Abstract
A new approach is introduced to simultaneously detect resolved glutamate (Glu), glutamine (Gln), and γ-aminobutyric acid (GABA) using a standard STEAM localization pulse sequence with the optimized sequence timing parameters. This approach exploits the dependence of the STEAM spectra of the strongly coupled spin systems of Glu, Gln, and GABA on the echo time TE and the mixing time TM at 4 Tesla to find an optimized sequence parameter set, i.e., {TE, TM}, where the outer-wings of the Glu C4 multiplet resonances around 2.35 ppm, the Gln C4 multiplet resonances around 2.45 ppm, and the GABA C2 multiplet resonance around 2.28 ppm are significantly suppressed and the three resonances become virtual singlets simultaneously and thus resolved. Spectral simulation and optimization were conducted to find the optimized sequence parameters, and phantom and in vivo experiments (on normal human brains, one patient with traumatic brain injury, and one patient with brain tumor) were carried out for verification. The results have demonstrated that the Gln, Glu, and GABA signals at 2.2–2.5 ppm can be well resolved using a standard STEAM sequence with the optimized sequence timing parameters around {82 ms, 48 ms} at 4T, while the other main metabolites, such as N-acetyl Aspartate (NAA), Choline (tCho), and Creatine (tCr), are still preserved in the same spectrum. The technique can be easily implemented and should prove to be a useful tool for the basic and clinical studies associated with metabolism of Glu, Gln, and/or GABA.
Keywords: glutamate, glutamine, γ-aminobutyric acid (GABA), 1H MR spectroscopy, STEAM
1. Introduction
Glutamate (Glu) is the major excitatory neurotransmitter in the central nervous system (CNS). Closely coupled to Glu, glutamine (Gln) is mostly located in glial cells as the end product of Glu catabolism and is a reservoir for Glu production in the neuron [1]. Due to a defective reuptake, excess Glu in the synaptic space will induce neurotoxicity, which has been linked to many degenerative diseases, including multiple sclerosis (MS) [2], amyotrophic lateral sclerosis (ALS) [3], Huntington’s disease (HD) [4], and Alzheimer’s disease (AD) [5], and to neuronal apoptosis after CNS injury, such as traumatic brain injury (TBI) and stroke [6]. Moreover, Glu and Gln are also directly and/or indirectly involved in tumor proliferation and apoptosis [7]. γ-aminobutyric acid (GABA) is the major inhibitory neurotransmitter. As a product of Glu metabolism, GABA is synthesized from astroglial Gln and contributes to total Glu/Gln cycling [8]. Like Glu, the role of GABA has also been emphasized in many neurological and psychiatric disorders, such as epilepsy and depression [9;10]. Consequently, reliably detecting these neurochemicals in vivo is valuable to both basic research and clinical applications.
Reliable detection of Glu, Gln, or GABA, however, remains challenging using proton MR spectroscopy (1H-MRS) at clinically available field strengths. At 1.5 Tesla, the resonances of these metabolites in the region of 2.2–2.5 ppm completely collapse and are mostly assigned to a mixture of Glu, Gln, and GABA, designated as Glx. At 3 or 4 Tesla, short-echo STEAM or PRESS localization pulse sequence is typically employed for better signal-to-noise ratio (SNR). In these short-echo spectra, the Glu C4 multiplet resonance around 2.35 ppm overlaps with the Gln C4 multiplet resonance around 2.45 ppm and the GABA C2 multiplet resonance around 2.28 ppm, the Gln C4 multiplet resonance overlaps with the C3 multiplet resonance of the aspartyl group of N-acetyl aspartate (NAA) around 2.48 ppm, and the C3 multiplet resonances of Gln, Glu, and GABA overlap with the singlet resonance of N-acetyl group of NAA at 2.02 ppm. The above mentioned spectral overlaps impair accurate quantification of Gln, Glu, and GABA, and consequently increase the thresholds for detecting the metabolite changes between the patient and control groups [11]. This overlap-dependent interference with spectral quantification can not be simply solved by post-processing [12;13]. A number of methods have been proposed for in vivo detection of Glu, Gln, and/or GABA, including spectral editing techniques [14–17], multiple quantum filtering [18–20], and J-resolved spectroscopy [21–23]. Although significant progress has been achieved, simultaneous detection of well-resolved Glu, Gln, and GABA remains to be accomplished, particularly at middle magnetic fields or below. Here we report an approach to simultaneously detect resolved Glu, Gln, and GABA at 4 Tesla.
2. Materials and methods
2.1. The concept
Thompson and Allen have made a detailed study of the responses of weakly (such as lactate) and strongly coupled spin systems (such as the aspartyl group of NAA and Glu) to the STEAM sequence in a typical short-echo range, namely, TE and TM less than 50 ms [24]. Their study suggested that these metabolites exhibit significant variability in response to the STEAM sequence parameters. The variability arises from the time-dependent anti-phase coherences (APC) and zero quantum coherences (ZQC). Both types of coherences influence the magnitude of spectral components that ultimately contribute to the observable line shape, and the amplitude of each type of these coherences can be influenced by sequence design, such as the TE and TM parameters. Interesting observations on the response of Glu to the STEAM sequence are: 1) the dominant C4 multiplet resonance of Glu (between 2.25 and 2.45 ppm, centered at 2.35 ppm) maintains a relatively stable triple-peak structure; and 2) the central peak of the pseudo-triplet remains relatively stable in the {TE, TM} parameter space while the intensities of two outer-wings exhibit marked variability. Due to the similarity of spin system [25;26], the spectral characteristics of Gln should remain as similar as those of Glu. Different from the “pseudo-triplet” C4 resonances of Glu and Gln, the GABA C2 resonance is a rigorous triplet. However, its outer-wings and central peak were found to have similar variation characteristics with TE and TM as observed in the Glu C4 resonance (see 3.1. Spectral simulations and parameter optimization section). With the above knowledge in mind, we approached the problem of detecting resolved Glu, Gln, and GABA from a different aspect – by exploiting the similar dependence of the target resonances on TE and TM to find an optimal parameter set where the target resonances change from multiplets to virtual singlets simultaneously. The concept that the C4 resonances of Glu and Gln and the C2 resonance of GABA can be resolved in the spectral range of 2.2–2.5 ppm comes from the following analyses. Since the central peaks of the pseudo-triplet resonances at 2.45 ppm for Gln, 2.35 ppm for Glu, and 2.28 ppm for GABA, respectively, are readily separate and the overlap exclusively exists over the outer-wings at 4T (see Fig. 1 and 2.3. Sequence parameter optimization section), resolving the overlap is possible if the outer-wings are significantly suppressed for Glu, Gln, and GABA simultaneously while their central peaks maintain a sufficient intensity. In other words, we intend to optimize the TE and TM parameters of a STEAM sequence to achieve virtual singlets for the C4 multiplet resonances of Glu and Gln and the C2 multiplet resonance of GABA simultaneously.
Fig. 1.
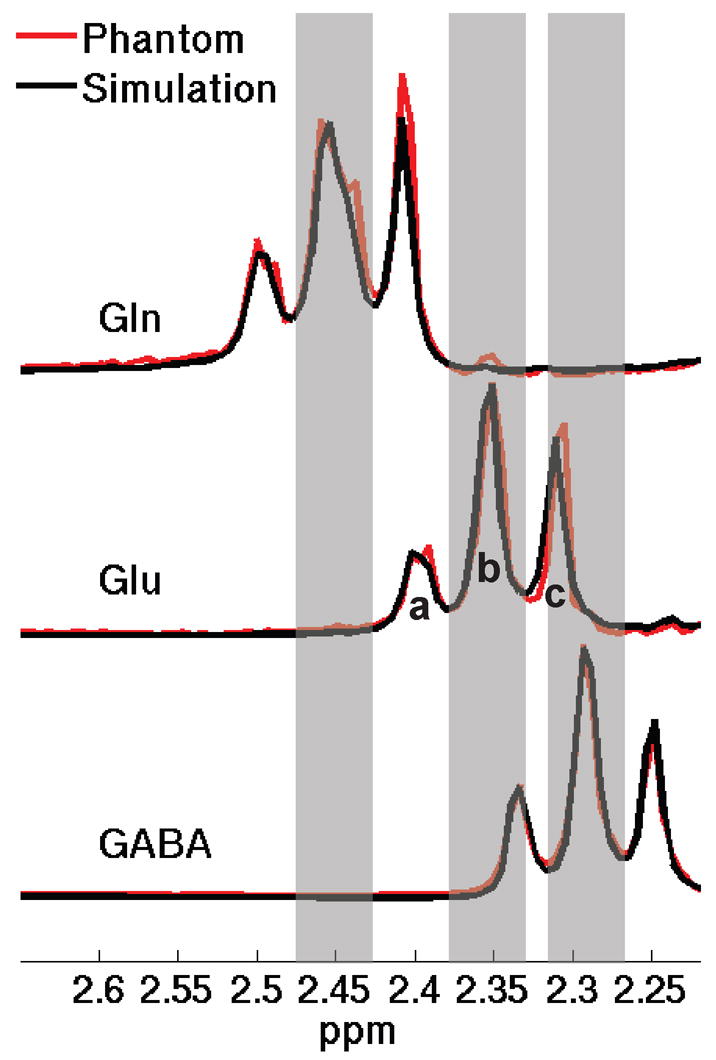
Simulated STEAM spectra (black line) of Gln, Glu, and GABA and their corresponding in vitro spectra (red line) at short-echo parameters of TE = 10 ms/TM = 10 ms. For simplicity, only the spectra in the 2.2-2.7 ppm region are shown. The shaded bars represent the spectral regions of central peaks of the Gln C4 pseudo-triplet resonance around 2.45 ppm, the Glu C4 pseudo-triplet resonance around 2.35 ppm, and the GABA C2 triplet resonance around 2.28 ppm, respectively. In the schematic diagram, a and c represent the outer-wings of a pseudo-triplet and b the central peak (for simplicity, only shown in the Glu spectrum).
2.2. Spectral simulation
Numerical simulation of the spectral response of a metabolite spin system to a pulse sequence has been proved an effective tool for facilitating prospective sequence design in MRS for optimal spectral yield of a target metabolite and/or minimization of co-resonant background signals [24;27–29]. For simulation of spectral response in this report, we employed the density matrix representation of metabolite spin systems and numerically solved the Liouville-von Neumann equation of motion of the density matrix using the publicly available GAMMA NMR simulation library [30]. The proton NMR chemical shifts and coupling constants of the brain metabolites were obtained from the literature [25]. The essential elements of a STEAM sequence, i.e., the RF pulse, the gradients, and the inter-pulse timings, were considered in our simulation, similar to those in previous studies [24;27;31;32]. The spectral responses of all the contributing metabolites in the target spectral region were evaluated in the two-dimensional (2D) {TE, TM} parameter space. The contour diagrams representing variation of a specified measure of the spectral response in the {TE, TM} parameter space were used to derive the optimized sequence timing parameters (see Fig. 2). To ensure the target multiplet resonances would turn into virtual singlets simultaneously, we chose relatively large ranges for TE and TM. A range of 0 to 200 ms was covered for TE with a step size of 2 ms and a range of 0 to 140 ms for TM with the same step size. The simulated spectra were multiplied by a 5-Hz exponential decay function to render them representative of typical in vivo line widths [24;28;29].
Fig. 2.
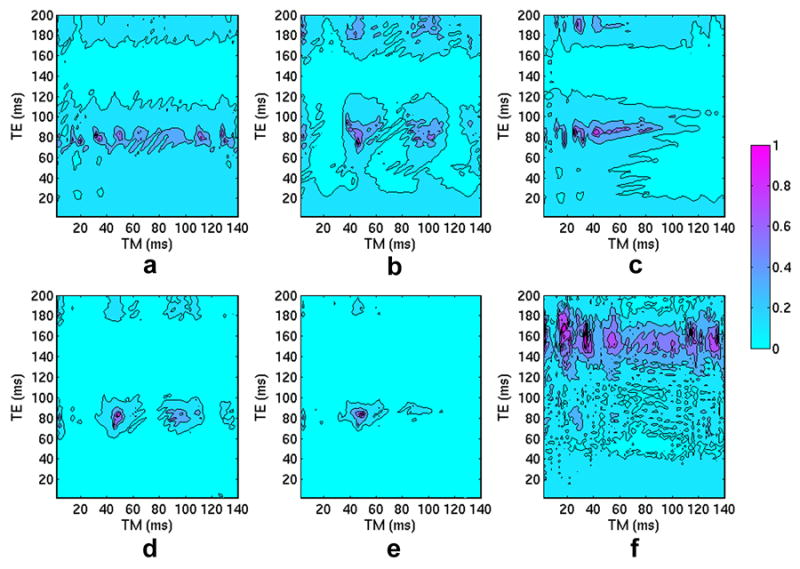
Contour diagrams, in the {TE, TM} parameter space, of the index calculated from Eq. (1) for (a) Gln, (b) Glu, and (c) GABA and the contour diagrams of the ultimate index calculated from Eq. (2) for (d) two target metabolites, Gln and Glu, and (e) three target metabolites, Gln, Glu, and GABA. The indices were individually normalized to the maximum value of each data set. A color bar shows the data range of 0 ~ 1. The maximum indices reside at {82 ms, 48 ms} in (d) and at {84 ms, 48 ms} in (e), respectively. Another index, the ratio of the peak area of NAA spectrum in the central-peak region of the Gln C4 resonance, i.e., 2.419-2.488 ppm, to the peak area of the central peak of Gln C4 resonance, was defined to evaluate the contamination from NAA to Gln. A contour diagram of this index is shown in (f), and the contribution of NAA in the central-peak region of the Gln C4 pseudo-triplet is rather low in the optimized parameter region, centered at {82 ms, 48 ms}.
2.3. Sequence parameter optimization
Simulated spectral responses (black line) of Gln, Glu, and GABA to a STEAM sequence at TE = 10 ms/TM = 10 ms are illustrated in Fig. 1. The individual in vitro spectra of each target metabolites (red line) are also included to demonstrate excellent agreement between the simulated and experimental phantom spectra. For simplicity, only the Gln C4 pseudo-triplet resonance around 2.45 ppm, the Glu C4 pseudo-triplet resonance around 2.35 ppm and the GABA C2 triplet resonance around 2.28 ppm are shown in the figure. The shaded bars represent spectral regions of the central peaks of these pseudo-triplets. In the schematic diagram of three spectral regions of a pseudo-triplet, a and c represent the outer-wings and b the central peak (for simplicity, only shown in the Glu spectrum). It has been found in the previous study [24] and in our own spectral simulation that the central peaks of the target resonances maintain relatively stable amplitudes while the outer-wings exhibit marked variation with TE and TM, and at some TE and TM parameters the amplitudes of these outer-wings are reduced significantly. We hypothesized that it is possible to find the optimized TE and TM parameters with which the outer-wings of the two (Gln and Glu) or three (Gln, Glu, and GABA) target pseudo-triplets can be significantly suppressed simultaneously. For a complete search in the {TE, TM} parameter space, an index or cost function was defined for each target metabolite. The function measures the product of the peak-area ratio of the central peak to its respective outer-wings and the peak area of the central peak of the pseudo-triplet, expressed as follows
| (1) |
where [b] stands for the peak area of the central peak of the pseudo-triplet, and [a] and [c] are the peak areas of the downfield and upfield outer-wings, respectively, as shown in Fig. 1.
As the spectral ranges of these triple-peak structures are quite stable [24], we defined the spectral ranges of [a] , [b], and [c] based on the simulated spectra at TE = 10 ms/TM = 10 ms, and the boundaries were chosen at the local minima in the simulated spectrum. The spectral ranges were 2.178–2.259 ppm, 2.259–2.327 ppm, and 2.327–2.408 ppm, respectively, for GABA; 2.247–2.327 ppm, 2.327–2.385 ppm, and 2.385–2.465 ppm, respectively, for Glu; and 2.350–2.419 ppm, 2.419–2.488 ppm, and 2.488–2.557 ppm, respectively, for Gln. Since two (Gln and Glu) or three (Gln, Glu, and GABA) target metabolites need to be resolved simultaneously, the ultimate index that we used in evaluation of the optimized TE and TM parameters was the product of the individual indices for Gln, Glu, and/or GABA, respectively, i.e.,
| (2) |
The ultimate index, F , measures the combined contribution from the peak areas of the central peaks and the outer-wing suppression of all the target resonances. The TE and TM parameters that yielded the maximum ultimate indices were chosen as the optimized STEAM sequence timing parameters for the purpose of this study.
2.4. In vitro and in vivo experiments
Phantom experiments were conducted on a Bruker/Siemens 4T scanner to verify the optimized TE and TM parameters predicted by simulation, using a quadrature head coil and a standard 1H STEAM localization pulse sequence. The sequence parameters, besides the optimized TE and TM parameters (see 3.1. Spectral simulations and parameter optimization section), were TR = 2000 ms, averages = 32, voxel-size = 4x4x2 cm3. The phantom set consisted of three phantoms containing either (a) 20mM Gln, (b) 20mM Glu and (c) 20mM GABA, respectively, in a buffered solution (pH = 7.2).
In addition to phantom validation, one healthy volunteer, one TBI patient, and one brain tumor patient were recruited for in vivo experiments. Subjects gave written informed consent for participation in this IRB-approved study and were financially compensated for their participation. A chemical shift imaging (CSI) sequence using STEAM to select a volume of interest, i.e., CSI-STEAM, was employed to acquire 2D spectroscopic data on a TBI patient and a healthy volunteer as a reference. The parameters of this CSI-STEAM sequence, besides the optimized TE and TM parameters, were TR = 1500 ms, FOV = 20x20 cm2, matrix size = 16x16, slice thickness = 17 mm, NA = 16, and total acquisition time = 21 minutes. An elliptical and Hamming-filter weighted k-space sampling scheme was applied to improve sampling efficiency, and the resulting real voxel size was 1.9x1.9x1.7 cm3. The spectra of a brain tumor patient were acquired using the same CSI-STEAM sequence with the optimized TE and TM parameters. Other sequence parameters were TR = 3000 ms, FOV = 20x20 cm2, matrix size = 16x16, slice thickness = 20 mm, NA = 8, and total acquisition time = 22.8 minutes. After applying a Hamming filter, the resulting real voxel size was 1.9x1.9x2.0 cm3. The patient, treated by Gamma Knife previously, was diagnosed with recurrent/persistent astrocytoma. In addition, biopsy examination showed the evidence of tumor proliferation.
3. Results
3.1. Spectral simulations and parameter optimization
A contour diagram, in the {TE, TM} parameter space, of the index defined as in Eq. (1) for Gln is shown in Fig. 2a. The data were normalized to the maximum value of the data set, so the maximum value in the normalized data set is 1 and the minimum is close to 0. A color bar beside the contour diagrams shows the range. The contour diagrams of the indices for Glu and GABA are shown in Figs. 2b and 2c, respectively. The contour diagrams of the ultimate index are shown in Figs. 2d and 2e, which are the results for two target metabolites (Gln and Glu) and three target metabolites (Gln, Glu, and GABA), respectively. Figs. 2d and 2e show very similar distribution of the ultimate indices in the {TE, TM} parameter space, and the optimized TE and TM parameters in both contour diagrams are located in the range of 40 – 60 ms for TM and 70 - 90 ms for TE. The maximum indices reside at {82 ms, 48 ms} in Fig. 2d and at {84 ms, 48 ms} in Fig. 2e, respectively. We defined another index to evaluate the contamination of NAA to the central peak of the C4 pseudo-triplet resonance of Gln. A contour diagram of the index (with the same normalization processing as in Figs. 2a–2e) in the {TE, TM} parameter space is illustrated in Fig. 2f. A more detailed description of the index and Fig. 2f will be given later in this section.
Fig. 3 shows the simulated spectra of several possible contributing metabolites in the 2.2–2.5 ppm region (indicated by the shaded bars) at typical short-echo parameters of TE = 10 ms/TM = 10 ms (Fig. 3a) and at the optimized timing parameters of TE = 82 ms/TM = 48 ms (Fig. 3b), both under the same amplitude scale. Compared with the spectra in Fig. 3a, the outer-wings of the pseudo-triplet resonances of Gln, Glu, and GABA are significantly suppressed at the optimized TE and TM parameters and virtual singlets are achieved simultaneously for the three target resonances as shown in Fig. 3b. The quantitative results of the outer-wings of target resonances and the spectral overlap between the target resonances at short-echo and optimized sequence timing parameters are summarized in Table 1. Table 1 clearly demonstrate that the target pseudo-triplet resonances of Gln, Glu, and GABA at the short-echo parameters of TE = 10 ms/TM = 10 ms turn into virtual singlets at the optimized sequence timing parameters of TE = 82 ms/TM = 48 ms, and the overlap between these metabolites are greatly suppressed.
Fig. 3.
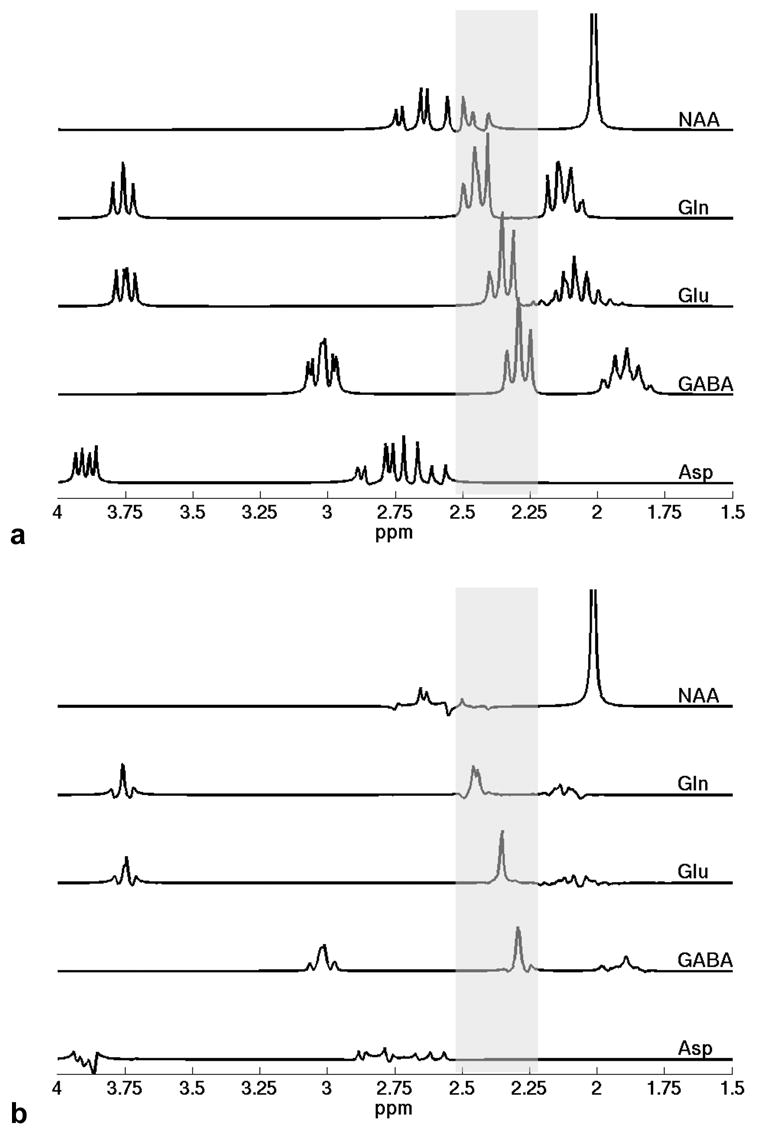
Simulated spectra of several possible contributing metabolites in the 2.2–2.5 ppm region, indicated by the shaded bars, at (a) typical short-echo parameters of TE = 10 ms/TM = 10 ms and (b) the optimized timing parameters of TE = 82 ms/TM = 48 ms, both under the same amplitude scale. The C3 multiplet resonance of the aspartyl group of NAA overlaps with the entire Gln C4 multiplet resonance from 2.37 to 2.51 ppm. Fortunately, the spectral contamination of NAA to Gln is rather low at the optimized sequence timing parameters (also shown in Fig. 2f). The spectral contamination of NAA to the Glu C4 multiplet resonance around 2.35 ppm and the GABA C2 multiplet resonance around 2.28 ppm is negligible, and the spectral resonances of Asp are completely outside the target region. The nonnegligible C3 multiplet resonances of Gln, Glu, and GABA overlap with the singlet resonance of the N-acetyl group of NAA at 2.02 ppm at short echo time in (a). In contrast, these C3 multiplet resonance signals decay significantly at the optimized sequence timing parameters in (b), thus greatly reducing the interference with quantification of NAA.
Table 1.
Outer-Wing Levels of Target Resonances and Spectral Overlap between Target Resonances at Short-Echo and Optimized Sequence Timing Parameters
| Outer-Wing Level1 | Spectral Overlap2 | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gln | Glu | GABA | Gln-Glu | Glu-GABA | Glu→GABA | GABA→Glu | |||
| Ratio reference | /Gln | /Glu | /GABA | /Gln | /Glu | /Glu | /GABA | /GABA | /Glu |
| TE=10ms /TM=10ms | 82% | 102% | 95% | 83% | 98% | 57% | 47% | 27% | 24% |
| TE=82ms /TM=48ms | 9% | 5% | 8% | 1% | 1% | 3% | 3% | 3% | 0% |
The ratio of the peak areas of the outer-wings to those of the respective central peaks was used to evaluate the outer-wing levels of the target resonance of Gln, Glu, and GABA.
Two types of spectral overlap (see Fig. 1) were analyzed: the overlap between the outer-wings of different target resonances, such as the C4 resonances of Gln and Glu, denoted as Gln-Glu, and the overlap between the outer-wing of one target resonance and the central peak of another target resonance, such as the outer-wing of Glu C4 resonance overlapping with the central peak of GABA C2 resonance, denoted as Glu→GABA. For the former (e.g., Gln-Glu), the ratio was taken from the peak area of the overlapped outer-wings of all the involved target resonances to the peak area of the central peak of each involved target resonance; for the latter (e.g., Glu→GABA), the ratio was taken from the peak area of one outer-wing of one target resonance in the overlapped region to the peak area of the central peak of the other target resonance.
In all the ratio calculation, the peak area of central-peak of the target resonance was first chosen as a reference, indicated by /Gln, /Glu, or /GABA. All the results were calculated under the assumption of the same concentrations of Glu, Gln, and GABA as well as the identical relaxtation times.
It should be noted that to finalize the optimized sequence timing parameters, it is necessary to consider two additional issues. The first issue is the spectral overlap or contamination from other metabolites to the target resonances. Potential contamination sources in the target 2.2–2.5 ppm region include NAA and aspartate (Asp). To assess these potential contamination, the simulated STEAM spectra of NAA and Asp at TE/TM = 10/10 ms are also presented in Fig. 3a, along with those of Gln, Glu, and GABA. The C3 multiplet resonance of the aspartyl group of NAA overlaps with the entire Gln C4 multiplet resonance from 2.37 to 2.51 ppm, including both the central peak and the outer-wings [27]. Therefore, the contamination from NAA may critically impair the detection of Gln. As the outer-wings of the Gln C4 resonance are suppressed at the optimized sequence timing parameters, we mainly evaluated the contamination of NAA to the central peak of the Gln C4 resonance. Another index, the ratio of the peak area of NAA spectrum in the central-peak region of the Gln C4 resonance, i.e., 2.419–2.488 ppm, to the peak area of the central peak of the Gln C4 resonance, was defined to evaluate the contamination from NAA to Gln. A contour diagram of the index in the {TE, TM} parameter space is illustrated in Fig. 2f. Fortunately, the contribution of NAA signal in the central-peak region of the Gln C4 resonance, as shown in Fig. 2f and Fig. 3b, is rather low in the optimized parameter range, centered at TE = 82 ms/TM = 48 ms. This result mitigated our initial concern about the possible strong contamination from NAA to Gln at the optimized sequence timing parameters. Besides the above overlap, the C3 multiplet resonance of the aspartyl group of NAA, as shown in Fig. 3a, overlaps with the downfield outer-wing of Glu C4 resonance, and just barely touches the boundary of the central peak of Glu C4 resonance. As the outer-wings of the target Glu C4 resonance would be significantly suppressed at the optimized sequence timing parameters, the contamination of NAA to the Glu C4 resonance is negligible. No overlap was observed between the NAA spectrum and the GABA C2 triplet resonance. Although some minor overlap between the resonances of Asp and the downfield outer-wing of the Gln C4 resonance in Fig. 3a, no overlap exists between the Asp resonances and the central peak of the Gln C4 resonance. Similarly, no overlap exists between the Asp resonances and the Glu C4 resonance and the GABA C2 resonance. Therefore, evaluation of the contamination of Asp can be omitted. Besides the above mentioned metabolites, the C2 resonance (2.37 ppm) of the GABA moiety of homocarnosine does overlap with the Glu C4 central-peak resonance. However, its concentration is 20–50 times lower than Glu [25], therefore possessing negligible contamination to the target resonances. One of the other resonances closely adjacent to the target 2.2–2.5 ppm region comes from the Glu moiety of Glutathione (GSH). At 4 Tesla, however, only the upfield outer-wing of the C4 resonance of Glu moiety of GSH overlaps the downfield outer-wing, but not the central peak, of the Gln C4 resonance (data not shown). Therefore, evaluation of the contamination of GSH, similarly, can be omitted at 4 Tesla. Mobile lipid and macromolecules are usually visible in short-echo 1H MR spectra. At the optimized timing parameters of TE = 82 ms/TM = 48 ms, the STEAM signals of mobile lipid and macromolecules may decrease considerably, due to the T2-relaxation decay in TE and the T1-relaxation decay in TM according to Eq. (1) in [33]. Nevertheless, there still might be some residual signals from mobile lipid, common macromolecules, and/or some special macromolecules with short T1 and long T2 values. Although none of their chemical shifts (generally 0.89 ppm, 1.30 ppm, 2.05 ppm, 2.24 ppm, 2.81 ppm for lipids and 0.89 ppm, 1.51 ppm, 2.10 ppm, 3.00 ppm for macromolecules [34]) are coincide with the three target resonances, the spectra of these residual signals are broad and might still interfere with the target resonances. As demonstrated in [34], an extended basis set can be incorporated exclusively to account for these residual background signals and their influence can thus be eliminated in the spectral analysis. The latest version of LCModel even provides the basis set of the macromolecule and lipid by default as one of the new features.
Another important issue in determining the optimized TE and TM parameters is the interference of the target metabolites with other main metabolites, such as NAA, tCr, and tCho. As reported in the previous study [12], a major concern is the overlap of the C3 multiplet resonances of Gln, Glu, and/or GABA with the singlet resonance of the N-acetyl group of NAA at 2.02 ppm, which may impair accurate NAA quantification, especially at short echo time and in the presence of increased Gln, Glu, and/or GABA signals. In Fig. 3b the amplitudes of the C3 multiplet resonances of Gln, Glu, and GABA at the optimized TE and TM parameters are much lower than those in the short-echo spectra in Fig. 3a. Thus, the STEAM sequence with the optimized TE and TM parameters not only brought about virtual singlets for Gln, Glu, and GABA simultaneously, but also had an additional advantage in reducing the interference with the N-acetyl resonance of NAA. Moreover, at such comparably longer echo times, the spectral baseline, although still possibly with some level of residual background signals, will be much simpler than those in the commonly used short-echo spectra [35].
3.2. In vitro and in vivo experiments
In vitro spectra were acquired on the Gln , Glu, and GABA phantoms using a standard STEAM pulse sequence with the short-echo and optimized sequence timing parameters. The short-echo in vitro spectra have already been shown in Fig. 1, with a good agreement with the simulated counterparts. At the optimized sequence timing parameters (data not shown for simplicity), the outer-wings of the target pseudo-triplet resonances were significantly suppressed, and the resonances, as expected, turned into virtual singlets simultaneously.
Fig. 4 shows examples of the in vivo STEAM spectra on a voxel in the brain of a healthy volunteer (Fig. 4a), a voxel away from the traumatic brain lesion of a TBI patient (Fig. 4b), and a voxel within the traumatic brain lesion of the same TBI patient (Fig. 4c), respectively. The selected voxel in the brain of the healthy volunteer was in a similar brain region with the traumatic lesion region of the TBI patient. In Fig. 4a, the Glu C4 resonance at 2.35 ppm is a dominant single peak, while the amplitudes of the Gln C4 resonance and the GABA C2 resonance are much lower. Similar spectral characteristics are observed in Fig. 4b. This is consistent with previous studies that the concentration of Glu in the healthy human brain is significantly higher than those of Gln and GABA [36]. Resolved Gln, Glu, and GABA signals in the target 2.2–2.5 ppm region are shown in Fig. 4c. The spectrum in Fig. 4c clearly demonstrates the elevated Gln and GABA levels related to traumatic brain injury, which are consistent with the previous reports of increased Glx signals in TBI patients [37].
Fig. 4.
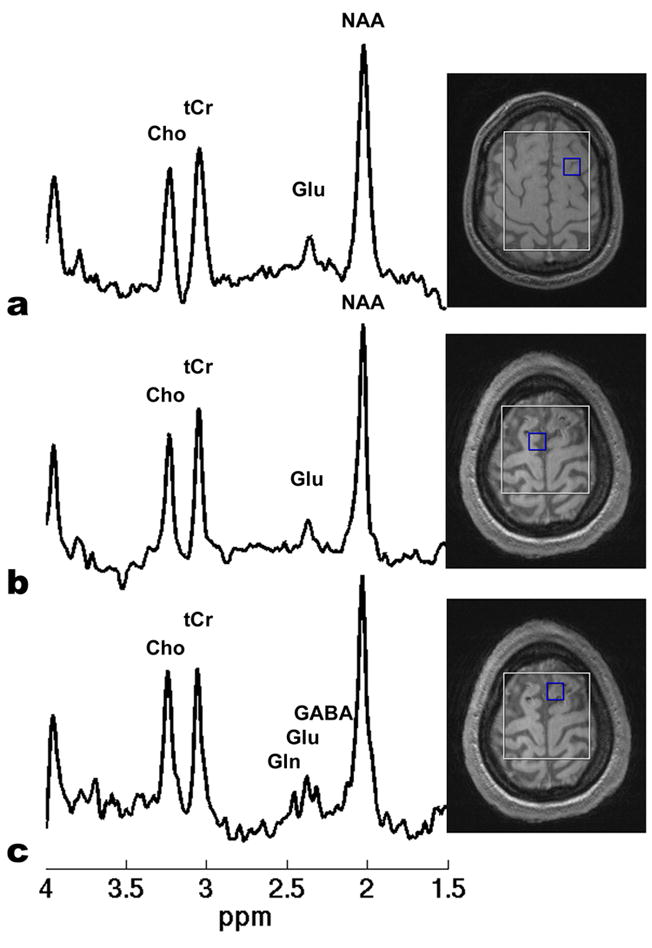
Examples of in vivo STEAM spectra on (a) a voxel in the brain of a healthy volunteer, (b) a voxel outside the traumatic brain lesion of a TBI patient, and (c) a voxel within the traumatic brain lesion of the same TBI patient, respectively. The selected voxel in the healthy volunteer brain was in a similar brain region with the traumatic lesion region of the TBI patient. All the spectra were extracted from the CSI-STEAM data acquired using the optimized sequence timing parameters. Other sequence parameters were TR = 1500 ms, FOV = 20x20 cm2, matrix size = 16x16, slice thickness = 17 mm, NA = 16, and total acquisition time = 21 minutes. An elliptical, Hamming-filter weighted k-space sampling scheme was applied and the resulting real voxel size was 1.9x1.9x1.7 cm3. In (a) and (b), the C4 resonance of Glu at 2.35 ppm is a dominant single peak, while the C4 resonance of Gln and the C2 resonance of GABA is much lower. Resolved Gln, Glu, and GABA signals in the target 2.2-2.5 ppm region are shown clearly in (c).
Fig. 5 illustrates two spectra acquired from a voxel in a T2 hyperintense brain region mesial to the cystic portion of the astrocytoma (Fig. 5a) and a voxel outside the lesion area (Fig. 5b) of the same patient. While the Glu C4 resonance is the only dominant signal in the 2.2–2.5 ppm region in the control brain tissue (Fig. 5b), the C4 resonances of Glu at 2.35 ppm and Gln at 2.45 ppm are clearly visible and well resolved in the lesion region (Fig. 5a). Since in primary high-grade glioma the T2 hyperintense brain region comprises not only vasogenic edema but also infiltrating tumor cells [38], the spectral signal in Fig. 5a is most likely from a mixture of tumor and edematous tissues. The dramatic increase in Gln as demonstrated in Fig. 5a is consistent with the biochemistry studies that Gln plays an important role in tumor proliferation [39;40]. The capability of simultaneous detection of well-separated Glu, Gln, and/or GABA signals in vivo offers potential for the study of some neurological and psychiatric disorders using 1H-MRS.
Fig. 5.
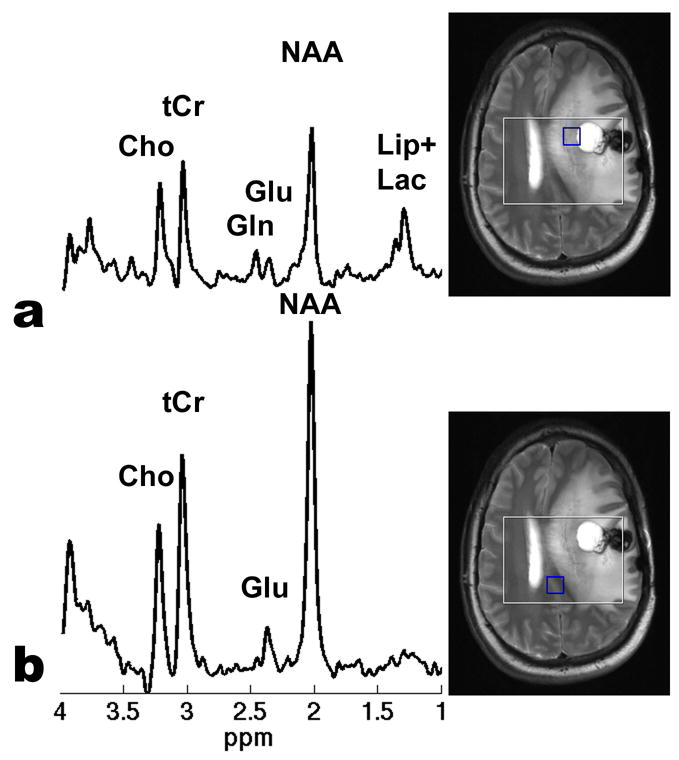
Examples of in vivo STEAM spectra on (a) a voxel in a T2 hyperintense brain region mesial to the cystic portion of the astrocytoma and (b) a voxel outside the lesion area, respectively, of the same patient. Both spectra were extracted from the CSI-STEAM data acquired using the optimized sequence timing parameters. Other sequence parameters were TR = 3000 ms, FOV = 20x20 cm2, matrix size = 16x16, slice thickness = 20 mm, NA = 8, and total acquisition time = 22.8 minutes. An elliptical, weighted k-space sampling scheme was used. After applying a Hamming filter, the resulting real voxel size was 1.9x1.9x2.0 cm3. In (a) “Lip+Lac” stand for the possible lipid and/or lactate signals. While the Glu C4 resonance is the only dominant signal in the 2.2–2.5 ppm region in (b), the C4 resonances of Glu at 2.35 ppm and Gln at 2.45 ppm are clearly visible and well resolved in the lesion area associated with the brain tumor in (a).
4. Discussion
In this report, we demonstrated a method for simultaneous detection of unobstructed Glu, Gln, and GABA at 4 Tesla by optimizing the timing parameters, i.e., {TE, TM}, of a standard STEAM localization pulse sequence. The essential idea is to convert the Gln C4 multiplet resonance around 2.45 ppm, the Glu C4 multiplet resonance around 2.35 ppm, and the GABA C2 multiplet resonance around 2.28 ppm to virtual singlets, by suppressing the outer-wings of the target pseudo-triplet resonances while maintaining the central peaks in sufficient amplitudes. As illustrated in the numerical simulation, in vitro, and in vivo experiments, the outer-wings of the target resonances were substantially suppressed at the optimized parameters of TE = 82 ms/TM = 48 ms, and virtual singlet spectral patterns were successfully achieved.
The direct and most important consequence of above sequence parameter optimization is to ensure sufficient spectral separation between the target resonances in the 2.2–2.5 ppm region, and thus to resolve the spectral overlap among Gln, Glu, and GABA, which is a common problem in the short-echo spectra. These resolved and simplified spectral patterns may generally facilitate quantification of individual target metabolites. We also evaluated potential contaminations to Gln, Glu, and GABA from other metabolites in the 2.2–2.5 ppm region. At the optimized TE and TM parameters the contribution of NAA in the central peak of Gln C4 resonance is rather low (see Fig. 2f and Fig. 3). Except for Asp and the Glu moiety of GSH, which overlap the downfield outer-wing, but not the central peak, of the Gln C4 resonance, no other metabolites in the 2.2-2.5 ppm region have significant interference with the three target resonances. The STEAM signals from most of the mobile lipid and macromolecules will decrease considerably at the optimized timing parameters of TE = 82 ms/TM = 48 ms. For those decreased, but still residual background signals and for the signals from certain special macromolecules with short T1 but long T2, an extended parameterized basis set can be incorporated exclusively, as demonstrated in [34], to account for them, and their influence can thus be eliminated in the spectral analysis, such as using latest version of LCModel.
One recent report addressed an important issue, i.e., interference of the Gln and Glu resonances with the quantification of NAA [12]. Besides the solution provided at 1.5T in the report, another technique, TE-averaged PRESS [23], which was originally proposed to reliably detect Glu, results in cancellation of the C3 multiplet resonances of Gln and Glu that contaminate the singlet resonance of the N-acetyl group of NAA at 2.02 ppm. At the optimized TE and TM parameters, the C3 resonances of Gln, Glu, and GABA decrease significantly (see Fig. 3b), compared with those resonances in the short-echo spectra in Fig. 3a. The residual C3 resonances of the target metabolites around 2.02 ppm are very small, with similarly low levels as those observed in the TE-averaged PRESS spectra. This is an additional advantage of using the standard STEAM sequence with the optimized TE and TM parameters. Moreover, the proposed method does not sacrifice other main metabolites, such as NAA, tCho, and tCr, in contrast to many editing techniques. The method also provides relatively smooth and simple baseline in the spectra.
Although the central peaks of the target pseudo-triplet resonances remained while the outer-wings were significantly suppressed, the amplitudes of the central peaks decreased at the optimized TE and TM parameters, compared with those in the short-echo spectra. Two factors contribute to the signal loss. The first one arises from the J-evolution of strongly-coupled spins with echo time(s). Although J-modulation did not produce monotonical amplitude-damping of the central peaks [24], these peaks of the target resonances at TE = 82 ms/TM = 48 ms, as shown in Fig. 3b, did decrease with J-evolution, compared with those at TE = 10 ms/TM = 10 ms in Fig. 3a. We calculated this type of signal loss between the two sets of timing parameters. The signal losses of the central peaks are 52% for Gln, 42% for Glu, and 48% for GABA, respectively. Because relatively long TE and TM are used, the second factor for the signal loss comes from T1 and T2 relaxation. Measurements of T1 and/or T2 values of Glu, Gln, and/or GABA in vivo are rarely reported in the literature until very recently [36;41], probably due to low SNR and other difficulties (e.g., J-modulation and spectral overlap) in the measurements. Based on the reported T1 (1.23 s) and T2 (168 ms) values of Glu [41] and Eq. (1) in [33], the estimated signal losses are 39% due to T2 and 12% due to T1 (assuming TR = 3s) at optimized TE = 82 ms/TM = 48 ms, resulting in a total signal reduction of 46%, compared with that without relaxation effects. The Gln relaxation times were assumed to be similar to those of Glu and the values for GABA are not reported in the literature. If other conditions remain the same, reduced signal intensity may lead to increased errors in spectral quantification. As stated before, however, the background contamination from macromolecule and lipids also decreases greatly at long echo time. This is sometimes also critical in quantification of low sensitivity of metabolite signals such as Glu, Gln, or GABA. In a recent paper [35], for instance, MRS acquisition at long echo time was exploited to reduce baseline contamination, while a Carr-Purcell (CP) train of refocusing pulses was implemented to maintain the strongly coupled target resonances with sufficient amplitudes. In brief, the advantages are simplified and resolved spectral patterns for Gln, Glu, and/or GABA, a reasonable flat baseline, and low interference with NAA at 2.02 ppm, while the disadvantages mainly come from the decreased signal intensity of the central peaks of the target resonances as well as the increased necessity for correction of the relaxation effects in spectral quantification. To fully address the quantification issue for in vivo data and compare the proposed approach with short-echo PRESS or STEAM and other existing methods, a more comprehensive study is needed, but beyond the scope of this proof-of-concept report.
In summary, we have demonstrated that the Glu C4 multiplet resonance around 2.35 ppm, the Gln C4 multiplet resonance around 2.45 ppm, and the GABA C2 multiplet resonance around 2.28 ppm can become virtual singlets simultaneously at the optimized timing parameters of a STEAM sequence and these target resonances can be well-resolved at 4 Tesla. Implementation of the technique is easy and only requires the widely available standard STEAM sequence. Therefore, the technique should prove to be a useful tool for the basic and clinical studies associated with metabolism of Glu, Gln, and/or GABA.
Acknowledgments
This work was supported by MTTC05024 grant and by the Intramural Research Program of the National Institute on Drug Abuse (NIDA), NIH. The authors thank Dr. Scott A. Smith (Florida State University) for discussion about density operator scaling in GAMMA and Dr. Varanavasi Govindaraju (University of Miami) for discussion about the chemical shifts and coupled constants.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Schousboe A, Westergaard N, Sonnewald U, Petersen SB, Huang R, Peng L, Hertz L. Glutamate and glutamine metabolism and compartmentation in astrocytes. DevNeurosci. 1993;15:359–366. doi: 10.1159/000111356. [DOI] [PubMed] [Google Scholar]
- 2.Pitt D, Werner P, Raine CS. Glutamate excitotoxicity in a model of multiple sclerosis. NatMed. 2000;6:67–70. doi: 10.1038/71555. [DOI] [PubMed] [Google Scholar]
- 3.Pioro EP, Majors AW, Mitsumoto H, Nelson DR, Ng TC. 1H-MRS evidence of neurodegeneration and excess glutamate + glutamine in ALS medulla. Neurology. 1999;53:71–79. doi: 10.1212/wnl.53.1.71. [DOI] [PubMed] [Google Scholar]
- 4.Taylor-Robinson SD, Weeks RA, Bryant DJ, Sargentoni J, Marcus CD, Harding AE, Brooks DJ. Proton magnetic resonance spectroscopy in Huntington's disease: evidence in favour of the glutamate excitotoxic theory. Mov Disord. 1996;11:167–173. doi: 10.1002/mds.870110209. [DOI] [PubMed] [Google Scholar]
- 5.Moats RA, Ernst T, Shonk TK, Ross BD. Abnormal cerebral metabolite concentrations in patients with probable Alzheimer disease. Magn ResonMed. 1994;32:110–115. doi: 10.1002/mrm.1910320115. [DOI] [PubMed] [Google Scholar]
- 6.Zipfel GJ, Babcock DJ, Lee JM, Choi DW. Neuronal apoptosis after CNS injury: the roles of glutamate and calcium. JNeurotrauma. 2000;17:857–869. doi: 10.1089/neu.2000.17.857. [DOI] [PubMed] [Google Scholar]
- 7.Mates JM, Perez-Gomez C, Nunez dC, Asenjo IM, Marquez J. Glutamine and its relationship with intracellular redox status, oxidative stress and cell proliferation/death. IntJBiochemCell Biol. 2002;34:439–458. doi: 10.1016/s1357-2725(01)00143-1. [DOI] [PubMed] [Google Scholar]
- 8.Patel AB, de Graaf RA, Mason GF, Rothman DL, Shulman RG, Behar KL. The contribution of GABA to glutamate/glutamine cycling and energy metabolism in the rat cortex in vivo. ProcNatlAcadSciUSA. 2005;102:5588–5593. doi: 10.1073/pnas.0501703102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petroff OA, Rothman DL, Behar KL, Lamoureux D, Mattson RH. The effect of gabapentin on brain gamma-aminobutyric acid in patients with epilepsy. AnnNeurol. 1996;39:95–99. doi: 10.1002/ana.410390114. [DOI] [PubMed] [Google Scholar]
- 10.Sanacora G, Mason GF, Rothman DL, Behar KL, Hyder F, Petroff OA, Berman RM, Charney DS, Krystal JH. Reduced cortical gamma-aminobutyric acid levels in depressed patients determined by proton magnetic resonance spectroscopy. ArchGenPsychiatry. 1999;56:1043–1047. doi: 10.1001/archpsyc.56.11.1043. [DOI] [PubMed] [Google Scholar]
- 11.Bartha R, Drost DJ, Menon RS, Williamson PC. Comparison of the quantification precision of human short echo time (1)H spectroscopy at 1.5 and 4.0 Tesla. Magn ResonMed. 2000;44:185–192. doi: 10.1002/1522-2594(200008)44:2<185::aid-mrm4>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 12.Clementi V, Tonon C, Lodi R, Malucelli E, Barbiroli B, Iotti S. Assessment of glutamate and glutamine contribution to in vivo N-acetylaspartate quantification in human brain by (1)H-magnetic resonance spectroscopy. Magn ResonMed. 2005;54:1333–1339. doi: 10.1002/mrm.20703. [DOI] [PubMed] [Google Scholar]
- 13.Ratiney H, Sdika M, Coenradie Y, Cavassila S, van Ormondt D, Graveron-Demilly D. Time-domain semi-parametric estimation based on a metabolite basis set. NMR Biomed. 2005;18:1–13. doi: 10.1002/nbm.895. [DOI] [PubMed] [Google Scholar]
- 14.de Graaf RA, Rothman DL. Detection of gamma-aminobutyric acid (GABA) by longitudinal scalar order difference editing. JMagn Reson. 2001;152:124–131. doi: 10.1006/jmre.2001.2371. [DOI] [PubMed] [Google Scholar]
- 15.Hetherington HP, Newcomer BR, Pan JW. Measurements of human cerebral GABA at 4.1 T using numerically optimized editing pulses. Magn ResonMed. 1998;39:6–10. doi: 10.1002/mrm.1910390103. [DOI] [PubMed] [Google Scholar]
- 16.Keltner JR, Wald LL, Christensen JD, Maas LC, Moore CM, Cohen BM, Renshaw PF. A technique for detecting GABA in the human brain with PRESS localization and optimized refocusing spectral editing radiofrequency pulses. Magn ResonMed. 1996;36:458–461. doi: 10.1002/mrm.1910360319. [DOI] [PubMed] [Google Scholar]
- 17.Lee HK, Yaman A, Nalcioglu O. Homonuclear J-refocused spectral editing technique for quantification of glutamine and glutamate by 1H NMR spectroscopy. Magn ResonMed. 1995;34:253–259. doi: 10.1002/mrm.1910340217. [DOI] [PubMed] [Google Scholar]
- 18.Thompson RB, Allen PS. A new multiple quantum filter design procedure for use on strongly coupled spin systems found in vivo: its application to glutamate. Magn ResonMed. 1998;39:762–771. doi: 10.1002/mrm.1910390514. [DOI] [PubMed] [Google Scholar]
- 19.Shen J, Rothman DL, Brown P. In vivo GABA editing using a novel doubly selective multiple quantum filter. Magn ResonMed. 2002;47:447–454. doi: 10.1002/mrm.10104. [DOI] [PubMed] [Google Scholar]
- 20.Choi C, Coupland NJ, Hanstock CC, Ogilvie CJ, Higgins AC, Gheorghiu D, Allen PS. Brain gamma-aminobutyric acid measurement by proton double-quantum filtering with selective J rewinding. Magn ResonMed. 2005;54:272–279. doi: 10.1002/mrm.20563. [DOI] [PubMed] [Google Scholar]
- 21.Jensen JE, Frederick BD, Wang L, Brown J, Renshaw PF. Two-dimensional, J-resolved spectroscopic imaging of GABA at 4 Tesla in the human brain. Magn ResonMed. 2005;54:783–788. doi: 10.1002/mrm.20644. [DOI] [PubMed] [Google Scholar]
- 22.Mayer D, Spielman DM. Detection of glutamate in the human brain at 3 T using optimized constant time point resolved spectroscopy. Magn ResonMed. 2005;54:439–442. doi: 10.1002/mrm.20571. [DOI] [PubMed] [Google Scholar]
- 23.Hurd R, Sailasuta N, Srinivasan R, Vigneron DB, Pelletier D, Nelson SJ. Measurement of brain glutamate using TE-averaged PRESS at 3T. Magn ResonMed. 2004;51:435–440. doi: 10.1002/mrm.20007. [DOI] [PubMed] [Google Scholar]
- 24.Thompson RB, Allen PS. Response of metabolites with coupled spins to the STEAM sequence. Magn ResonMed. 2001;45:955–965. doi: 10.1002/mrm.1128. [DOI] [PubMed] [Google Scholar]
- 25.Govindaraju V, Young K, Maudsley AA. Proton NMR chemical shifts and coupling constants for brain metabolites. NMR Biomed. 2000;13:129–153. doi: 10.1002/1099-1492(200005)13:3<129::aid-nbm619>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 26.Choi C, Coupland NJ, Bhardwaj PP, Malykhin N, Gheorghiu D, Allen PS. Measurement of brain glutamate and glutamine by spectrally-selective refocusing at 3 Tesla. Magn ResonMed. 2006;55:997–1005. doi: 10.1002/mrm.20875. [DOI] [PubMed] [Google Scholar]
- 27.Hanstock CC, Coupland NJ, Allen PS. GABA X2 multiplet measured pre- and post-administration of vigabatrin in human brain. Magn ResonMed. 2002;48:617–623. doi: 10.1002/mrm.10265. [DOI] [PubMed] [Google Scholar]
- 28.Thompson RB, Allen PS. Sources of variability in the response of coupled spins to the PRESS sequence and their potential impact on metabolite quantification. Magn ResonMed. 1999;41:1162–1169. doi: 10.1002/(sici)1522-2594(199906)41:6<1162::aid-mrm12>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 29.Kim H, Thompson RB, Hanstock CC, Allen PS. Variability of metabolite yield using STEAM or PRESS sequences in vivo at 3.0 T, illustrated with myoinositol. Magn ResonMed. 2005;53:760–769. doi: 10.1002/mrm.20434. [DOI] [PubMed] [Google Scholar]
- 30.Smith SA, Levante TO, Meier BH, Ernst RR. Computer Simulations in Magnetic Resonance. An Object-Oriented Programming Approach. Journal of Magnetic Resonance, Series A. 1994;106:75–105. [Google Scholar]
- 31.Maudsley AA, Govindaraju V, Young K, Aygula ZK, Pattany PM, Soher BJ, Matson GB. Numerical simulation of PRESS localized MR spectroscopy. JMagn Reson. 2005;173:54–63. doi: 10.1016/j.jmr.2004.11.018. [DOI] [PubMed] [Google Scholar]
- 32.Young K, Matson GB, Govindaraju V, Maudsley AA. Spectral simulations incorporating gradient coherence selection. JMagn Reson. 1999;140:146–152. doi: 10.1006/jmre.1999.1809. [DOI] [PubMed] [Google Scholar]
- 33.Ernst T, Kreis R, Ross BD. Absolute Quantitation of Water and Metabolites in the Human Brain .1. Compartments and Water. Journal of Magnetic Resonance Series B. 1993;102:1–8. [Google Scholar]
- 34.Seeger U, Klose U, Mader I, Grodd W, Nagele T. Parameterized evaluation of macromolecules and lipids in proton MR spectroscopy of brain diseases. Magn ResonMed. 2003;49:19–28. doi: 10.1002/mrm.10332. [DOI] [PubMed] [Google Scholar]
- 35.Soher BJ, Pattany PM, Matson GB, Maudsley AA. Observation of coupled 1H metabolite resonances at long TE. Magn ResonMed. 2005;53:1283–1287. doi: 10.1002/mrm.20491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schubert F, Gallinat J, Seifert F, Rinneberg H. Glutamate concentrations in human brain using single voxel proton magnetic resonance spectroscopy at 3 Tesla. Neuroimage. 2004;21:1762–1771. doi: 10.1016/j.neuroimage.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 37.Shutter L, Tong KA, Holshouser BA. Proton MRS in acute traumatic brain injury: role for glutamate/glutamine and choline for outcome prediction. JNeurotrauma. 2004;21:1693–1705. doi: 10.1089/neu.2004.21.1693. [DOI] [PubMed] [Google Scholar]
- 38.Oh J, Cha S, Aiken AH, Han ET, Crane JC, Stainsby JA, Wright GA, Dillon WP, Nelson SJ. Quantitative apparent diffusion coefficients and T2 relaxation times in characterizing contrast enhancing brain tumors and regions of peritumoral edema. JMagn ResonImaging. 2005;21:701–708. doi: 10.1002/jmri.20335. [DOI] [PubMed] [Google Scholar]
- 39.Chang WK, Yang KD, Shaio MF. Lymphocyte proliferation modulated by glutamine: involved in the endogenous redox reaction. ClinExpImmunol. 1999;117:482–488. doi: 10.1046/j.1365-2249.1999.01009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mates JM, Perez-Gomez C, Nunez dC, Asenjo IM, Marquez J. Glutamine and its relationship with intracellular redox status, oxidative stress and cell proliferation/death. IntJBiochemCell Biol. 2002;34:439–458. doi: 10.1016/s1357-2725(01)00143-1. [DOI] [PubMed] [Google Scholar]
- 41.Choi C, Coupland N, Kalra S, Malykhin N, Bhardwaj P, Gheorghiu D, Allen P. Brain Glutamate Proton Transverse Relaxation Time as measured by Spectrally-Selective Refocusing. Proceedings 14th.Scientific.Meeting., International Society.for Magnetic.Resonance.in Medicine; 2006; p. 3064. [Google Scholar]


