be further stabilized by six hydrogen bonds within each peptide chain that connect the N and C termini. The B2.1 CBRs, Tyr P5, Cys P14, and Ile 15, are probably required to stabilize the peptide structure since they do not directly contact the antibody.
The N-terminal residues of B2.1 do not contact b12 until Ser P4, where each peptide chain begins to descend down along one side of CDR-H3 towards CDR-L1. Each chain then forms a hairpin, reverses direction, and extends across CDR-L3 to terminate at CDR-H2 (Figure 3(a)). The most significant region of peptide interaction is within the depression between CDRs L1, L3 and the light chain N terminus (Figure 3(b)). Here, Phe P7 forms a side-chain hydrophobic contact with Arg L29 (Figure 3 (c)) as well as hydrogen bonds between its main-chain carbonyl oxygen to the Arg L32 guanidinium nitrogens of CDR-L1 (Supplementary Table 1). Ser P8 contacts Ala L93 and Ser L94 indirectly via a water molecule. Asp P9 contacts Ile L28a, Arg L29 and Ser L30 of CDR-L1. Leu P10 contacts the N terminus of the light chain, although these contacts differ between peptide chains P and R due to structural differences between the flexible N termini of the antibody light chains. In peptide chain P, Leu P10 interacts with a side-chain carbon of Ile L2 (Figure 3(c)), whereas in peptide chain R, Leu R10 contacts a side-chain carbon of Glu M1 (Supplementary Table 1). N-terminal residues of the peptide mediate interaction with CDR-H3, where Met P6 contacts Gln H100e and Ser P4 abuts Pro H100d (Figure 3(d)). In addition, the side chain of Arg P13 hydrogen bonds to the main chain of Asp H100a and Asp H100b in CDR-H3, although this interaction is only observed for one of the two peptide chains. Various C, Cα, and Cβ atoms of the Orn-Lys-Lys C-terminal extension of B2.1 make hydrophobic contact with CDR-H2. However, these contacts are likely to result from crystal packing rather than specific binding, as they differ for each peptide chain and involve residues that were not part of the original peptide sequence selected by b12. Furthermore, a well-ordered 3-(cyclohexylamino)-1 propanesulfonic acid (CAPS) buffer molecule is also observed bound to CDR-H3 of Fab LH (Suplementary Data, Figure 1). This interaction is of interest as the H3 loop of the antibody is key for recognition of the CD4 binding site on gp120.10
Figure 3.
B2.1-b12 interactions. In all panels, only one chain (P) of the B2.1 peptide is shown bound to its respective Fab b12 (LH), with the light chain colored pink and the heavy chain blue in panels a-d. (a) Top view of the Fab b12 antigen-binding site with the bound B2.1 peptide chain illustrated in yellow ball-and-stick representation with the N and C termini of the chain indicated. The b12 combining site is illustrated as a molecular surface with the locations of the six CDRs (L1, L2, L3, H1, H2, and H3) indicated. (b)Side view of the b12 antibody-combining site. The B2.1 chain (P) dips into a canyon formed between CDRs L1, L3 and H3. (c) Specific contacts between B2.1 chain (P) and the Fab light chain (L) involve the residues Ser 8, Asp 9 and Leu 10 of the originally-selected, SDLX3CI consensus motif. Main chain carbonyls are not pictured for clarity. (d) Side view of the b12 antibody-combining site illustrating interactions of the B2.1 chain (P) (yellow) with b12 heavy chain (H) residues Pro H100d, Gln H100e, Trp 50, Asn H56, Lys 57, and Glu H58. Heavy chain contact residues are illustrated in blue in ball and stick representation. (e) Molecular surface representation of b12 showing the antibody areas important for binding to B2.1 and b12. Shaded areas indicate substitutions that: (i) decrease binding to both B2.1 and gp120, red; (ii) have opposite effects on B2.1 and gp120 binding, red and white striped; (iii) affect gp120 binding only, blue; (iv) affect B2.1 binding only, green; and (v) have no effect on either antigen, black. Mutagenesis data are from Zwick et al. (11).
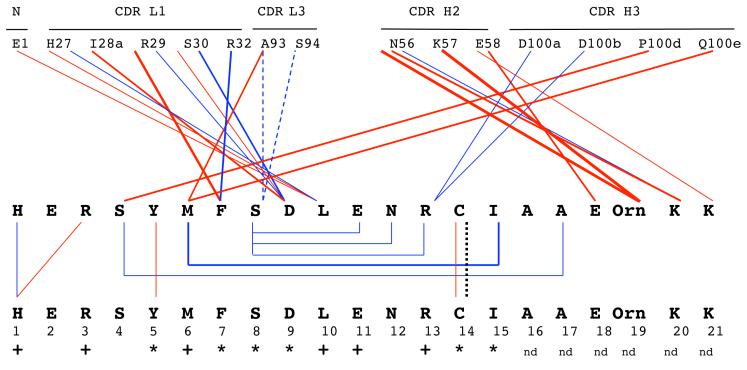
The Ala scanning data revealed that Tyr P5, Phe P7, Ser P8, Asp P9, Cys P14 and Ile P15 are critical for b12 binding, whereas other residues (Met P6, Leu P10, Glu P11 and Arg P13) were important. The principal contacts observed in the crystal structure are schematically represented in Figure 4, along with the classification of each B2.1 residue as critical, important or unimportant to b12 binding, according to the Ala substitution data in Figure 1(a). Taken together, the structural and Ala substitution results reveal the importance of both intra- and inter-peptide interactions for the functional activity of B2.1. Tyr P5 and Cys P14, which form peptide interchain contacts, and Ile P15, which forms peptide intrachain contacts, are all critical for binding to b12, even though they do not directly interact with the antibody. Likewise, His 1, Arg 3 and Glu 11 are important for maintaining the antigenic and dimeric structure of B2.1. Phe 7, Asp 9, and Leu 10 form critical or ‘important’ contacts with the antibody paratope, and Ser 8 does so via a water-mediated contact, whereas Met 6 and Arg 13 are important because of their antibody contact and/or because they contribute to the peptide structure.
Figure 4.
Schematic representation of direct contacts between one B2.1 peptide chain, R and b12 Fab, MK (top panel), and contacts between the two chains of a single B2.1 dimer (bottom panel). Atomic contact data are taken from Supplementary Table 2. Red lines represent hydrophobic contacts, blue lines represent hydrogen bonds, blue dashed lines represent interactions through a water molecule, and the dotted black line represents the disulfide bridge within each B2.1 molecule. Thinner lines represent a single pair of contacting atoms whereas thicker lines represent 2-4 atomic contacts per residue. Peptide residues critical for binding to b12 are indicated by *; residues important (but not critical) for binding are indicated by +, and nd is not determined.
Antibodies generated against B2.1 peptide do not cross-react with gp120
The B2.1 peptide is a specific marker for b12.21 Thus, it was of interest to determine if immunization with B2.1 could elicit a b12-like antibody response that would cross-react with gp120. BALB/c mice were subcutaneously immunized seven times with synthetic B2.1 peptide conjugated to ovalbumin (OVA). B2.1 was immunogenic in this context, producing an average half-maximum serum titer of ∼73,000 (Table 3), yet the sera reacted poorly with gp120Ba-L, even at low serum dilutions (i.e., titers 1:50). Immunization of BALB/c mice with recombinant B2.1 displayed on the surface of phage, or with synthetic B2.1 conjugated to f1-K phage, also produced a range of very strong anti-peptide titers with averages ranging from ∼4,000 to ∼100,000, respectively. However, these immunogens failed to elicit gp120 cross-reactivity (Table 3). Similar results were obtained using C57BL/6 mice (data not shown). One mouse, immunized with B2.1-expressing phage, produced a titer of ∼13,000 against the B2.1 peptide, yet failed to produce significant gp120 reactivity (data not shown). BALB/c mice immunized with the B2.1-f1.K conjugate produced somewhat higher anti-B2.1 peptide titers than those immunized with phage displaying the B2.1 sequence (Table 3), probably because of the increased B2.1 copy number (∼1200 copies per B2.1-f1.K phage compared to ∼200 copies per B2.1 phage). In conclusion, the B2.1 sequence did not elicit antibodies in mice that are functionally similar to b12.
Table 3.
Anti-gp120 and anti-B2.1 antibody responses in mice immunized with synthetic B2.1 peptide conjugated to ovalbumin (B2.1-OVA), peptide conjugated to f1.K phage (B2.1-f1.K) or recombinant phage bearing the B2.1 sequence (B2.1 phage).
| Immunogen | Anti-B2.1 peptide antibody titer1 |
Plate-bound Antigen | ||||
|---|---|---|---|---|---|---|
| BSA 1,2 (O.D.405-490) |
gp120 (O.D.405-490) |
OVA (O.D.405-490) |
||||
| dil. in BSA | dil. in milk | dil. in BSA | dil. in milk | |||
|
B2.1-OVA OVA |
73,510 (13,022)3 0 |
0.08 (0.02) 0.08 (0.02) |
0.18 (0.06) 0.16 (0.04) |
0.06 (0.00) 0.05 (0.00) |
n.d. n.d. |
n.d. n.d. |
|
B2.1-f1.K f1.K |
106,298 (34,564) 0 |
0.05 (0.01) 0.03 (0.00) |
0.14 (0.03) 0.11 (0.03) |
0.10 (0.02) 0.11 (0.03) |
0.23 (0.07) 0.05 (0.00) |
0.17 (0.09) 0.02 (0.00) |
|
B2.1-f1.K-Adj f1.K-Adj. |
112,935 (33,812) 0 |
0.09 (0.03) 0.08 (0.02) |
0.20 (0.02) 0.16 (0.05) |
0.12 (0.01) 0.11 (0.00) |
0.48 (0.17) 0.13 (0.03) |
0.36 (0.19) 0.04 (0.03) |
| B2.1 phage | 4,682 (2,294) | 0.17 (0.02) | 0.17 (0.02) | n.d. | n.d. | n.d. |
Serum samples were diluted 1:50 in TBS/Tween containing 1% BSA or 5% (w/v) non-fat dried milk.
Serum reactivities against BSA, gp120 and OVA are expressed as OD.405-490 since they were too weak to calculate a titer.
Standard errors are in parentheses.
A comparison of the binding of the anti-B2.1 sera and antibody b12 to the panel of B2.1 Ala substituted phage (Figure 1b) also supports this interpretation. Whereas Phe P7 seems to be important for binding to both b12 and the murine anti-B1.2 antibodies, significant differences in the effect of Ala substitutions were observed for critical residues, Tyr P5 and Cys P14, which maintain the B2.1's dimeric structure (the Tyr P5 residues of the B2.1 chains stack on each other, whereas its Cys P14 residues form a disulfide bridge that covalently connects the chains). Substitution of these two residues with Ala ablated binding to b12, but did not significantly affect the anti-B2.1 peptide reactivity of the immune mouse sera, indicating that most antibodies elicited by the B2.1/OVA conjugate in mice differ in their fine specificity from b12.
Discussion
The concept of molecular mimicry holds that an antigen mimic reproduces some or all of the important molecular contacts that an antigen makes in binding to its “cognate” antibody. In such instances, a common assumption is made that there is a connection between molecular mimicry and immunogenic mimicry, which supposes that the closer an immunogen comes to emulating the three-dimensional structure of the original epitope, the greater the likelihood that it will elicit antibodies that cross-react with that epitope. However, this assumption has largely been untested by structural comparisons of immunogenic mimics and their cognate epitopes in complex with their corresponding antibodies.
With the advent of peptide library technology, the concept of immunogenic mimicry has been applied to vaccine development with limited success. For example, immunogenic mimicry has been reported for cross-reactive peptides that bind to antibodies against carbohydrate20 and protein linear epitopes.17,18,19 However, the structural basis of such mimicry has not been analyzed. Importantly, most anti-protein antibodies recognize discontinuous epitopes,29,30,31,32 but as yet, unrelated, cross-reactive peptides for such antibodies have not been clearly shown to act as immunogenic mimics of their cognate antigens.
The structure presented here is the first example of a peptide bound to an antibody elicited against a discontinuous protein epitope. We have characterized the mode of peptide binding to b12, and have compared it to models proposed for b12-gp120 interaction based on extensive mutagenesis of b1211 and gp120.8 Our previous hypothesis was that B2.1 might resemble the D-loop of gp120, based on sequence homology,21 and the requirement of Asp 279 of the D-loop for binding to b12.9,11 However, our analysis of the structure of B2.1 synthetic peptide bound to Fab b12, together with the Ala substitution data on phage-displayed B2.1 indicate that, besides Arg 13, only a single cluster of residues, whose core comprises three contiguous CBRs (Phe 7, Ser 8, Asp 9), is in direct contact with b12 Fab. Thus, B2.1 “mimic” at most one to two sub-sites on gp120, and not the entire b12 epitope. Our immunization studies with the B2.1 peptide also support this conclusion in showing that B2.1 does not elicit detectable cross-reactivity with gp120. The recently-determined co-crystal structure of Fab b12 bound to a gp120 core has revealed that gp120 binds a site removed from the B2.1 binding site, and provides proof that the B2.1 peptide and gp120 interact with b12 by very different mechanisms (pers. comm., P. Kwong and T. Zhou, Vaccine Research Center, NIH, Bethesda, MD).
Ala substitution studies on Fab b12, particularly for light-chain CDR L1 residues Arg L29, Arg L31, and Arg L32, also suggested a shared mechanism of binding between B2.1 and gp12011 (see Fig. 3e and Supplementary Table 2). These residues are required for b12 binding to B2.1 via critical contacts with Phe P7 and Asp P9. The involvement of these residues in binding to gp120 is also now unclear, as CDR-L1 does not appear to contact gp120 (pers. comm., P. Kwong and T. Zhou). Our study has also revealed that Ala substitution of residues in b12 that do not contact B2.1, such as Asp100f, Asn100g, Tyr100h and Tyr100i of CDR-H3, can also affect binding of b12 to both B2.1 and gp12011 (Supplementary Table 2). These residues are most likely required to maintain the b12 structure, at least, for B2.1 peptide binding. We previously supposed that there must be some overlap in the binding sites for B2.1 and gp120, since there is reciprocal cross-reactivity between gp120 and the B2.1; each antigen can block 100% of b12 binding to the other.21 However, Figure 2(b) shows that the structure of CDR-H3 changes somewhat on Fab binding to B2.1; perhaps this or some other structural change induced by peptide binding affects the ability of b12 to bind gp120. Alternatively, the two antigen binding sites on b12 may be close enough or overlap sufficiently to sterically hinder the approach of the alternate antigen when the other ligand is already bound. Our present conclusion, that B2.1 does not mimic the b12-epitope on gp120 and binds to b12 by a different mechanism, is consistent with our recent results with three other MAbs, for which crystal structures of the original Fab-cognate antigen complexes showed that they also bind to discontinuous epitopes33,34,35 (unpublished data M. I., L. Craig, A. M., M. M. and J.K.S.). Nevertheless, in all four cases, the peptide ligands selected from phage display peptide libraries competed with the original cognate antigen for antibody binding. However, none of the peptide mimics elicited antibodies that were cross-reactive with their cognate antigens. The crystal structure of the Fab-peptide complex described here has allowed us to differentiate contact residues from those required to maintain the structure of the peptide. The structure has also elucidated that only a highly restricted portion of the B2.1 sequence directly contacts b12; therefore, B2.1 cannot reproduce the many critical contacts that b12 very likely makes with the gp120 epitope.
Although other immunogenic-mimic peptides have been described,13,14,15,16,17,18,19,20 nostructures of these peptides in complex with their antibodies have been determined. Thus, the degree to which they faithfully mimic the contacts that the corresponding cognate epitopes make with the same antibodies is unknown. Only one published study has tested the hypothesis that a peptide can mimic the gross structure and antibody contacts made by a cognate epitope.36 In this study, which involves a peptide that cross-reacts with a carbohydrate epitope, the corresponding crystal structures of the peptide and oligosaccharide individually complexed with the Fab of an anti-carbohydrate antibody showed that both ligands bind in the same overall region of the Fab, but by different structural mechanisms, but the Fab uses different residues to contact each antigen. Interestingly, while the intrinsic affinities of the Fab for each antigen differed by only two-fold, entropic vs. enthalpic contributions of the carbohydrate and peptide to binding were very different. Furthermore, another study compared complexes of the HIV-1 neutralizing antibody 2G12 with a phage-display-selected, gp120-cross-reactive peptide and with oligosaccharide fragments corresponding to the 2G12 epitope on gp120; these studies revealed only slight overlap in the binding footprints on 2G12 for the two antigens (A.M., D.A. Calarese, C.N. Scanlan, K.C. Chow, R. Kunert, R.L. Stanfield, H. Katinger, D.R.B., I.AW. and J.K.S., manuscript submitted). Thus, in those cases, as in the results presented here, it appears that, in screening peptide libraries, antibodies select peptides to fit their cognate paratope, or part of it, but may accomplish binding through very different contacts for peptide and cognate epitope. While these two studies did not test the peptide ligands for immunogenic mimicry, our immunization studies here with the B2.1 peptide clearly showed its failure to elicit gp120-binding antibodies.
In contrast, a recent study by Dorgham et al.37 describe b12-binding peptides, similar to B2.1, that behave as immunogenic mimics of gp120. Using the b12 antibody to screen a phage-displayed peptide library, the authors identified peptides bearing a binding motif, V(W/F)SD, which is similar to the one we previously described,21 and is shared by B2.1. Although b12 binding to free, or KLH-conjugated peptide was not observed, immunization of mice with whole phage bearing the peptide elicited weak gp120 cross-reactivity, but no HIV-1-neutralizing activity; moreover, the anti-peptide antibodies in the anti-phage sera were neither measured, nor tested for competition with the b12 antibody. These results37 contrast with the immunization results reported here, for which strong anti-peptide titers were, in fact, obtained, yet no appreciable gp120 cross-reactivity was observed. Moreover, this study showed that the critical binding residues in B2.1 that were recognized by the highest titer, anti-peptide sera were quite different from those recognized by the b12 antibody (Figure 1b). It is possible that the difference in gp120 reactivity of anti-peptide antibodies between the two studies is due to sequence difference that flank the shared, ‘(Aromatic) SD’ motif in these immunogens . Another possibility is that the contribution of the protein scaffold to the peptide structure is different for the two coat proteins to which the peptides are fused (pIII vs. pVIII). Nevertheless, the work of Dorgham et al.37 is of interest as it suggests that b12-binding peptides may be able to elicit some, albeit weak, gp120 cross-reactivity. However, in order to conclude that the serum antibodies, or cloned scFvs, produced by phage-immunized mice specifically bind the b12 epitope on gp120, it should be demonstrated that b12 blocks their cross-reactivity with gp120.
Our experience with b12 and several other antibodies that are known to bind discontinuous protein epitopes highlights the difficulties in using peptides to mimic this type of epitope. While cross-reactive, neutralizing and even protective antibody responses have been elicited by peptide “mimics” of linear and carbohydrate epitopes, the work presented here illustrates the necessity and value of direct structural evidence to elucidate the basis of antigenic and/or immunogenic mimicry of discontinuous protein epitopes.
Materials and Methods
Protein expression and peptide synthesis
IgG1 b12 was expressed in CHO-K1 cells as previously described.38 Fabs were obtained by papain cleavage and purified with protein A and protein G affinity chromatography.39 The B2.1 synthetic peptide comprises two identical disulfide-bridged polypeptide chains. The peptide used in crystallization experiments is > 95% pure as assessed by HPLC (AnaSpec, Inc., San Jose, CA), and its sequence is: (NH3+)-HERSYMFSDLENRCIAAE-Orn(biotin)-KK-(CONH2+). The biotinylated ornithine was incorporated into this peptide for detection in ELISAs, and the C-terminal two Lys residues for chemical conjugation. The B2.1 synthetic peptide used for conjugation to OVA and f1-K has the sequence: (NH3+)-HERSYMFSDLENRCIAAEGK-(CONH2+).57 The C-terminal Gly-Lys sequence was added for chemical conjugation.
Crystallization and data collection
Plate-like crystals were obtained from 17 mg/ml Fab, using a five-fold molar excess of B2.1 peptide, 1.7 M ammonium sulfate, 200 mM lithium sulfate, and 100 mM CAPS buffer, pH 10.5. The crystals are monoclinic P21 with two Fabs and one dimeric peptide per asymmetric unit (VM= 2.46 Å3/Da, 50% solvent) (Table 2). For cryoprotection, crystals were soaked in a solution of mother liquor containing increasing concentrations of glycerol (from 1-16%) and flash-cooled in liquid nitrogen. Data were collected at 100K to 1.75 Å resolution at SSRL beamline 11-1 (Table 2), and integrated and scaled with DENZO40 and SCALEPACK.40
Structure determination and refinement
Fab structures were determined by molecular replacement in AMoRe41 using the Fab fragments of the intact IgG1 b12 structure9 as search models. The Rcryst after molecular replacement was 37.4 for 15-4 Å data. Peptide and ligand molecules were built using the modeling programs TOM/FRODO42,43 and O,44 and the structure was refined to 1.8 Å using CNS45 (Table 2). The electron density was clearly interpretable for residues L1-L214 of antibody light chain L, residues H1-H128 and H137-H228 of antibody heavy chain H, and in the second Fab for residues M1-M214 of light chain M and residues K1-K127 and K136-K230 of heavy chain K. Heavy chain residues 127-137 comprise a solvent exposed loop in the constant region of the Fab that is frequently disordered in antibody structures.46 Clear electron density was observed in the antibody-combining site for the first 18 residues of the 21-mer-peptide sequence, whereas the density for Lys P21 (peptide chain P), Orn R19 and Lys R20 (peptide chain R) was only interpretable for the polypeptide backbone. These residues are not present on the original phage-displayed sequence, and have been built into the crystal structure as alanines. The biotin moiety is not defined in the electron density maps. Coordinates and structure factors have been deposited in the Protein Data Bank (http://www.rcsb.org/pdb/, accession code 1N0X). Figures 2 was created using Molscript.47 Figure 3 was created in PMV,48 with molecular surfaces generated using a 1.4 Å probe radius in msms.49
Site-directed mutagenesis of B2.1 phage
All residues of the B2.1 peptide, with the exception of the cysteine, were singly replaced by alanine in the context of B2.1. Alanine substitution of the Cys 14 residue on B2.1 phage has been previously described.21 Single-stranded DNA from phage B2.1 was used as template for site-directed mutagenesis using a set of fourteen oligonucleotides as previously described.50
Direct ELISA
To assess binding of b12 to B2.1 mutant phage in an ELISA,21 microwells were coated overnight at 4 °C or for 4 hours at room temperature with 1010 phage particles in Tris-buffered saline, pH 7.4 (TBS), blocked for one hour at room temperature with TBS containing 2% (w/v) bovine serum albumin (BSA), washed six times with TBS containing 0.1% (v/v) Tween 20 (TBS/Tween), and incubated with either 0.1 nM IgG b12 or 10 nM biotinylated Fab b1221 in TBS/Tween containing 1% BSA for two hours at room temperature. After washing, microwells were incubated for one hour at room temperature with one of two horseradish peroxidase (HRP) conjugates diluted 1:1000 in TBS/Tween: bound IgG was incubated with Protein A/G-HRP (Pierce Chemical Co., Rockford, IL) whereas bound biotinylated Fab was incubated with NeutrAvidin-HRP (Pierce). Plates were washed six times, and bound HRP was detected by the addition of ABTS solution (400 μg/ml 2'2'-azino-bis (3-ethylbenzthiazoline-6-sulfonic acid) in citrate/phosphate buffer containing 0.03% (v/v) H2O2), followed by incubation at room temperature for periods up to 45 minutes. Absorbance at 405 nm and 490 nm was measured using a Versamax microplate reader (Molecular Devices, Sunnyvale, CA) and reported as OD405-490. To verify the amount of phage bound to wells, replicate phage-coated wells were reacted with 1 ug/ml purified rabbit anti-phage IgG in TBS/Tween containing 1% BSA. After six washes, bound rabbit IgG was incubated for one hour with Protein A/G-HRP conjugate, and after six washes, bound HRP was detected with ABTS solution. The relative binding of b12 IgG and Fab to phage mutants bearing alanine substitutions was calculated as percent binding with respect to B2.1 wild-type phage (100%).
Sera from immunized mice were diluted in TBS/Tween and reacted with immobilized antigen: B2.1 synthetic peptide, phage-bearing the B2.1 sequence or single amino-acid substitutions in B2.1, gp120 or OVA, or BSA-blocked wells as a negative control. ELISAs were performed essentially as described above, except that 1 μg OVA, 200 ng B2.1 synthetic peptide, or 100 ng gp120 (Ba-L isolate, kindly provided by T. Fouts, Institute of Human Virology, MD) were among the antigens adsorbed to microplate wells. Mouse sera were serially diluted in TBS/Tween containing 1% BSA, added to BSA-blocked microwells, and incubated for two hours at room temperature. The wells were washed six times with TBS/Tween and incubated with goat (anti-mouse IgG (Fc))-HRP conjugate (Pierce) diluted 1:2000 in TBS/Tween. Serum reactivity with B2.1 synthetic peptide and for OVA was reported as an IgG titer (i.e., the inverse of the dilution producing the half-maximal OD405-490 signal on antigen). Serum IgG reactivity with BSA, gp120 and OVA was weak, and therefore reported as the OD405-490 produced by a 1:50 serum dilution. All ELISA data are representative of two or more independent experiments.
B2.1-MBP production and affinity measurements
The MBP-fusion protein was prepared as described,51 and dimer was isolated and further purified by preparative SDS-PAGE as described by Castellanos-Serra et. al.52 The affinity at equilibrium of IgG1 b12 for B2.1-MBP was measured using a KinExA 3000 instrument (Sapidyne Instruments, Boise, ID) as described,53 and by SPR using a Biacore 3000 instrument (Biacore AB, Uppsala, Sweden; instrument located at the Biothermodynamics Hub, Laboratory of Molecular Biophysics, Univ. of BC, Vancouver). Several methods for SPR analysis were used to validate the results. The affinity of b12 for B2.1 synthetic peptide in solution was determined by a binding-inhibition procedure54,55 (which measures Kd in solution at equilibrium) as described.56 For solution measurements using the Biacore and KinExA instruments, independent reactions comprising a fixed concentration of antibody and different concentrations of antigen were allowed to reach equilibrium for 12-16 hours; the free antibody concentration was then determined for each reaction, and the Kd calculated using the in-solution affinity models of KinExA and BiaEvaluation 3.1 software. For the SPR kinetic method, several analyte concentrations were injected over immobilized b12 antibody. The B2.1-MBP data were fitted to the 1:1 Langmuir binding model. The B2.1 peptide binding curves plateaued quickly to steady-state and, thus, this Kd was calculated using the BiaEvaluation 3.1 steady-state model.
Coupling of synthetic B2.1 peptide to f1.K phage, and OVA
A detailed description of: i) f1.K (an engineered f1 filamentous phage carrying an additional Lys near the N-terminus of every copy of the pVIII major coat protein), ii) the cross-linking protocol for coupling B2.1 synthetic peptide to f1.K phage or OVA, and iii) the immunization protocols, are provided in van Houten et al.57 Briefly, the B2.1-f1.K phage conjugate was prepared by mixing 8.6 × 1012 particles/ml f1.K phage, 2 mg/ml synthetic B2.1 peptide, and 3 mg/ml Bis(sulfosuccinimidyl)suberate (BS3) cross-linker (Pierce) in a total volume of 1.4 ml PBS, pH 7.4. Conjugation mixtures were rotated slowly at room temperature for one hour, and quenched with 0.1 vol 1M Tris-HCl, pH 7.4. The phage/peptide conjugates were precipitated with 0.15 vol PEG/NaCl, incubated overnight at 4°C, and centrifuged at 13,000 × g for 40 min at 4°C. To remove PEG/NaCl, conjugate pellets were resuspended in 12 ml sterile PBS, and transferred to a 12 ml polyallomer quick-seal tube (Beckman Coulter Inc., Fullerton, CA) that was placed in a 70 Ti.1 rotor. Following ultracentrifugation at 50,000 rpm in a L8-80 ultracentrifuge (Beckman Coulter), the conjugate pellet was resuspended in 1 ml PBS. Aliquots were removed and analyzed for DNA content by electrophoresis on a 0.8% agarose gel in 4 × GBB.58
The B2.1-OVA conjugate was prepared by mixing 298 μg OVA with 2.0 mg B2.1 peptide, and 2.8 mg BS3 cross-linker in 1 ml PBS, pH 7.4. The conjugation reaction was treated as described above, except that the conjugates were washed three times using a Centricon-30 ultrafiltration device (Amicon, Inc., Beverly, MA) rather than being precipitated by PEG, then pelleted by ultracentrifugation.
Immunization of mice with B2.1 peptide-bearing immunogens
As described in van Houten et al.,57 groups of five eight-week old female BALB/c mice were immunized by intraperitoneal injection with either 100 μg recombinant B2.1 phage, 10 μg B2.1-OVA conjugate or 10 μg OVA. All samples were diluted in a total volume of 100 μl PBS. No adjuvant was used. For all groups, the mice were immunized on days 0, 14, 28, 42, 63, 98 and 119. The mice were bled from the tail vein on days 0, 14, 28, 42, and 63 just prior to immunization and also on day 77. Subsequent tail bleeds were not performed due to tail scarring. The final bleeds for mice immunized with the B2.1-OVA conjugate or OVA were performed on day 140 by cardiac puncture under CO2anesthesia. The final bleed for the mice immunized with B2.1 phage was performed on day 77. After collection, each blood sample was heated to 37°C for 60 min, allowed to clot overnight at 4°C, and centrifuged at 12,000 × g for 15 min. Serum samples were then transferred to fresh microfuge tubes and stored at −20°C.
In a separate study, groups of four eight-week old female BALB/c mice received a total of 10 μg B2.1-f1.K conjugate or untreated f1.K phage via subcutaneous injection at two different sites. Immunogens were injected in a total volume of 100 μl PBS with or without adjuvant. Immunizations were given on days 0, 21, 42, 63, 92, 147, and 210. Tail bleeds were conducted on day 0 and two weeks after each immunization (days 14, 35, 56, and 105). Bleeds were not taken after the fourth and the sixth immunizations due to tail scarring. The final bleed was taken on day 224. After collection, sera were prepared as described above.
Supplementary Material
Acknowledgements
We thank the staff of SSRL Beamline 11-1, and A. Hessell, O. Pan, S. Bahr and B. Vanderkist for excellent technical assistance, and T. Fouts (Institute of Human Virology, Baltimore, MD) for providing HIV-1 Ba-L gp120. This work was supported by NIH grants GM46192 to IAW, AI33292 to DRB, AI49111 to JKS, and AI40377 to PWHIP, the University wide AIDS Research Program (EOS), a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund (EOS), the Elizabeth Glaser Pediatric AIDS Foundation (MBZ), The Michael Smith Foundation for Health Research (MM and AM) and the Natural Sciences and Engineering Research Council of Canada (MM and AM), the Skaggs Institute for Chemical Biology (IAW and EOS), and the International AIDS Vaccine Initiative (IAW and DRB) and the Canada Research Chair Program (JKS) for support. We are grateful to Peter Kwong and Tongqing Zhou (Vaccine Research Center, NIH) for kindly sharing their structural results with us before publication. This is publication No. 15160-MB from The Scripps Research Institute.
Abbreviations used
- HIV-1
human immunodeficiency virus type 1
- Env
viral envelope protein
- MAb
monoclonal antibody
- CDR
complementarity determining region
- MBP
maltose binding protein of E. coli
- CBRs
critical binding residues
- Kd
dissociation constant at equilibrium
- SPR
surface plasmon resonance
- rmsd
root mean square deviation
- CAPS
3-(cyclohexylamino)-1-propanesulfonic acid
- OVA
ovalbumin
- TBS
Tris-buffered saline, pH 7.4
- BSA
bovine serum albumin
- TBS/Tween
TBS containing 0.1% Tween 20
- HRP
horseradish peroxidase
- ABTS
2'2'-azino-bis (3-ethylbenzthiazoline-6-sulfonic acid)
- PEG/NaCl
16.7% (w/v) polyethylene glycol 8000, 3.3 M sodium chloride
Footnotes
Data deposition: The atomic coordinates for the b12-B2.1 complex have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1NOX).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Burton DR, Pyati J, Koduri R, Sharp SJ, Thornton GB, Parren PW, Sawyer LS, Hendry RM, Dunlop N, Nara PL, Lamacchia M, Garratty E, Richard ES, Bryson YJ, Cao Y, Moore JP, Ho DD, Barbas CF., 3rd Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- 2.Kessler JA, 2nd, McKenna PM, Emini EA, Chan CP, Patel MD, Gupta SK, Mark GE, Barbas CF, 3rd, Burton DR, Conley AJ. Recombinant human monoclonal antibody IgG1b12 neutralizes diverse human immunodeficiency virus type 1 primary isolates. AIDS Res. Hum. Retroviruses. 1997;13:575–582. doi: 10.1089/aid.1997.13.575. [DOI] [PubMed] [Google Scholar]
- 3.Trkola A, Pomales AB, Yuan H, Korber B, Maddon PJ, Allaway GP, Katinger H, Barbas CF, 3rd, Burton DR, Ho DD, Moore JP. Cross-clade neutralization of primary isolates of human immunodeficiency virus type 1 by human monoclonal antibodies and tetrameric CD4-IgG. J. Virol. 1995;69:6609–6617. doi: 10.1128/jvi.69.11.6609-6617.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parren PWHI, Ditzel HJ, Gulizia RJ, Binley JM, Barbas CF, 3rd, Burton DR, Mosier DE. Protection against HIV-1 infection in hu-PBL-SCID mice by passive immunization with a neutralizing human monoclonal antibody against the gp120 CD4-binding site. AIDS. 1995;9:F1–6. doi: 10.1097/00002030-199506000-00001. [DOI] [PubMed] [Google Scholar]
- 5.Gauduin MC, Parren PW, Weir R, Barbas CF, 3rd, Burton DR, Koup RA. Passive immunization with a human monoclonal antibody protects hu-PBL-SCID mice against challenge by primary isolates of HIV-1. Nat. Med. 1997;3:1389–1393. doi: 10.1038/nm1297-1389. [DOI] [PubMed] [Google Scholar]
- 6.Parren PWHI, Marz PA, Hessell AJ, Luckay A, Harouse J, Cheng-Mayer C, Moore JP, Burton DR. Antibody protects macaques against vaginal challenge with a pathogenic R5 simian/human immunodeficiency virus at serum levels giving complete neutralization in vitro. J. Virol. 2001;75:8340–8347. doi: 10.1128/JVI.75.17.8340-8347.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Veazey RS, Shattock RJ, Pope M, Kirijan JC, Jones J, Hu Q, Ketas T, Marx PA, Klasse PJ, Burton DR, Moore JP. Prevention of virus transmission to macaque monkeys by a vaginally applied monoclonal antibody to HIV-1 gp120. Nat. Med. 2003;9:343–346. doi: 10.1038/nm833. [DOI] [PubMed] [Google Scholar]
- 8.Pantophlet R, Saphire EO, Poignard P, Parren PWHI, Wilson IA, Burton DR. Fine mapping of the interaction of neutralizing and non-neutralizing monoclonal antibodies with the CD4 binding site of human immunodeficiency virus type I gp120. J. Virol. 2003;77:642–658. doi: 10.1128/JVI.77.1.642-658.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saphire EO, Parren PWHI, Pantophlet R, Zwick MB, Morris GM, Stanfield RL, Rudd PM, Dwek RA, Burton DR, Wilson IA. Crystal structure of an intact human IgG with broad and potent activity against primary HIV-1 isolates: A template for HIV vaccine design. Science. 2001;293:1155–1159. doi: 10.1126/science.1061692. [DOI] [PubMed] [Google Scholar]
- 10.Saphire EO, Stanfield RL, Crispin MD, Rudd PM, Dwek RA, Parren PWHI, Burton DR, Wilson IA. Contrasting IgG structures reveal extreme asymmetry and flexibility. J. Mol. Biol. 2002;309:9–18. doi: 10.1016/S0022-2836(02)00244-9. [DOI] [PubMed] [Google Scholar]
- 11.Zwick MB, Parren PWHI, Saphire EO, Church S, Wang M, Scott JK, Dawson PE, Wilson IA, Burton DR. Molecular features of the broadly neutralizing antibody b12 required for recognition of HIV-1 gp120. J. Virol. 2003;77:6965–6978. doi: 10.1128/JVI.77.10.5863-5876.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Irving MB, Pan O, Scott JK. Random-peptide libraries and antigen-fragment libraries for epitope mapping and the development of vaccines and diagnostics. Curr. Opin. Chem. Biol. 2001;5:314–324. doi: 10.1016/S1367-5931(00)00208-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Puntoriero G, Meola A, Lahm A, Zucchelli S, Ercole BB, Tafi R, Pezzanera M, Mondelli MU, Cortese R, Tramontano A, Galfre G, Nicosia A. Towards a solution for hepatitis C virus hypervariability: mimotopes of the hypervariable region 1 can induce antibodies cross-reacting with a large number of viral variants. EMBO J. 1998;17:3521–3533. doi: 10.1093/emboj/17.13.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stoute JA, Ballou WR, Kolodny N, Deal CD, Wirtz RA, Lindler LE. Induction of humoral immune response against Plasmodium falciparum sporozoites by immunization with a synthetic peptide mimotope whose sequence was derived from screening a filamentous phage epitope library. Infect. Immun. 1995;63:934–939. doi: 10.1128/iai.63.3.934-939.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mattioli S, Imberti L, Stellini R, Primi D. Mimicry of the immunodominant conformation-dependent antigenic site of hepatitis A virus by motifs selected from synthetic peptide libraries. J. Virol. 1995;69:5294–5299. doi: 10.1128/jvi.69.9.5294-5299.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chargelegue D, Obeid OE, Hsu SC, Shaw MD, Denbury AN, Taylor G, Steward MW. A peptide mimic of a protective epitope of respiratory syncytial virus selected from a combinatorial library induces virus-neutralizing antibodies and reduces viral load in vivo. J. Virol. 1998;72:2040–2046. doi: 10.1128/jvi.72.3.2040-2046.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scala G, Chen X, Liu W, Telles JN, Cohen OJ, Vaccarezza M, Igarashi T, Fauci AS. Selection of HIV-specific immunogenic epitopes by screening random peptide libraries with HIV-1-positive sera. J. Immunol. 1999;162:6155–6161. [PubMed] [Google Scholar]
- 18.Steward MW, Stanley CM, Obeid OE. A mimotope from a solid-phase peptide library induces a measles virus-neutralizing and protective antibody response. J. Virol. 1995;69:7668–7673. doi: 10.1128/jvi.69.12.7668-7673.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu MW, Scott JK, Fournier A, Talbot PJ. Characterization of murine coronavirus neutralization epitopes with phage-displayed peptides. Virology. 2000;271:182–196. doi: 10.1006/viro.2000.0310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prinz DM, Smithson SL, Westerink MA. Two different methods result in the selection of peptides that induce a protective antibody response to Neisseria meningitidis serogroup C. J. Immunol. Methods. 2004;285:1–14. doi: 10.1016/j.jim.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 21.Zwick MB, Bonnycastle LL, Menendez A, Irving MB, Barbas CF, 3rd, Parren PW, Burton DR, Scott JK. Identification and characterization of a peptide that specifically binds the human, broadly neutralizing anti-human immunodeficiency virus type 1 antibody b12. J. Virol. 2001;75:6692–6699. doi: 10.1128/JVI.75.14.6692-6699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dodson E, Kleywegt GJ, Wilson K. Report of a workshop on the use of statistical validators in protein x-ray crystallography. Acta Crystallogr. sect. D. 1996;52:228–234. doi: 10.1107/S0907444995010638. [DOI] [PubMed] [Google Scholar]
- 23.Brünger AT. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–474. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 24.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 25.Connolly ML. Solvent-accessible surfaces of proteins and nucleic acids. Science. 1983;221:709–713. doi: 10.1126/science.6879170. [DOI] [PubMed] [Google Scholar]
- 26.Wilson IA, Stanfield RL. Antibody-antigen interactions. Curr. Opin. Struct. Biol. 1993;3:113–118. doi: 10.1016/0959-440x(94)90267-4. [DOI] [PubMed] [Google Scholar]
- 27.Stanfield RL, Wilson IA. X-ray crystallographic studies of antibody-peptide complexes. Immunomethods. 1993;3:211–221. [Google Scholar]
- 28.MacCallum RM, Martin AC, Thornton JM. Antibody-antigen interactions: contact analysis and binding site topography. J. Mol. Biol. 1996;262:732–745. doi: 10.1006/jmbi.1996.0548. [DOI] [PubMed] [Google Scholar]
- 29.Benjamin DC, Berzofsky JA, East IJ, Gurd FR, Hannum C, Leach SJ, Margoliash E, Michael JG, Miller A, Prager EM, Reichlin M, Sercarz EE, Smith-Gill SJ, Todd PE, Wilson AC. The antigenic structure of proteins: a reappraisal. Ann. Rev. Immunol. 1984;2:67–101. doi: 10.1146/annurev.iy.02.040184.000435. [DOI] [PubMed] [Google Scholar]
- 30.Corcoran A, Mahon BP, Doyle S. B cell memory is directed toward conformational epitopes of parvovirus B19 capsid proteins and the unique region of VP1. J. Infect. Dis. 2004;189:1873–1880. doi: 10.1086/382963. [DOI] [PubMed] [Google Scholar]
- 31.Manaresi E, Gallinella G, Morselli Labate AM, Zucchelli P, Zaccarelli D, Ambretti S, Delbarba S, Zerbini M, Musiani M. Seroprevalence of IgG against conformational and linear capsid antigens of parvovirus B19 in Italian blood donors. Epidemiol. Infect. 2004;132:857–862. doi: 10.1017/s0950268804002389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moore JP, Ho DD. Antibodies to discontinuous or conformationally sensitive epitopes on the gp120 glycoprotein of human immunodeficiency virus type 1 are highly prevalent in sera of infected humans. J. Virol. 1993;67:863–875. doi: 10.1128/jvi.67.2.863-875.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fischmann TO, Bentley GA, Bhat TN, Boulot G, Mariuzza RA, Phillips SE, Tello D, Poljak RJ. Crystallographic refinement of the three-dimensional structure of the FabD1.3-lysozyme complex at 2.5-A resolution. J. Biol. Chem. 1991;266:12915–12920. [PubMed] [Google Scholar]
- 34.Cohen GH, Silverton EW, Padlan EA, Dyda F, Wibbenmeyer JA, Willson RC, Davies DR. Water molecules in the antibody-antigen interface of the structure of the Fab HyHEL-5-lysozyme complex at 1.7 Å resolution: comparison with results from isothermal titration calorimetry. Acta Crystallogr. sect. D. 2005;61:628–633. doi: 10.1107/S0907444905007870. [DOI] [PubMed] [Google Scholar]
- 35.Mylvaganam SE, Paterson Y, Getzoff ED. Structural basis for the binding of an anti-cytochrome c antibody to its antigen: crystal structures of Fab E8-cytochrome c complex to 1.8 Å resolution and Fab E8 to 2.26 Å resolution. J. Mol. Biol. 1998;281:301–322. doi: 10.1006/jmbi.1998.1942. [DOI] [PubMed] [Google Scholar]
- 36.Vyas NK, Vyas MN, Chervenak MC, Bundle DR, Pinto BM, Quiocho FA. Structural basis of peptide-carbohydrate mimicry in an antibody-combining site. Proc Natl Acad Sci U S A. 2003;100:15023–15028. doi: 10.1073/pnas.2431286100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dorgham K, Dogan I, Bitton N, Parizot C, Cardona V, Debre P, Hartley O, Gorochov G. Immunogenicity of HIV type 1 gp120 CD4 binding site phage mimotopes. AIDS Res. Hum. Retroviruses. 2005;21:82–92. doi: 10.1089/aid.2005.21.82. [DOI] [PubMed] [Google Scholar]
- 38.Saphire EO, Parren PWHI, Barbas CF, 3rd, Burton DR, Wilson IA. Crystallization and preliminary structure determination of an intact human immunoglobulin b12: an antibody that broadly neutralizes primary isolates of HIV-1. Acta Crystallogr. sect. D. 2001;57:168–171. doi: 10.1107/s0907444900017376. [DOI] [PubMed] [Google Scholar]
- 39.Wilson IA, Rini JM, Fremont DH, Fieser GG, Stura EA. X-ray crystallographic analysis of free and antigen-complexed Fab fragments to investigate structural basis of immune recognition. Methods Enzymol. 1991;203:153–176. doi: 10.1016/0076-6879(91)03009-6. [DOI] [PubMed] [Google Scholar]
- 40.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 41.Navaza J. AMoRe: An automated package for molecular replacement. Acta Crystallogr sect. A. 1994;50:157–163. [Google Scholar]
- 42.Jones AT. A graphics model building and refinement system for macromolecules. J. Appl. Crystallogr. 1978;11:268–272. [Google Scholar]
- 43.Jones TA. In: Computational Crystallography. Sayre D, editor. Oxford University Press; 1982. pp. 303–317. [Google Scholar]
- 44.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. sect. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 45.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. sect. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 46.Stanfield RL, Fieser TM, Lerner RA, Wilson IA. Crystal structures of an antibody to a peptide and its complex with peptide antigen at 2.8 Å. Science. 1990;248:712–719. doi: 10.1126/science.2333521. [DOI] [PubMed] [Google Scholar]
- 47.Esnouf RM. Further additions to MolScript version 1.4, including reading and contouring of electron-density maps. Acta Crystallogr. sect. D. 1999;55:938–940. doi: 10.1107/s0907444998017363. [DOI] [PubMed] [Google Scholar]
- 48.Sanner MF. Python: A programming language for software integration and development. J. Mol. Graphics and Modeling. 1999;17:57–61. [PubMed] [Google Scholar]
- 49.Sanner MF, Olson AJ, Spehner JC. Reduced surface: an efficient way to compute molecular surfaces. Biopolymers. 1996;38:305–320. doi: 10.1002/(SICI)1097-0282(199603)38:3%3C305::AID-BIP4%3E3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 50.Kunkel TA, Bebenek K, McClary J. Efficient site-directed mutagenesis using uracil-containing DNA. Methods Enzymol. 1991;204:125–139. doi: 10.1016/0076-6879(91)04008-c. [DOI] [PubMed] [Google Scholar]
- 51.Zwick MB, Bonnycastle LL, Noren KA, Venturini S, Leong E, Barbas CF, 3rd, Noren CJ, Scott JK. The maltose-binding protein as a scaffold for monovalent display of peptides derived from phage libraries. Anal. Biochem. 1998;264:87–97. doi: 10.1006/abio.1998.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Castellanos-Serra LR, Fernandez-Patron C, Hardy E, Huerta V. A procedure for protein elution from reverse-stained polyacrylamide gels applicable at the low picomole level: An alternative route to the preparation of low abundance proteins for microanalysis. Electrophoresis. 1996;17:1564–1572. doi: 10.1002/elps.1150171012. [DOI] [PubMed] [Google Scholar]
- 53.Craig L, Sanschagrin PC, Rozek A, Lackie S, Kuhn LA, Scott JK. The role of structure in antibody cross-reactivity between peptides and folded proteins. J. Mol. Biol. 1998;281:183–201. doi: 10.1006/jmbi.1998.1907. [DOI] [PubMed] [Google Scholar]
- 54.Karlsson R. Real-time competitive kinetic analysis of interactions between low-molecular-weight ligands in solution and surface-immobilized receptors. Anal. Biochem. 1994;221:142–151. doi: 10.1006/abio.1994.1390. [DOI] [PubMed] [Google Scholar]
- 55.Nieba L, Krebber A, Pluckthun A. Competition BIAcore for measuring true affinities: large differences from values determined from binding kinetics. Anal. Biochem. 1996;234:155–165. doi: 10.1006/abio.1996.0067. [DOI] [PubMed] [Google Scholar]
- 56.Menendez A, Chow KC, Pan OC, Scott JK. Human immunodeficiency virus type 1-neutralizing monoclonal antibody 2F5 is multispecific for sequences flanking the DKW core epitope. J. Mol. Biol. 2004;338:311–327. doi: 10.1016/j.jmb.2004.02.051. [DOI] [PubMed] [Google Scholar]
- 57.van Houten NE, Zwick MB, Menendez A, Scott JK. Filamentous phage as an immunogenic carrier to elicit focused antibody responses against a synthetic peptide. Vaccine. 2006;24:4188–4200. doi: 10.1016/j.vaccine.2006.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bonnycastle LLC, Menendez A, Scott JK. General Phage Methods. In: Barbas CF 3rd, Burton DR, Scott JK, Silverman GJ, editors. Phage Display: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Plainview, NY: 2001. pp. 15.14–15-16. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.