Abstract
We have synthesized and assessed the ability of symmetrical fluorobenzoins and fluorobenzils to inhibit mammalian carboxylesterases (CE). The majority of the latter were excellent inhibitors of CEs however unexpectedly, the fluorobenzoins were very good enzyme inhibitors. Positive correlations were seen with the charge on the hydroxyl carbon atom, the carbonyl oxygen, and the Hammett constants for the derived Ki values with the fluorobenzoins.
Keywords: Fluorobenzil, fluorobenzoin, carboxylesterase, inhibitor
1. INTRODUCTION
Carboxylesterases (CEs) are ubiquitous enzymes responsible for the detoxification of xenobiotics1. It has been reported that CEs can metabolize a wide variety of ester containing compounds including clinical drugs such as, meperidine, flumazenil, procaine, oxybutynin, and the anticancer prodrugs capecitabine and CPT-111-4. Consistent with their proposed function, CEs are expressed in high levels in human tissues such as the liver, lung, small intestine and kidney. All CEs examined to date maintain a catalytic triad of amino acids (serine, histidine and glutamic acid) that are essential for hydrolytic activity.
As CEs are responsible for the metabolism and activation of a host of different clinically useful agents, we have hypothesized that selective inhibitors of these enzymes may be useful in modulating the biological activity of these drugs. For example, for compounds that are inactivated by CEs (e.g. flestolol), addition of an inhibitor may prolong the period of time for which the drug is active5. Conversely for drugs that are activated by these enzymes (e.g. CPT-11), specific CE inhibitors may be useful in ameliorating the toxicity associated with these agents6. Therefore, we have screened for compounds that have selective inhibitory activity towards CEs. This was performed using Telik's Target Related Affinity Profiling (TRAP) technology7-9. Following the identification of compounds that demonstrated activity toward three mammalian CEs, (human intestinal CE (hiCE), human liver CE (hCE1) and rabbit liver CE (rCE)), these chemicals were then assessed for their inhibition of human acetylcholinesterase (AChE). Molecules that inhibited the latter enzyme were discarded. This process identified benzil as a general, selective inhibitor of mammalian CEs10. More recent studies have shown that the characteristics for a good CE inhibitor are the presence of: (i) aromatic rings or increasing hydrophobicity, (ii) a 1,2-dione moiety, and (iii) substitution which does not impede access of the compound reaching the active site of the enzyme10, 11. These studies also demonstrated that benzoin (2-hydroxy-1,2-diphenylethanone), an intermediate in the synthesis of benzil from benzaldehyde, was a poor inhibitor of CEs, consistent with the hypothesis that the 1,2-dione chemotype is important in enzyme inhibition.
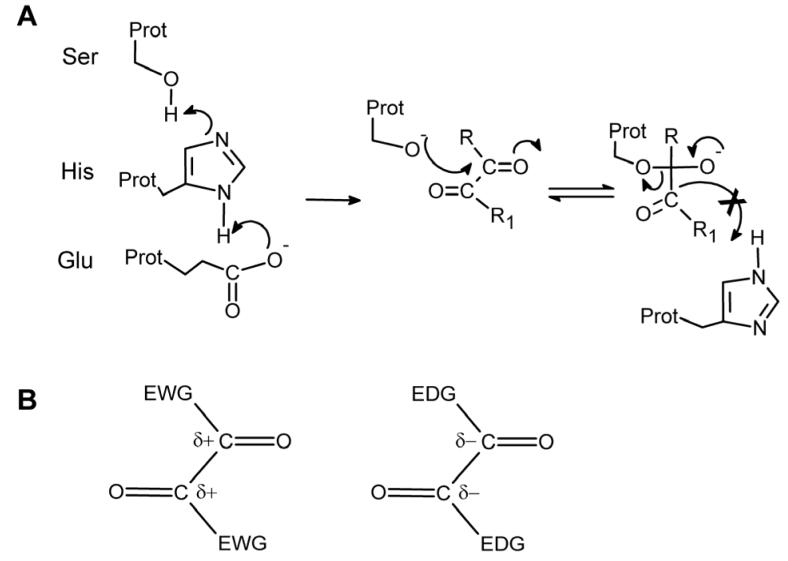
We believe that the benzils are potent inhibitors, in part, because the 1,2-dione structure mimics the ester chemotype allowing for the initiation of the nucleophilic addition-elimination reactions that are observed for this class of compounds (Figure 1A). Abortive nucleophilic attack by the active site serine on one of the carbonyl carbons would yield a tetrahedral intermediate that would be unlikely to undergo C-C cleavage, the next step in the reaction. Therefore, in the presence of benzil, repetitive attack and release by the serine residue on the carbonyl groups would occur, resulting in enzyme inhibition. This hypothesis suggests that decreasing the electron density around the carbonyl carbon atom would make this atom more susceptible to nucleophilic attack by the serine oxygen. Therefore appropriate inclusion of electron withdrawing groups (EWG) should increase the likelihood of attack and presumably the potency of the inhibitors (Figure 1B). In this series of studies, we also assessed the ability of the benzoin to inhibit the mammalian CEs. Benzoins (α-hydroxy ketones) were chosen because they are key intermediates in the synthesis of benzils from the aldehydes and they possess similar structural characteristics to benzil i.e. aromatic rings, carbonyl groups, etc (see Tables 1 and 2). While we have previously demonstrated that the benzoins are poor inhibitors of CEs10, modification of the electron density associated with the dione carbon atoms, might have the potential to produce compounds that demonstrate inhibitory activity.
Figure 1.
(A) Proposed mechanism of interaction of the benzil analogues with the catalytic amino acids of CEs. A serine nucleophile is generated by proton transfer to a glutamic acid via a histidine residue, and the resulting oxygen atom attacks one of the carbonyl groups within the dione moiety. The tetrahedral intermediate that is generated is relatively stable, due to the increased strength of the C-C bond as compared to the C-O bond present in esters. Therefore the former bond is not cleaved resulting in inhibition of the enzyme. (B) Increasing or decreasing the electron density surrounding the carbonyl carbon atoms by introducing either electron withdrawing groups (EWG) or electron donating groups (EDG) within the molecules, should make the compounds better and poorer enzyme inhibitors, respectively.
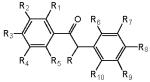
Table 1.
Structure of the fluorobenzoins used in this study
| ID | Name | R | R1 | R2 | R3 | R4 | R5 | R6 | R7 | R8 | R9 | R10 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Benzoin (1,2-diphenyl-2-hydroxy- ethanone) |
OH | ||||||||||
| 3 | 1,2-bis-(2-fluorophenyl)-2- hydroxyethanone |
OH | F | F | ||||||||
| 4 | 1,2-bis-(3-fluorophenyl)-2- hydroxyethanone |
OH | F | F | ||||||||
| 5 | 1,2-bis(4-fluorophenyl)-2-2- hydroxyethanone |
OH | F | F | ||||||||
| 6 | 1,2-bis(2,4-difluorophenyl)-2- hydroxyethanone |
OH | F | F | F | F | ||||||
| 7 | 1,2-bis-(2,6-difluorophenyl)-2- hydroxyethanone |
OH | F | F | F | F | ||||||
| 8 | 1,2-bis(3,4-difluorophenyl)-2- hydroxyethanone |
OH | F | F | F | F | ||||||
| 9 | 1,2-bis(2,3-difluorophenyl)-2- hydroxyethanone |
OH | F | F | F | F | ||||||
| 10 | 1,2-bis(2,5-difluorophenyl)-2- hydroxyethanone |
OH | F | F | F | F | ||||||
| 11 | 1,2-bis(3,5-difluorophenyl)-2- hydroxyethanone |
OH | F | F | F | F | ||||||
| 12 | 1,2-bis-(2,3,5-trifluorophenyl)-2- hydroxyethanone |
OH | F | F | F | F | F | F | ||||
| 13 | 1,2-bis-(2,3,4-trifluorophenyl)-2- hydroxyethanone |
OH | F | F | F | F | F | F | ||||
| 14 | 1,2-bis-(3,4,5-triflurophenyl)-2- hydroxyethanone |
OH | F | F | F | F | F | F | ||||
| 15 | 1,2-bis-[4-(trifluoromethyl)phenyl]- 2-hydroxyethanone |
OH | CF3 | CF3 | ||||||||
| 16 | 1,2-bis-[3-(trifluoromethyl)phenyl]- 2-hydroxyethanone |
OH | CF3 | CF3 | ||||||||
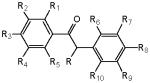
Table 2.
Structure of the fluorobenzils used in this study
| ID | Name | R | R1 | R2 | R3 | R4 | R5 | R6 | R7 | R8 | R9 | R10 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | Benzil (diphenylethane-1,2-dione) | =O | ||||||||||
| 17 | 1,2-bis-(2-fluorophenyl)- ethane- 1,2-dione |
=O | F | F | ||||||||
| 18 | 1,2-bis-(3-fluorophenyl)-ethane- 1,2-dione |
=O | F | F | ||||||||
| 19 | 1,2-bis(4-fluorophenyl)-ethane- 1,2-dione |
=O | F | F | ||||||||
| 20 | 1,2-bis(2,4-difluorophenyl)- ethane-1,2-dione |
=O | F | F | F | F | ||||||
| 21 | 1,2-bis(2,6-difluorophenyl)- ethane-1,2-dione |
=O | F | F | F | F | ||||||
| 22 | 1,2-bis(3,4-difluorophenyl)- ethane-1,2-dione |
=O | F | F | F | F | ||||||
| 23 | 1,2-bis(2,3-diflurophenyl)-ethane- 1,2-dione |
=O | F | F | F | F | ||||||
| 24 | 1,2-bis(2,5-difluorophenyl)- ethane-1,2-dione |
=O | F | F | F | F | ||||||
| 25 | 1,2-bis(3,5-difluorophenyl)- ethane-1,2-dione |
=O | F | F | F | F | ||||||
| 26 | 1,2-bis(2,3,6-trifluophenyl)- ethane-1,2-dione |
=O | F | F | F | F | F | F | ||||
| 27 | 1,2-bis(2,3,5-trifluorophenyl) ethane-1,2-dione |
=O | F | F | F | F | F | F | ||||
| 28 | 1,2-bis(2,3,4-trifluorophenyl) ethane-1,2-dione |
=O | F | F | F | F | F | F | ||||
| 29 | 1,2-bis(3,4,5-triflurophenyl) ethane-1,2-dione |
=O | F | F | F | F | F | F | ||||
| 30 | 1,2-bis[4-(trifluoromethyl)phenyl] ethane-1,2-dione |
=O | CF3 | CF3 | ||||||||
| 31 | 1,2-bis[3(trifluoromethyl)phenyl] ethane-1,2-dione |
=O | CF3 | CF3 | ||||||||
| 32 | 1,2-bis[2,4- bis(trifluoromethyl)phenyl] ethane- 1,2-dione |
=O | CF3 | CF3 | CF3 | CF3 | ||||||
| 33 | 1,2-bis[3,5- bis(trifluoromethyl)phenyl] ethane- 1,2-dione |
=O | CF3 | CF3 | CF3 | CF3 | ||||||
| 34 | 1,2-bis[2,5- bis(trifluoromethyl)phenyl] ethane-1,2-dione |
=O | CF3 | CF3 | CF3 | CF3 | ||||||
To test these hypotheses, we have synthesized a panel of fluorobenzoins and their analogous fluorobenzils, and assessed them for CE inhibition using hiCE, hCE1 and rCE. Results presented here indicate that fluorine substitution within the benzene rings generates benzoin analogs that are potent inhibitors of mammalian CEs.
2. RESULTS
2.1. Analysis of carboxylesterase inhibition by fluorobenzils
In a previous paper, we hypothesized that nucleophilic attack at one of the carbonyl groups within the 1,2-dione moiety of benzil (2) by the catalytic serine was in part, responsible for enzyme inhibition (Figure 1A)10. We therefore surmised that substituted benzene rings, which would withdraw electron density from these carbon atoms, might improve the potency of CE inhibition (Figure 1B). To assess the validity of this hypothesis, we synthesized a panel of fluorobenzoins and fluorobenzils for use in enzyme inhibition studies. The structures of the compounds used for these assays are shown in Tables 1 and 2.
The ability of these compounds to inhibit hiCE, hCE1, rCE, human AChE and human BChE was then determined and the Ki values for enzyme inhibition are reported in Tables 3 and 4. As indicated, the fluorobenzil analogs were all relatively good inhibitors of CEs with Ki values ranging from 3nm to 2.8μM. The most potent inhibitor was 1,2-bis(2,3-difluorophenyl)ethane-1,2-dione (23), yielding a Ki value of 3.3nM with rCE. Interestingly, the inhibition constants were as high as 2.84μM with 1,2-bis[2,4-bis(trifluoromethyl)phenyl]ethane-1,2-dione (32), with hiCE. In general, inhibitors that were potent for one enzyme demonstrated similar levels of activity towards the other two CEs. Overall, these results demonstrate that these fluorobenzils are good inhibitors; however, fluorine substitution did not dramatically increase the inhibitory potency of these compounds as compared to benzil. The latter compound demonstrates Ki values of 15.1nM, 45.1nM and 103nM for hiCE, hCE1 and rCE, respectively.
Table 3.
Ki values for enzyme inhibition using the benzoin analogs. Values for CEs were determined using o-NPA as a substrate, and those for AChE and BChE using AcTCh or BuTCh as a substrate, respectively.
| ID | hiCE Ki ± SE (nM) |
hCE1 Ki ± SE (nM) |
rCE Ki ± SE (nM) |
AChE Ki (nM) | BChE Ki (nM) |
|---|---|---|---|---|---|
| 1 | 2,670 ± 1,010 | 7,210 ± 2,410 | >100,000 | >100,000 | >100,000 |
| 3 | 250 ± 23 | 990 ± 100 | 91.4 ± 44.0 | >100,000 | >100,000 |
| 4 | 640 ± 80 | 3,130 ± 470 | 660 ± 45 | >100,000 | >100,000 |
| 5 | >100,000a | >100,000 | >100,000 | >100,000 | >100,000 |
| 6 | 390 ± 44 | 480 ± 30 | 140 ± 18 | >100,000 | >100,000 |
| 7 | 120 ± 9 | 190 ± 5 | 61.8 ± 2.9 | >100,000 | >100,000 |
| 8 | 150 ± 17 | 1,300 ± 210 | 220 ± 22 | >100,000 | >100,000 |
| 9 | 260 ± 15 | 730 ± 60 | 200 ± 39 | >100,000 | >100,000 |
| 10 | 55.2 ± 3.7 | 230 ± 7 | 48.3 ± 1.7 | >100,000 | >100,000 |
| 11 | 71.0 ± 9.0 | 170 ± 16 | 18.4 ± 1.0 | >100,000 | >100,000 |
| 12 | 99.5 ± 9.2 | 665 ± 30 | 31.7 ± 3.9 | >100,000 | >100,000 |
| 13 | 25.7 ± 3.1 | 260 ± 13 | 8.3 ± 0.4 | >100,000 | >100,000 |
| 14 | 1,040 ± 50 | 1,000 ± 50 | 85.7 ± 4.6 | >100,000 | >100,000 |
| 15 | 400 ± 35 | 870 ± 390 | 12.9 ± 2.9 | >100,000 | >100,000 |
| 16 | 220 ± 17 | >100,000 | 42.4 ± 4.4 | >100,000 | >100,000 |
>100,000 indicates less than 50% enzyme inhibition at an inhibitor concentration of 100μM, i.e. essentially no inhibition.
Table 4.
Ki values for enzyme inhibition using the benzil analogs. Values for CEs were determined using o-NPA as a substrate, and those for AChE and BChE using AcTCh or BuTCh as a substrate, respectively.
| ID | hiCE Ki ± SE (nM) |
hCE1 Ki ± SE (nM) |
rCE Ki ± SE (nM) |
AChE Ki (nM) | BChE Ki (nM) |
|---|---|---|---|---|---|
| 2 | 15.1 ± 2.0 | 45.1 ± 3.4 | 103 ± 20 | >100,000 | >100,000 |
| 17 | 55.9 ± 3.4 | 185 ± 1 | 47.8 ± 1.4 | >100,000 | >100,000 |
| 18 | 25.9 ± 5.1 | 100 ± 27 | 36.2 ± 7.3 | >100,000 | >100,000 |
| 19 | 170 ± 12 | 230 ± 11.8 | 400 ± 59 | >100,000 | >100,000 |
| 20 | 270 ± 41 | 370 ± 109 | 98.0 ± 28.8 | >100,000 | >100,000 |
| 21 | 830 ± 63 | 5,650 ± 420 | 1,170 ± 90 | >100,000 | >100,000 |
| 22 | 150 ± 17 | 300 ± 8 | 49.3 ± 6.3 | >100,000 | >100,000 |
| 23 | 4.98 ± 0.16 | 34.7 ± 2.7 | 3.34 ± 0.02 | >100,000 | 1,270 ± 100 |
| 24 | 61.4 ± 2.7 | 260 ± 24 | 60.8 ± 9.0 | >100,000 | >100,000 |
| 25 | 30.6 ± 3.4 | 97.2 ± 8.5 | 22.8 ± 3.5 | >100,000 | >100,000 |
| 26 | 40.7 ± 3.8 | 320 ± 33 | 33.8 ± 8.2 | >100,000 | >100,000 |
| 27 | 31.9 ± 8.8 | 140 ± 28 | 28.9 ± 10.1 | >100,000 | >100,000 |
| 28 | 13.6 ± 3.4 | 91.3 ± 12.3 | 3.3 ± 0.6 | >100,000 | >100,000 |
| 29 | 360 ± 62 | 470 ± 113 | 67.9 ± 4.5 | >100,000 | >100,000 |
| 30 | 180 ± 91 | 170 ± 45 | 12.7 ± 6.0 | >100,000 | >100,000 |
| 31 | 250 ± 55 | >100,000 | 150 ± 38 | >100,000 | >100,000 |
| 32 | 2,840 ± 950 | >100,000 | >100,000 | >100,000 | >100,000 |
| 33 | 550 ± 170 | >100,000 | >100,000 | >100,000 | >100,000 |
| 34 | 2,370 ± 630 | >100,000 | >100,000 | >100,000 | >100,000 |
The trifluoromethyl analogs were less effective CE inhibitors than the fluorobenzils. For example, only the para- substituted compound (30) inhibited all three mammalian enzymes. Indeed, none of the other trifluoromethyl analogues that we analyzed inhibited hCE1 and compounds 32-34 also failed to inhibit rCE. We believe that this is likely due to steric constraints afforded by the CF3 group(s), Since we have previously demonstrated that the entrance to the active site of hCE1 is considerably smaller and less flexible the other CEs12. As the trifluoromethyl compounds are more bulky than the other benzils assayed in these studies, it is likely that steric hinderance prevents access to hCE1 and rCE active sites, and hence they do not inhibit these proteins. The benzoin analogs of compounds 32 - 34 were not produced due to their direct oxidation to the benzil derivative under the reaction conditions employed. This is likely due to the electron-withdrawing effects of the trifluoromethyl groups resulting in increased reactivity of the oxygen atoms and hence facile oxidation to the corresponding benzil.
2.2. Analysis of carboxylesterase inhibition by fluorobenzoins
Benzoin, 1, is not a good inhibitor of mammalian CEs; it demonstrates Ki values of 2.7μM and 7.2μM for hiCE and hCE1, respectively, and is inactive towards rCE10. However the addition of fluorine atoms to the benzene rings in the benzoins resulted in compounds that were very potent inhibitors of CEs (Table 4). The majority of the fluorobenzoin analogs had Ki values ranging from 8nM to 1.3μM, with the most potent inhibitor being 1,2-bis(2,3,4-trifluorophenyl)-2-hydroxyethanone (13), yielding a Ki value of 8.3nM with rCE. With some exceptions, the fluorobenzoins were more potent inhibitors than benzoin. In contrast to the benzil analogs, fluorine substitutions on the benzene rings resulted in marked increases in the inhibitory potency of the benzoins. For example, compound 11, demonstrated Ki values of 71.1nM, 170nM, and 18.4nM for hiCE, hCE1, and rCE, respectively. These values are 37- to 5400-fold lower than that observed for the unsubstituted benzoin (1) for the same enzymes. However, there were examples where the patterns of enzyme inhibition were very different. As an example, compounds 3 and 4 inhibited all three enzymes, but 5, did not inhibit any of the enzymes (see Table 3). The only difference between these analogs is the position of the single fluorine atom in the benzene ring. Our results suggest therefore, that bond polarization plays a major role in mono-substituted aromatic rings causing inductive effects to prevail when influencing the electron density associated with the α carbon atom. This inductive effect is likely responsible for the differential inhibition of the three mammalian CEs across the mono-substituted series.
In the disubstituted analogues, it was apparent that resonance effects could account for the differences in the inhibition constants for the mammalian CEs. For example compound 11 demonstrated lower Ki values than 6 or 9, probably due to withdrawal of electrons from the dione moiety via the fluorine atoms at the 3- and 5-positions on the benzene ring. Due to the distances involved, this is unlikely to be mediated by inductive effects, but rather due to the stabilization of resonance structures where the π electrons are conjugated via the benzene rings. Substitution of fluorine atoms at the 3- and 5- positions will enhance the stability of the resonance structures, yielding a more electron deficient environment at the dione carbon atoms. This would make them more susceptible to attack by the serine nucleophile and therefore better CE inhibitors.
2.3. Assessment of electronic parameters for the fluorobenzils and fluorobenzoins
Having demonstrated that fluorine substitution increased the potency of the fluorobenzoin analogs, we decided to evaluate the electron distribution within these compounds using computational methods. Therefore, we calculated a variety of electronic parameters for the fluorobenzoins and fluorobenzils using density functional theory (Table 5). These included the atomic charges of the oxygen and carbon atoms within the dione (or the hydroxy-ethanone) group, the pKa of the hydroxyl group of the benzoins, and Hammett substituent constants. The results were then analyzed and compared to the Ki values for each compound, with each mammalian CEs.
Table 5.
Calculated electronic parameters for the compounds used in this study.
| ID | Chargea C–OH |
Chargea C=O |
Chargea C–OH |
Chargea C=O |
Benzoin OH pKa |
Hammett δ |
|---|---|---|---|---|---|---|
| 1 | 0.019 | 0.411 | −0.531 | −0.444 | 7.909 | 0 |
| 2 | 0.342 | −0.462 | 0 | |||
| 3 | 0.029 | 0.389 | −0.531 | −0.444 | 7.487 | 1.060 |
| 4 | 0.022 | 0.412 | −0.539 | −0.442 | 7.214 | 0.674 |
| 5 | 0.017 | 0.411 | −0.539 | −0.452 | 7.209 | 0.124 |
| 6 | 0.034 | 0.410 | −0.545 | −0.444 | 6.668 | 0.592 |
| 7 | 0.041 | 0.386 | −0.534 | −0.426 | 7.230 | 1.060 |
| 8 | 0.020 | 0.414 | −0.539 | −0.445 | 7.713 | 0.399 |
| 9 | 0.027 | 0.395 | −0.529 | −0.439 | 7.052 | 0.867 |
| 10 | 0.026 | 0.373 | −0.544 | −0.394 | 6.719 | 0.867 |
| 11 | 0.028 | 0.410 | −0.538 | −0.436 | 8.435 | 0.674 |
| 12 | 0.025 | 0.401 | −0.526 | −0.433 | 6.855 | 1.204 |
| 13 | 0.034 | 0.412 | −0.544 | −0.438 | 7.079 | 0.929 |
| 14 | 0.022 | 0.418 | −0.538 | −0.439 | 8.342 | 0.736 |
| 15 | 0.029 | 0.347 | −0.539 | −0.452 | 10.764 | 1.080 |
| 16 | 0.021 | 0.418 | −0.538 | −0.440 | 10.784 | 0.860 |
| 17 | 0.317 | −0.416 | 1.060 | |||
| 18 | 0.348 | −0.456 | 0.674 | |||
| 19 | 0.343 | −0.465 | 0.124 | |||
| 20 | 0.318 | −0.419 | 0.867 | |||
| 21 | 0.337 | −0.400 | 1.060 | |||
| 22 | 0.281 | −0.376 | 0.399 | |||
| 23 | 0.321 | −0.411 | 0.867 | |||
| 24 | 0.321 | −0.410 | 0.867 | |||
| 25 | 0.353 | −0.449 | 0.674 | |||
| 26 | 0.341 | −0.395 | 1.397 | |||
| 27 | 0.326 | −0.405 | 1.024 | |||
| 28 | 0.354 | −0.445 | 0.929 | |||
| 29 | 0.352 | −0.453 | 0.736 | |||
| 30 | 0.346 | −0.452 | 1.080 | |||
| 31 | 0.349 | −0.453 | 0.860 | |||
| 32 | 0.318 | −0.414 | 0.750 | |||
| 33 | 0.357 | −0.447 | 0.860 | |||
| 34 | 0.298 | −0.400 | 0.640 | |||
The charge on the respective atom is indicated in bold.
As can be seen in Table 6, no correlations were observed between the charges present on the carbonyl carbon or oxygen atoms with the benzils and the observed Ki values for CE inhibition. In addition, poor Spearman r correlation coefficients were seen with the predicted charge on the hydroxyl oxygen atom in the benzoins, when compared to the enzyme inhibition data. However, we observed significant P values (<0.05) for the correlation analyses when the charge on the carbonyl oxygen atom of the fluorobenzoins was compared to the Ki values for the mammalian CEs (Table 6). Additionally, a correlation (p= 0.077 with hiCE at the 10% level) was seen between the charge on the hydroxyl carbon atom and the Ki values for the inhibition of hCE1 and rCE with these compounds. Overall, these results indicate that the relative efficiency of inhibition by the benzoins is related to the electron density surrounding these atoms within the hydroxy-ethanone moiety. Perhaps not surprisingly, no obvious correlations were apparent when using linear regression analysis of the datasets (Table 6). No correlations were seen between the Ki values for enzyme inhibition and the pKa value of the hydroxyl proton in the benzoins (Table 7). Interestingly, a significant P value was seen for the correlation between the Hammett constants and the Ki values for rCE with the benzoins, but not, however, with the benzils (Table 8). This is likely a consequence of the lack of polarizability in the C-OH bond relative to the C=O bond.
Table 6.
Correlation coefficients for the atomic charges on the hydroxyl and the carbonyl oxygen and carbon atoms in the fluorobenzoins and the fluorobenzils, with the observed Ki values for CE inhibition.
| Compounds | Atoma | Linear regression (r2) |
Spearman correlation coefficient (Spearman r) |
P value for Spearman correlationb | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| hiCE | hCE1 | rCE | hiCE | hCE1 | rCE | hiCE | hCE1 | rCE | ||
| Benzoins | C–OH | 0.16 | 0.22 | 0.25 | −0.470 | −0.757 | −0.533 | 0.077 | 0.001 | 0.041 |
| C=O | 0.02 | 0.08 | 0.04 | 0.176 | 0.533 | 0.223 | 0.53 | 0.04 | 0.43 | |
| C–OH | 0.02 | 0.03 | 0.00 | −0.198 | −0.146 | −0.013 | 0.48 | 0.60 | 0.96 | |
| C=O | 0.23 | 0.11 | 0.20 | −0.672 | −0.665 | −0.518 | 0.006 | 0.007 | 0.05 | |
| Benzils | C=O | 0.07 | 0.0 | 0.03 | −0.118 | −0.152 | −0.166 | 0.63 | 0.53 | 0.50 |
| C=O | 0.03 | 0.0 | 0.02 | 0.115 | 0.245 | −0.041 | 0.64 | 0.31 | 0.87 | |
Charge on specific atom is indicated in bold.
P values ≤ 0.05 are indicated in bold.
Table 7.
Correlation coefficients for the pKa values for the hydroxyl proton in the fluorobenzoins with the observed Ki values for CE inhibition.
| Compound | Linear regression (r2) |
Spearman correlation coefficient (Spearman r) |
P value for Spearman correlation | ||||||
|---|---|---|---|---|---|---|---|---|---|
| hiCE | hCE1 | rCE | hiCE | hCE1 | rCE | hiCE | hCE1 | rCE | |
| Benzoins | 0.02 | 0.13 | 0.01 | 0.232 | 0.336 | −0.170 | 0.41 | 0.22 | 0.55 |
Table 8.
Correlation coefficients for the Hammett substituents constants for the fluorobenzoins and the fluorobenzils, with the observed Ki values for CE inhibition.
| Compound | Linear regression (r2) |
Spearman correlation coefficient (Spearman r) |
P value for Spearman correlationa | ||||||
|---|---|---|---|---|---|---|---|---|---|
| hiCE | hCE1 | rCE | hiCE | hCE1 | rCE | hiCE | hCE1 | rCE | |
| Benzoins | 0.26 | 0.11 | 0.63 | −0.489 | −0.462 | −0.703 | 0.065 | 0.083 | 0.004 |
| Benzils | 0.0 | 0.0 | 0.0 | −0.041 | 0.006 | 0.407 | 0.87 | 0.98 | 0.08 |
P values ≤ 0.05 are indicated in bold.
2.4. QSAR analysis of fluorobenzoin- and fluorobenzil-mediated inhibition of carboxylesterases
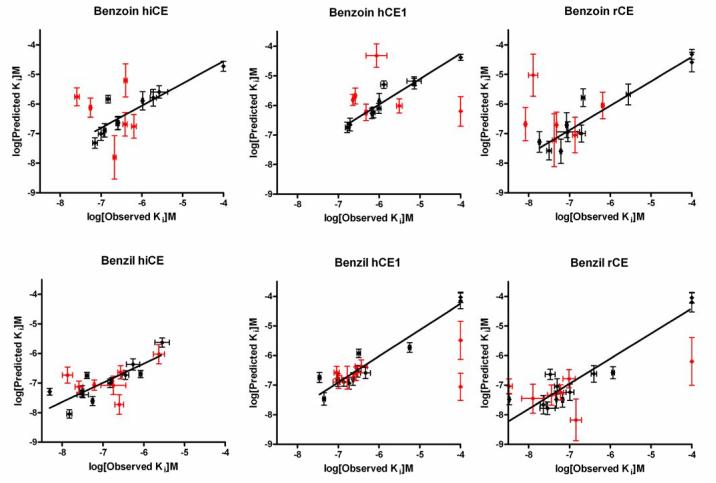
Since the results obtained from the comparisons of the enzyme inhibition data and the electronic parameters indicated that the charge distribution within the central carbon atoms may be an important factor in CE inhibition, we performed 3D-QSAR analysis of the data using Quasar software. These experiments generated linear correlation coefficients (r2) values ranging from 0.65 - 0.89 for the observed versus predicted Ki values for CE inhibition (Table 9; Figure 2). In addition, excellent cross correlation coefficients (q2) were obtained from these analyses. Since these values, and q2/r2 ratios, are all close to unity, these results suggest that the models have considerable predictive power in assessing CE inhibition by the benzoins and the benzils.
Table 9.
Correlation coefficients for the QSAR models.
| Compound | Enzyme | Observed versus predicted Ki values (r2) |
Cross correlation coefficient (q2) |
q2/r2 |
|---|---|---|---|---|
| Benzoins | hiCE | 0.816 | 0.751 | 0.92 |
| hCE1 | 0.914 | 0.861 | 0.94 | |
| rCE | 0.898 | 0.826 | 0.92 | |
| Benzils | hiCE | 0.686 | 0.648 | 0.94 |
| hCE1 | 0.918 | 0.887 | 0.97 | |
| rCE | 0.885 | 0.848 | 0.96 | |
Figure 2.
Graphs depicting the observed versus the predicted Ki values for the inhibition of the CEs following QSAR analysis. Data points used to construct the model are shown in black, and the test values are indicted in red.
Graphic representations of the 3D-QSAR models for hCE1, hiCE, and rCE are depicted in Figure 3. No simple descriptor of the active site gorge would explain the inhibitory activity of the benzils and benzoins toward each enzyme. Rather, as we previously observed with other benzil analogs10, it is the overall hydrophobic and electrostatic milieu of the binding site that contributes to the affinity of each analog, and these characteristics are slightly different for each enzyme.
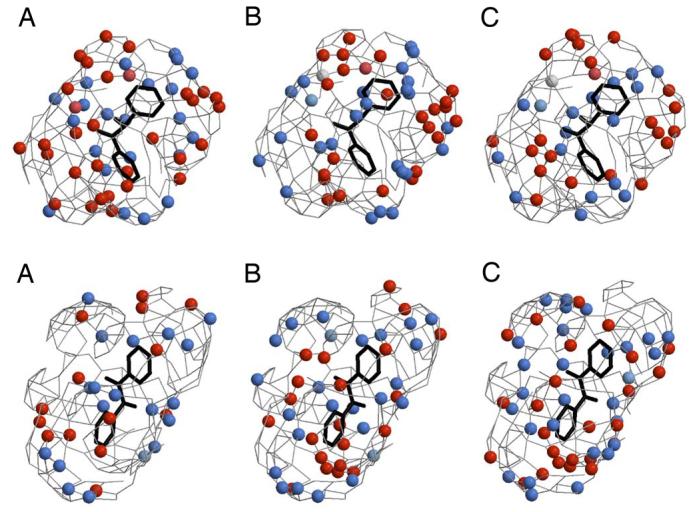
Figure 3.
3D-QSAR pseudoreceptor models for the inhibition of CEs by the fluorobenzoins (upper panel) and the fluorobenzils (lower panel). In each case, the models for hiCE (A), hCE1 (B), and rCE (C) are shown as colored spheres on a hydrophobic gray grid. Areas that are hydrophobic are indicated in gray, with blue spheres representing regions that are positively charged and hydrophobic (+0.1e), and light blue spheres corresponding to hydrogen bond donors. Orange spheres indicate hydrogen bond acceptors and orange-red spheres correspond to areas that are negatively charged and hydrophobic (−0.1e). The structure of benzil or benzoin is shown in black. The figure was created using Raster3D and Molscript32.
3. Discussion
In this paper, we have examined the fluorine substituent effects on the inhibitory potency of a panel of fluorobenzoins and fluorobenzils towards mammalian CEs. These studies were initiated since we had previously hypothesized that the electron density surrounding the carbonyl carbon atoms in the 1,2-dione moiety may be an important factor in inhibitor potency10. If nucleophilic attack on these atoms, by the catalytic serine in the active site of the CE, is necessary for enzyme inhibition (Figure 1), then reducing the electron density around the carbonyl carbons may make the inhibitors more potent. We reasoned that this could be achieved by introducing EWG (e.g. fluorine atoms) as substituents to the benzene rings of these compounds.
While inclusion of fluorine (or multiple fluorine atoms) within the benzil analogues did not dramatically improve their inhibitory potency, the corresponding benzoin compounds were considerably more potent CE inhibitors (Table 4). This effect was most pronounced when the fluorine atoms were either at the ortho- or the meta- positions. For example, compounds 3 (ortho-substituted) and 4 (meta-substituted), demonstrated inhibition towards all CEs; whereas 5 (para-substituted), displayed no inhibitory effect towards any of the enzymes (see Table 4). The lack of increase in the potency of the fluorobenzil analogues may be due to the fact that the electrons in both the α and the π orbitals in the carbonyl bonds are principally distributed towards the more electronegative O atom. Therefore, any attempt to reduce the electron density surrounding the carbon atom (by substituting EWG groups in the benzene ring) will actually redistribute the electrons within the polarizable π bond towards the carbon atom. This would reduce the overall positive charge on this atom (consistent with the results seen in Table 5) rendering it less susceptible to nucleophilic attack. This is also consistent with our experimental results (e.g. most of the fluorobenzils are poorer inhibitors of the human CEs than benzil; Table 4). The change in the electron density, due to polarization of the C-OH bond in the fluorobenzoins by addition of fluorine atoms, would be much less pronounced and hence this effect is unlikely to be observed with these compounds. Again, this is born out by both the MO calculations (Table 5) and the inhibition assays (Table 3).
The substituent effects exerted by the fluorine atoms could be achieved by any one (or a combination) of three different properties. These include resonance, inductive and/or field effects. Being highly electronegative, it is likely that the inductive effects produced by these atoms would be sufficient to modulate the inhibitory activity of the benzoin analogs. The results we obtained are consistent with the inductive effects exerted by the fluorine atoms via the sigma bonds in the molecule. Hence, when the fluorine groups are closer to the hydroxy-ethanone moiety (e.g. in the ortho- position), they have a more pronounced effect on the charge on the carbon atoms, and hence the Ki values are lower. In compounds where the fluorine group is further from the hydroxy-ethanone chemotype, (e.g. the meta- or para- substituted analogues) there is a considerable reduction in the potency of these molecules as CE inhibitors. As a consequence, an order for the Ki values for the single substituted fluorobenzoins was observed where ortho-<meta-<para- (3<4<5). Similarly, with the di-substituted analogs, the ortho-meta-substituted benzoin (10), was more potent at CE inhibition than the ortho-para-analogue (6). Again, these data are all consistent with the reduced inductive effects that would be exerted by the fluorine atoms in the para- position as compared to those in the meta- position.
Additionally, intramolecular hydrogen bonding between fluorine in the ortho-position and the benzoin hydroxyl hydrogen atom may influence inhibitor potency. Such a bond would generate a relatively stable 6 membered ring that may minimize or impede rotation of the benzene ring in the benzoin. Such an interaction might be favorable, e.g. if the inhibitor is locked in a conformation that is more potent at enzyme inhibition, or unfavorable, if the reverse is true. Comparison of the Ki values for different ortho- substituted benzoin analogues versus their meta- substituted isomers (e.g. 2 vs 3, 6 vs 8 or 7 vs 11) however, revealed no clear pattern of enzyme inhibition. This may be due to the fact that all of the biochemical assays are performed in aqueous solution (∼55M H2O), and hence the formation of a stable F■■■H–O–C intramolecular hydrogen bond would be unlikely.
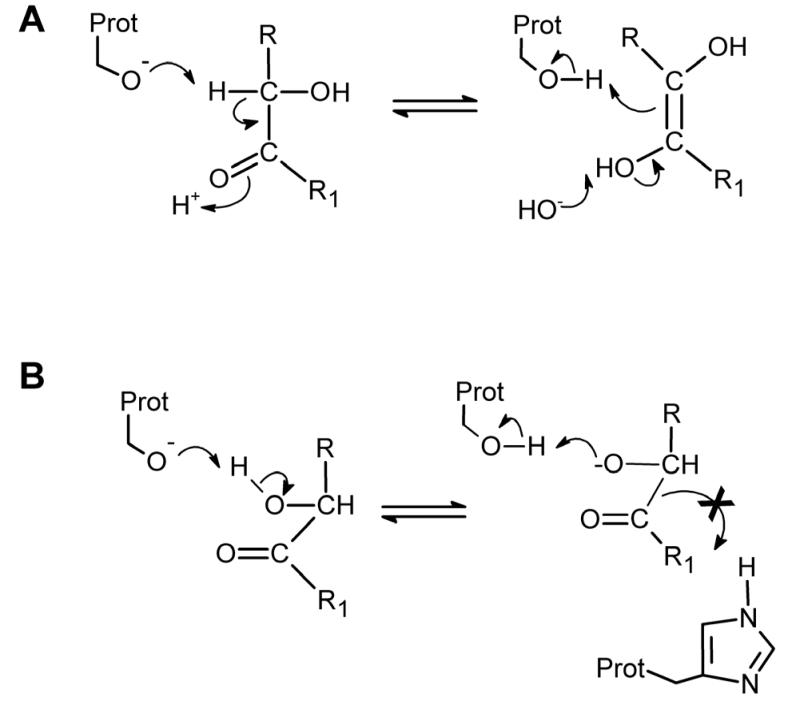
Attempts to correlate the predicted charge on the carbon atoms within the dione moiety of the benzils with the Ki values for CE inhibition proved unsuccessful (Table 6). However, similar analyses indicated that for the benzoins, the charge on the hydroxyl carbon was highly correlated with the inhibition constants. Indeed, Spearman r coefficients of −0.470, −0.757 and −0.533 and P values of 0.077, 0.001 and 0.041 were obtained for hiCE, hCE1 and rCE, respectively. Additionally, we observed similar correlation with the charges on the carbonyl oxygen atom and the Ki values (Table 6). This suggests that the initiating event in enzyme inhibition in the case of the benzoins is the interaction of the serine nucleophile with the hydroxyl carbon (Figure 4A). Since the inhibition is reversible, the formation of any diol intermediate (Figure 4A) is presumably transient and presumably repetitive removal and release of the proton on the hydroxyl carbon atom would occur. This mechanism would account for the inhibition of the CEs.
Figure 4.
Proposed mechanism of inhibition of CEs by the fluorobenzoins.
(A) The serine nucleophile (Prot-O−) removes the proton on the hydroxy carbon atom to yield an intermediate that rearranges to form the ethylene diol analogue. However, this species is probably in equilibrium with the benzoin and hence the reaction is readily reversible. This repetitive transfer of the proton likely inhibits the CE.
(B) In a reversible process, the proton on the oxygen atom of the hydroxyl group of the benzoin is transferred to the serine oxygen atom. This results in the formation of the hydroxyl moiety on the serine residue. However, this would likely be attacked by the oxygen atom in the benzoin to reform the starting materials.
Alternatively, since the acidity and hence the lability of the hydroxyl proton is mediated in part by the electronegativity of the carbon atom, it is possible that the O− present within the serine may remove this proton to regenerate the amino acid (Figure 4B). Again, this would likely be in rapid equilibrium such that upon removal of the inhibitor, free active protein would be obtained. It is currently unknown whether either of the above mechanisms is correct, but both chemical and structural studies are underway to assess the validity of these scenarios. Since we did not identify one single parameter that would reflect the biological activity of all of the benzoins and the benzils, we used 3D-QSAR analysis of the datasets, to derive suitable relationships that would correlate the chemical structures of the inhibitors with their biological potency. The advantage of the 3D approach is that multiple parameters can be simultaneously evaluated and hence models that more accurate represent the inhibition of CEs can be obtained. In addition, we have previously performed similar analyses for a variety of different CE inhibitors, generating highly predictive 3D-pseudoreceptor site models for the different mammalian proteins6, 10.
Analysis of the inhibition data for the benzoins and the benzils using Quasar software yielded good r2 values for the observed versus predicted Ki values (Table 9; Figure 2). In addition, cross correlation coefficients (q2) ranging from 0.65 – 0.89 were obtained for the datasets. Since q2 provides a measure of the predictive power of the model, and values greater than 0.4 are generally assumed to be suitable for use in biological systems13, our results suggest that the 3D-pseudoreceptor site models will be useful in the design of novel fluorine-based benzoin and benzil CE inhibitors.
Six inhibitors with known Ki values were used to test the ability of the benzoins models to predict Ki values, while 7 were used to test the benzils models. In general, prediction was accurate (as indicated in Figure 2). However, in all of the benzoin models, compound 15, having symmetric p-trifluoromethyl groups on the phenyl rings was systematically predicted to be a better inhibitor than was observed experimentally. Likewise, the inhibition constants for the benzil analog 31 were mis-predicted for hCE1 and rCE. Analysis of the models and data did not reveal any obvious reason why these inhibitors should have lower biological activity than was predicted. We hypothesize that these inhibitors may be interacting with sites on the surface of the CE's that cannot be accounted for in the Ki data, particularly the opening to the active site gorge12. We have demonstrated that the entrance to the CE active site can influence substrate turnover and hence it is likely that the same holds true for enzyme inhibitors. For example, 31 was correctly predicted by the QSAR model to have good inhibitory activity towards rCE. As rCE has a demonstrably larger opening to the entrance to the active site gorge on the surface of the enzyme, this presumably allows access of bulkier inhibitors to the catalytic amino acids. Since both hiCE and hCE1 have more constrained active site entrances12, potentially facile access of compound 31 to the catalytic amino acids would be impeded. This would not be predicted in the current models. In contrast, the biological activity of compound 30 was accurately predicted by the QSAR models. Since 30 and 31 are isomers, we believe that it is unlikely that the low experimental Ki values of 31 would be so significantly different from those obtained with analog 30 unless interactions outside the active site were responsible.
Consistent with out previous studies, none of the benzoins or the benzils demonstrated any inhibitory activity towards AChE or BChE, with the exception of compound 23. This fluorobenzil demonstrated reasonably good inhibition of BChE. The exact reason for this is unclear, especially since this compound failed to inhibit AChE. Interestingly, we have seen inhibition of both of the cholinesterases by some nitrogen-containing fused ring diones (Hyatt et al, manuscript in preparation). These results suggest that this heteroatom is important for interaction of such molecules with the active sites of these proteins. This is perhaps not surprising since the choline group that is hydrolyzed from the choline esters contains a quaternary nitrogen that interacts with negatively charged amino acid residues within the cholinesterase active sites. This so called anionic site is critical for efficient hydrolysis of these esters. Since none of the phenylethane-1,2-diones, or their hydroxy-ethanone analogues that we have assayed can form stable positively charged quaternary compounds, we believe it is unlikely that these inhibitors would demonstrate activity towards AChE or BChE. The data presented here, and in previously publications10, 11, support this hypothesis.
Overall our studies suggest that the mechanism of CE inhibition by benzil and its analogs occurs via abortive nucleophilic attack of the active site serine towards one of the carbonyl groups within the molecule (Figure 1). Substitution of fluorine within the benzene rings did not alter benzil potency, but resulted in a significant increase in the inhibitory activity of the corresponding benzoins. This is likely due to a decrease in the electron density surrounding the target carbon atom, resulting in more facile attack by the serine nucleophile. In summary, these studies should allow the design of more potent selective CE inhibitors.
4. EXPERIMENTAL SECTION
4.1. General
Thiamine, copper acetate, ammonium nitrate, and fluorobenzaldehydes were purchased from Sigma Aldrich (St. Louis, MO). Product formation was monitored by thin layer chromatography TLC using pre-coated silica gel GF plates (Analtech Inc., Newark, DE) and visualized using UV light (254nm). Melting points were determined using a Mel-temp (Barnstead International, Dubuque, IA) and are reported uncorrected. NMR spectra (1H and 13C) were obtained using a Bruker DPX- 400 (400 MHz) spectrometer using CDCl3 (Aldrich) as a solvent and chemical shifts are reported in δ units (ppm) using tetramethylsilane as an internal standard with coupling constants (J) indicated in Hertz (Hz). Elemental analyses were performed by either Atlantic Microlabs (Norcross, GA) or Galbraith Laboratories (Knoxville, TN). Mass spectra were recorded on a VG 70-VSE(B) instrument (UIUC, Urbana-Champagne, IL) using EI or CI techniques.
4.2. Synthesis of fluorobenzoins
Fluorinated benzoins (1,2-diphenyl-2-hydroxy-ethanones) were synthesized by condensation of the correspondingly substituted fluorobenzaldehyde (0.04 mol) in the presence of thiamine hydrochloride (0.002 mol) in ethanolic sodium hydroxide (0.005 mol)14. Routinely reactions were run at 50°C for 48 hr and following cooling on ice, crude product was filtered and washed with ice cold 75% ethanol. Material was re-crystallized from ethanol, and purity and structures were assessed by melting point, TLC, NMR, and total elemental analyses. All physical data for the synthesized compounds are shown in Table 10.
Table 3.
Physical and NMR parameters for the compounds synthesized in this paper.
| ID | mp | 1H NMR | 13C NMR | MS m/z |
Elemental analysis |
|---|---|---|---|---|---|
| 5 | 81-82 | δ 7.95 (m, J = 7.9 Hz, 2H), δ 7.15 (m, J = 7.3 Hz, 2H), δ 7.0 (m, J = 7.0 Hz, 4H), δ 5.95 (s, 1H) δ 4.2 (bs, 1H) |
δ 197.20 (C=O), δ 167.38 (Ar, C-F), δ 161.56 (Ar, C-F), δ 134.83 (Ar, C-C=O), δ 133.92 (Ar, C-C-OH), δ 131.70 (Ar, C-H), δ 129.26 (Ar, C-H), δ 116.37 (Ar, C-H), δ115.99 (Ar, C-H), δ 88.36 (C-OH) |
248 | Anal. (C14H10O2); calc., (%) C 67.74, H 4.06, F 15.31; found, (%) C 67.81, H 4.03, F 15.44. |
| 6 | 83-85 | δ 7.95 (m, J = 7.9 Hz, 1H), δ 7.75 (m, J = 7.5Hz, 1H), δ 7.15 (m, J = 7.2 Hz, 2H), δ 7.05 (m, J = 7.0 Hz, 2H), δ 6.1 (s, 1H), δ 4.3 (bs, 1H) |
δ 188.57 (C=O), δ 165.4 (Ar, C-F), δ 164.49 (Ar, C-F), δ 162.60 (Ar, C-F), δ 162.47 (Ar, C-F), δ 132.84 (Ar, C-H), δ 132.12 (Ar, C-H), δ 118.01 (Ar, C-C-OH), δ 117.90 (Ar, C-C=O), δ 112.69 (Ar, C- H), δ 111.99 (Ar, C-H), δ 111.68 (Ar, C- H), δ 105.15 (Ar, C-H), δ 78.62 (C-OH) |
284 | Anal. (C14H8F4O2); calc., (%) C 59.16, H 2.84, F 26.74; found, (%) C 59.22, H 2.73, F 26.78. |
| 7 | 71-73 | δ 7.51(m, J = 7.6 Hz, 1H),7.30 (m, J = 7.4 Hz, 1H), δ 7.20 (m, J = 7.2 Hz, 2H), δ 7.10 (m, J = 7.1 Hz, 2H), δ 6.01 (s, 1H) δ 4.2 (bs, 1H) |
δ 197.2 (C=O), δ 162.70 (Ar, C-F), δ 159.26 (Ar, C-F), δ 133.86 (Ar, C-H), δ 130.13 (Ar, C-H), δ 115.23 (Ar, C-C=O), δ 112.54 (Ar, C-H), δ 112.17 (Ar, C-H), δ 110.12 (Ar, C-C-OH), δ 76.31 (C-OH) |
284 | Anal. (C14H8F4O2); calc., (%) C 59.16, H 2.84, F 26.74; found, (%) C 59.23, H 2.63, F 26.75. |
| 8 | 62-64 | δ 7.85 (m, J = 7.9 Hz, 4H), δ 7.41 (m, J = 7.4 Hz, 2H), δ 6.00 (s, 1H), δ 4.2 (bs, 1H) |
δ 191.23 (C=O), δ 155.21 (Ar, C-F), δ 151.61 (Ar, C-F), δ 150.12 (Ar, C-F), δ 146.33 (Ar, C-F), δ 132.99 (Ar, C-C=O), δ 132.01 (Ar, C-C-OH), δ 127.54 (Ar, C-H), δ 126.92 (Ar, C-H), δ 124.12 (Ar, C-H), δ 121.36 (Ar, C-H), δ 119.62 (Ar, C-H), δ 117.35 (Ar, C-H), δ 82.41 (Ar, C-OH) |
284 | Anal. (C14H8F4O2); calc., (%) C 59.16, H 2.84, F 26.74; found, (%) C 59.15, H 2.66, F 26.67. |
| 9 | 54-56 | δ 7.60 (m, J = 7.6 Hz, 1H), δ 7.35 (m, J = 7.4 Hz, 1H), δ 7.10 (m, J = 7.1 Hz, 4H) δ 6.10 (s, 1H), δ 4.48 (bs, 1H) |
δ 195.74 (C=O), δ 156.96 (Ar, C-F), δ 150.14 (Ar, C-F), δ 147.23 (Ar, C-F), δ 139.85 (Ar, C-F), δ 127.66 (Ar, C-C-OH), δ 127.25 (Ar, C-C=O), δ 125.36 (Ar, C- H), δ 125.33 (Ar, C-H), δ 124.93 (Ar, C- H), δ 122.7 (Ar, C-H), δ 120.52 (Ar, C-H), δ 117.96 (Ar, C-H), δ 78.65 (C-OH) |
284 | Anal. (C14H8F4O2); calc., (%) C 59.16, H 2.84, F 26.74; found, (%) C 59.10, H 2.80, F 26.75. |
| 10 | 91-93 | δ 7.8 (m, J = 7.8 Hz, 2H) δ 7.50 (m, J = 7.5 Hz, 2H), δ 7.20 (m, J = 7.3 Hz, 2H), δ 6.2 (s, 1H), δ 4.55 (bs, 1H) |
δ 188.44 (C=O), δ 159.91(Ar, C-F), δ 157.73 (Ar, C-F), δ 157.70 (Ar, C-F), δ 156.82 (Ar, C-F), δ 123.87 (Ar, C-C-OH), δ 123.62 (Ar, C-C=O), δ 122.09 (Ar, C- H), δ 118.38 (Ar, C-H), δ 118.25 (Ar, C- H), δ 117.93 (Ar, C-H), δ 116.39 (Ar, C- H), δ 115.37 (Ar, C-H), δ 79.26 (C-OH) |
284 | Anal. (C14H8F4O2); calc., (%) C 59.16, H 2.84, F 26.74; found, (%) C 59.06, H 2.70, F 26.73. |
| 11 | 78-80 | δ 7.25 (m, J = 7.4 Hz, 2H), δ 7.05 (m, J = 7.0 Hz, 1H), δ 6.95 (m, J = 6.9 Hz, 2H), δ 6.98 (m, 6.8 Hz, 1H), δ 5.95 (s, 1H), δ 4.15 (bs, 1H) |
δ 192.3 (C=O), δ 164.50 (Ar, C-F), δ 161.3 (Ar, C-F), δ 139.23 (Ar, C-C=O), δ 138.86 (Ar, C-C-OH), δ 113.06 (Ar, C-H) δ 110.26 (Ar, C-H), δ 109.19 (Ar, C-H) δ 102.12 (Ar, C-H), δ 86.32 (C-OH) |
284 | Anal. (C14H8F4O2); calc., (%) C 59.16, H 2.84, F 26.74; found, (%) C 59.18, H 2.71, F 26.6. |
| 12 | 73-75 | δ 7.35 (d, J = 7.4 Hz, 1H), δ 7.15 (d, J = 7.2 Hz, 1H), δ 6.9 (d, J = 6.9 Hz, 1H), δ 6.65 (d, J = 6.7 Hz, 1H), δ 6.01 (s, 1H), δ 4.5 (bs, 1H) |
δ 192.23 (C=O), δ 160.10 (Ar, C-F), δ 157.89 (Ar, C-F), δ 157.32 (Ar, C-F), δ 149.65 (Ar, C-F), δ 140.23 (Ar, C-F), δ 133.26 (Ar, C-F), 125.26 (Ar, C-C-OH), δ 124.45 (Ar, C-C=O), δ 113.66 (Ar, C-H), δ 112.19 (Ar, C-H), δ 110.87(Ar, C-H), δ 102.70 (Ar, C-H), δ 81.23 (C-OH) |
320 | Anal. (C14H6F6O2); calc., (%) C 52.52, H 1.89, F 35.60; found, (%) C 52.56, H 1.71, F 35.43. |
| 13 | 63-65 | δ 7.75 (m, J = 7.7 Hz, 1H), δ 7.50(m, J = 7.6 Hz, 1H), δ 7.35 (m, J = 7.3 Hz, 1H) δ 7.25 (m, J = 7.3 Hz, 1H) δ 6.0 (s, 1H), δ 4.50 (bs, 1H) |
δ 191.23 (C=O), δ 157.32 (Ar, C-F), δ 151.56 (Ar, C-F), δ 148.0 (Ar, C-F), δ 143.65 (Ar, C-F), δ 140.95 (Ar, C-F), δ 139.99 (Ar, C-F), δ 130.26 (Ar, C-H), 129.26 (Ar, C-H), δ 123.45 (Ar, C-C-OH), δ 122.65 (Ar, C-C=O), δ 118.23 (Ar, C- H), δ 113.66 (Ar, C-H), δ 83.23 (C-OH) |
320 | Anal. (C14H6F6O2); calc., (%) C 52.52, H 1.89, F 35.60; found, (%) C 52.45, H 1.70, F 35.51. |
| 14 | 75-77 | δ 7.25 (s, 2H), δ 7.95 (s, 2H), δ 5.9 (s, 1H), δ 4.1 (bs, 1H) |
δ 197.67(C=O), δ 161.34 (Ar, C-F), δ 152.36 (Ar, C-F), δ 145.27 (Ar, C-F), δ 140.28 (Ar, C-F), δ 133.96 (Ar, C-C=O), δ 133.72 (Ar, C- C-OH), δ 115.09 (Ar, C-H), δ 113.36 (Ar, C-H), δ 85.62 (C-OH) |
348 | Anal. (C14H6F6O2); calc., (%) C 52.52, H 1.89, F 35.60; found, (%) C 52.44, H 1.71, F 35.51. |
| 15 | 87-89 | δ 8.0 (d, J = 8.0 Hz, 2H), δ 7.85 (d, J = 7.7 Hz, 2H), δ 7.30 (d, J = 7.5 Hz, 2H), δ 7.2 (d, J = 7.3 Hz, 2H), δ 6.01 (s, 1H), δ 4.42 (bs, 1H) |
δ 197.68 (C=O), δ 141.95 (Ar, C-C=O), δ 138.98 (Ar, C-C-OH), δ 135.61 (Ar, C- CF3), δ 129.40 (Ar, C-CF3), δ 128.10 (Ar, C-H), δ 126.84 (Ar, C-H), δ 125.94 (Ar, C-H), δ 124.12 (Ar, C-H), δ 123.69 (CF3), δ 85.12 (C-OH) |
348 | Anal. (C16H10F6O2); calc., (%) C 55.18, H 2.89, F 32.73; found, (%) C 55.20, H 2.84, F 33.00. |
| 16 | 56-58 | δ 8.0 (s, 2H), δ 7.70 (d, J = 8.0 Hz, 2H), δ 7.60 (m, J = 7.9 Hz, 2H), δ 7.4 (d, J = 7.7 Hz, 2H), δ 6.01 (s, 1H), δ 4.42 (bs, 1H) |
δ 191.32 (C=O), δ 136.89 (Ar, C-C=O), δ 136.55 (Ar, C-C-OH), δ 133.65 (Ar, C-H), δ 132.86 (Ar, C-H), δ 130.46 (Ar, C-CF3), δ 129.55 (Ar, C-CF3), δ 128.23 (Ar, C-H), δ 127. 56 (Ar, C-H), δ 126.71 (Ar, C-H), δ 125.91 (Ar, C-H), δ 124.0 (Ar, C-H), δ 123.99 (CF3) δ 123.92 (CF3), δ 85.18 (C-OH) |
348 | Anal. (C16H10F6O2); calc., (%) C 55.18, H 2.89, F 32.73; found, (%) C 55.38, H 2.85, F 32.92. |
| 17 | 103- 105 |
δ 8.05 (d, J = 8.0 Hz, 2H), δ 7.50 (m, J = 7.9 Hz, 2H), δ 7.05 (m, J = 7.7 Hz, 2H), δ 6.85 (m, J = 7.7 Hz, 2H) |
δ 190.21 (C=O), δ 163.0 (Ar, C-F), δ 136.72 (Ar, C-H), δ 130.89 (Ar, C-H), δ 125.00 (Ar, C-H), δ 122.32 (Ar, C-C=O) |
246 | Anal. (C14H8F2O2); calc., (%) C 68.30, H 3.28, F 15.43; found, (%) C 68.15, H 3.18, F 15.66. |
| 18 | 97-99 | δ 7.74 (d, J = 8.0 HZ, 2H), δ 7.71 (d, J = 7.8 Hz, 2H), δ 7.51 (m, J = 7.5 Hz, 2H) δ 7.39 (d, J = 7.4 Hz, 2H) |
δ 192.20 (C=O), δ 162.89 (Ar, C-F), δ 138.68 (Ar, C-C=O), δ 130.89 (Ar, C-H), δ 126.05 (Ar, C-H), δ 122.26 (Ar, C-H), δ 116.21 (Ar, C-H) |
246 | Anal. (C14H8F2O2); calc., (%) C 68.30, H 3.28, F 15.43; found, (%) C 68.17, H 3.25, F 15.59. |
| 19 | 118- 119 |
δ 8.05 (m, J = 7.9 Hz, 4H), δ 7.50 (m, J = 7.5 Hz, 4H) |
δ 192.23 (C=O), δ 168.17 (Ar, C-F), δ 132.82 (Ar, C-H), δ 128.36 (Ar, C-C=O), δ 116.47 (Ar, C-H) |
246 | Anal. (C14H8F2O2); calc., (%) C 68.30, H 3.28, F 15.43; found, (%) C 68.51, H 3.20, F 15.7. |
| 20 | 91-93 | δ 8.49 (d, J = 8.0 Hz, 2H), δ 7.40 (m, J = 7.7 Hz, 2H), δ 7.20 (m, J = 7.8 Hz, 2H) |
δ 188.45 (C=O), δ 169.48 (Ar, C-F), δ 165.32 (Ar, C-F), δ 134.38 (Ar, C-H), δ 118.38 (Ar, C-C=O), δ 115.71 (Ar, C-H), δ 110.67 (Ar, C-H) |
282 | Anal. (C14H6F4O2); calc., (%) C 59.59, H 2.14, F 26.93; found, (%) C 59.32, H 2.05, F 26.77. |
| 21 | 163- 165 |
δ 7.95 (m, J = 7.9 Hz, 2H), δ 7.45 (m, J = 7.4 Hz, 4H) |
δ 185.07 (C=O), δ 162.37 (Ar, C-F), δ 135.86 (Ar, C-H), δ 112.39 (Ar, C-H), δ 111.84 (Ar, C-C=O) |
282 | Anal. (C14H6F4O2); calc., (%) C 59.59, H 2.14, F 26.93; found, (%) C 59.59, H 2.05, F 27.13. |
| 22 | 107- 109 |
δ 8.05 (d, J = 8.1 Hz, 2H), δ 7.85 (d, J = 7.9 Hz, 2H), δ 7.70 (m, J = 7.7 Hz, 2H) |
δ 190.20 (C=O), δ 156.30 (Ar, C-F), δ 150.37 (Ar, C-F), δ 133.70 (Ar, C-C=O), δ 127.70 (Ar, C-H), δ 126.30 (Ar, C-H), δ 118.50 (Ar, C-H) |
282 | Anal. (C14H6F4O2); calc., (%) C 59.59, H 2.14, F 26.93; found, (%) C 59.32, H 2.08, F 26.63. |
| 23 | 91-93 | δ 7.85 (d, J = 8.0 Hz, 2H), δ 7.45 (m, J = 7.5 Hz, 2H), δ 7.25 (m, J = 7.4 Hz, 2H) |
δ 188.60 (C=O), δ 157.50 (Ar, C-F), δ 150.0 (Ar, C-F), δ 128.50 (Ar, C-H), δ 126.50 (Ar, C-H), δ 125.10 (Ar, C-C=O), δ 123.00 (Ar, C-H) |
282 | Anal. (C14H6F4O2); calc., (%) C 59.59, H 2.14, F 26.93; found, (%) C 59.45, H 2.03, F 26.98. |
| 24 | 110- 112 |
δ 8.49 (d, J = 8.1 Hz, 2H) δ 7.38 (m, J = 7.8 Hz, 2H), δ 7.39 (m, J = 7.7 Hz, 2H) |
δ 188.50 (C=O), δ 160.20 (Ar, C-F), δ 157.75 (Ar, C-F), δ 124.75 (Ar, C-C=O), δ 122.10 (Ar, C-H), δ 118.20 (Ar, C-H), δ 116.50 (Ar, C-H) |
282 | Anal. (C14H6F4O2); calc. (%) C 59.59, H 2.14, F 26.93; found, (%) C 59.42, H 2.08, F 27.13. |
| 26 | 124- 126 |
δ 7.45 (m, J = 7.4 Hz, 2H), δ 7.00 (m, J = 7.0 Hz, 2H) |
δ 183.55 (C=O), δ 158.50 (Ar, C-F), δ 155.00 (Ar, C-F), δ 148.55 (Ar, C-F), δ 122.80 (Ar, C-H), δ 113.30 (Ar, C-CH), δ 112.0 (Ar, C-C=O) |
318 | Anal. (C14H4F6O2); calc., (%) C 52.85, H 1.27, F 35.83; found, (%) C 52.62, H 1.15, F 35.49. |
| 27 | 98- 100 |
δ 7.55 (m, J = 7.5 Hz, 2H), δ 7.30 (m, 2H) |
δ 186.80 (C=O), δ 158.30 (Ar, C-F), δ 157.75 (Ar, C-F), δ 146.70 (Ar, C-F), δ 123.0 (Ar, C-C=O), δ 111.70 (Ar, C-H), δ 110.50 (Ar, C-H) |
318 | Anal. (C14H4F6O2); calc., (%) C 52.85, H 1.27, F 35.83; found, (%) C 52.68, H 1.16, F 35.72. |
| 28 | 94-96 | δ 7.86 (m, J = 7.9 Hz, 2H), δ 7.23 (m, J = 7.2 Hz, 2H) |
δ 194.38 (C=O), δ 157.32 (Ar, C-F), δ 151.32 (Ar, C-F), δ 138.39 (Ar, C-F), δ 127.29 (Ar, C-H), δ 121.25 (Ar, C-H), δ 120.42 (Ar, C-C=O) |
318 | Anal. (C14H4F6O2); calc., (%) C 52.85, H 1.27, F 35.83; found, (%) C 52.72, H 1.21, F 36.03. |
| 30 | 134- 136 |
δ 8.10 (d, J = 8.1 Hz, 4H), δ 7.78 (d, J = 7.8 Hz, 4H) |
δ 191.90 (C=O), δ 136.40 (Ar, C-CF3), δ 135.25 (Ar, C-C=O), δ 130.37 (Ar, C-H), δ 126.18 (Ar, C-H), δ 124.63 (CF3) |
346 | Anal. (C16H8F6O2); calc., (%) C 55.50, H 2.33, F 32.92; found, (%) C 55.28, H 2.20, F 33.2. |
| 31 | 101- 103 |
δ 8.30 (s, 2H), δ 8.18 (d, J = 8.2 Hz, 2H), δ 7.95 (d, J = 7.9 Hz, 2H), δ 7.78 (m, J = 7.7 Hz, 2H) |
δ 191.32 (C=O), δ 139.05 (Ar, C-C=O), δ 132.14 (Ar, C-CF3), δ 131.58 (Ar, C-H), δ 130.25 (Ar, C-H), δ 129.85 (Ar, C-H), δ 127.69 (Ar, C-H), δ 124.70 (CF3) |
346 | Anal. (C16H8F6O2); calc., (%) C 55.50, H 2.33, F 32.92; found, (%) C 55.31, H 2.23, F 33.21. |
| 32 | 103- 105 |
δ 8.10 (s, 2H), δ 8.0 (d, J = 8.0 Hz, 2H), δ 7.78 (d, J = 7.7 Hz, 2H) |
δ 188.45 (C=O), δ 136.68 (Ar, C-CF3), δ 133.70 (Ar, C-C=O), δ 130.58 (Ar, C-H), δ 129.57 (Ar, C-CF3), δ 128.79 (Ar, C-H), δ 124.29 (Ar, C-H), δ 123.78 (Ar, CF3), δ 121.43 (Ar, CF3) |
483 (M+H)+1 |
Anal. (C18H6F12O2); calc., (%) C 44.83, H 1.25, F 47.28; found, (%) C 44.76, H 1.18, F 47.55. |
| 33 | 130- 132 |
δ 8.00 (s, 4H), δ 7.80 (s, 2H) |
δ 188.11 (C=O), δ 135.23 (Ar, C-C=O), δ 132.42 (Ar, C-CF3), δ 129.03 (Ar, C-H), δ 128.10 (Ar, C-H), δ 124.11 (CF3) |
483 (M+H)+1 |
Anal. (C18H6F12O2); calc., (%) C 44.83, H 1.25, F 47.28; found, (%) C 44.83, H 1.14, F 47.47. |
| 34 | 135- 137 |
δ 8.08 (s, 2H), δ 7.71 (d, J = 8.0Hz, 2H), δ 7.79 (d, J = 7.7Hz, 2H) |
δ 188.09 (C=O), δ 134.38 (Ar, C-CF3), δ 132.41 (Ar, C-CF3), δ 131.33 (Ar, C-H), δ 130.01 (Ar, C-C=O), δ 127.31(Ar, C-H), δ 126.58 (Ar, C-H), δ 124.17(CF3), δ 120.36 (CF3) |
483 (M+H)+1 |
Anal. (C18H6F12O2); calc., (%) C 44.83, H 1.25, F 47.28; found, (%) C 44.86, H 1.09, F 47.35. |
4.2.1. 1,2-bis(2-fluorophenyl)-2-hydroxyethanone (3)
1,2-bis(2-fluorophenyl)-2-hydroxyethanone was synthesized from 2-fluorobenzaldehyde. Physical and NMR parameters of 3 were consistent with that found in the literature15.
4.2.2. 1,2-bis(3-fluorophenyl)-2-hydroxyethanone (4)
1,2-bis(3-fluorophenyl)-2-hydroxyethanone was synthesized the condensation of 3-fluorobenzaldehyde. Physical and NMR parameters of 4 were consistent with those previously reported15.
4.2.3. 1,2-bis(4-fluorophenyl)-2-hydroxyethanone (5)
1,2-bis(4-fluorophenyl)-2-hydroxyethanone was synthesized from 4-fluorobenzaldehyde to give white crystals in 75% yield.
4.2.4. 1,2-bis(2,4-difluorophenyl)-2-hydroxyethanone (6)
1,2-bis(2,4-difluorophenyl)-2-hydroxyethanone was synthesized from 2,4-difluorobenzaldehyde to give white crystals in 67% yield.
4.2.5. 1,2-bis(2,6-difluorophenyl)-2-hydroxyethanone (7)
1,2-bis(2,6-difluorophenyl)-2-hydroxyethanone was synthesized from 2,6-difluorobenzaldehyde to give white crystals in 72% yield.
4.2.6. 1,2-bis(3,4-difluorophenyl)-2-hydroxyethanone (8)
1,2-bis(3,4-difluorophenyl)-2-hydroxyethanone was synthesized from 3,4-difluorobenzaldehyde to give white crystals in 70% yield.
4.2.7. 1,2-bis(2,3-difluorophenyl)-2-hydroxyethanone (9)
1,2-bis(2,3-difluorophenyl)-2-hydroxyethanone was synthesized from 2,3-difluorobenzaldehyde to give white crystals in 73% yield.
4.2.8. 1,2-bis(2,5-difluorophenyl)-2-hydroxyethanone (10)
1,2-bis(2,5-difluorophenyl)-2-hydroxyethanone was synthesized from 2,5-difluorobenzaldehyde to give white crystals in 72% yield.
4.2.9. 1,2-bis(3,5-difluorophenyl)-2-hydroxyethanone (11)
1,2-bis(3,5-difluorophenyl)-2-hydroxyethanone was synthesized from 3,5-difluorobenzaldehyde to give white crystals in 72% yield.
4.2.10. 1,2-bis(2,3,5-trifluorophenyl)-2-hydroxyethanone (12)
1,2-bis(2,3,5-trifluorophenyl)-2-hydroxyethanone was synthesized from 2,3,5-trifluorobenzaldehyde to give white crystals in 74% yield.
4.2.11. 1,2-bis(2,3,4-trifluorophenyl)-2-hydroxyethanone (13)
1,2-bis(2,3,4-trifluorophenyl)-2-hydroxyethanone was synthesized from 2,3,4-trifluorobenzaldehyde to give white crystals in 72% yield.
4.2.12. 1,2-bis(3,4,5-trifluorophenyl)-2-hydroxyethanone (14)
1,2-bis(3,4,5-trifluorophenyl)-2-hydroxyethanone was synthesized from 3,4,5-trifluorobenzaldehyde to give white crystals in 70% yield.
4.2.13. 1,2-bis[(4-(trifluoromethyl)phenyl]-2-hydroxyethanone (15)
1,2-bis[(4-(trifluoromethyl)phenyl]- 2-hydroxyethanone was synthesized from 4-trifluoromethylbenzaldehyde to give white crystals in 68% yield.
4.2.14. 1,2-bis[3-(trifluoromethyl)phenyl]-2-hydroxyethanone (16)
1,2-bis[3-(trifluoromethyl)phenyl]-2-hydroxyethanone was synthesized from 3-trifluoromethylbenzaldehyde to give white crystals in 69% yield.
4.3. Synthesis of fluorobenzils
The substituted fluorobenzils were synthesized by oxidation of the corresponding benzoin using copper acetate (0.001mol) and ammonium nitrate (0.006mol) in 80% acetic acid.16 Briefly, the benzoin was refluxed for 90 mins and following cooling, the product appeared as a solid yellow mass. After washing extensively with water and cold 75% ethanol, the benzil was re-crystallized from ethanol. Product purity was assessed as described above for the benzoins.
4.3.1. 1,2-bis(2-fluorophenyl)ethane-1,2-dione (17)
1,2-bis(2-fluorophenyl)ethane-1,2-dione was synthesized from 3 to give yellow crystals in 95% yield.
4.3.2. 1,2-bis(3-fluorophenyl)ethane-1,2-dione (18)
1,2-bis(3-fluorophenyl)ethane-1,2-dione was synthesized from 4 to give yellow crystals in 98% yield.
4.3.3. 1,2-bis(4-fluorophenyl)ethane-1,2-dione (19)
1,2-bis(4-fluorophenyl)ethane-1,2-dione was synthesized from 5 to give yellow crystals in 98% yield.
4.3.4. 1,2-bis(2,4-difluorophenyl)ethane-1,2-dione (20)
1,2-bis(2,4-difluorophenyl)ethane-1,2-dione was synthesized from 6 to give yellow crystals in 95% yield.
4.3.5. 1,2-bis(2,6-difluorophenyl)ethane-1,2-dione (21)
1,2-bis(2,6-difluorophenyl)ethane-1,2-dione was synthesized from 7 to give yellow crystals in 98% yield.
4.3.6. 1,2-bis(3,4-difluorophenyl)ethane-1,2-dione (22)
1,2-bis(3,4-difluorophenyl)ethane-1,2-dione was synthesized from 8 to give yellow crystals in 98% yield.
4.3.7. 1,2-bis(2,3-fluorophenyl)ethane-1,2-dione (23)
1,2-bis(2,3-fluorophenyl)ethane-1,2-dione was synthesized from 9 to give yellow crystals in 93% yield.
4.3.8. 1,2-bis(2,5-difluorophenyl)ethane-1,2-dione (24)
1,2-bis(2,5-difluorophenyl)ethane-1,2-dione was synthesized from 10 to give yellow crystals in 96% yield.
4.3.9. 1,2-bis(3,5-difluorophenyl)ethane-1,2-dione (25)
1,2-bis(3,5-difluorophenyl)ethane-1,2-dione was synthesized from 11 to give yellow crystals in 97% yield. Physical and NMR parameters of 25 were as previously described10.
4.3.10. 1,2-bis(2,3,6-trifluorophenyl)ethane-1,2-dione (26)
1,2-bis(2,3,6-trifluorophenyl)ethane-1,2-dione was synthesized from 2,3,6-trifluorobenzaldehyde under the conditions described for the benzoin reactions. This was presumably due to immediate oxidation of the benzoin to the corresponding benzil.
4.3.11. 1,2-bis(2,3,5-trifluorophenyl)ethane-1,2-dione (27)
1,2-bis(2,3,5-trifluorophenyl)ethane-1,2-dione was synthesized from 12 to give yellow crystals in 90% yield.
4.3.12. 1,2-bis(2,3,4-trifluorophenyl)ethane-1,2-dione (28)
1,2-bis(2,3,4-trifluorophenyl)ethane-1,2-dione was synthesized from the oxidation of 13 to give yellow crystals in 95% yield.
4.3.13. 1,2-bis(3,4,5-trifluorophenyl)ethane-1,2-dione (29)
1,2-bis(3,4,5-trifluorophenyl)ethane-1,2-dione was synthesized from 14 to give yellow crystals in 94% yield. Physical and NMR parameters of 29 were as previously described10.
4.3.14. 1,2-bis[4-(trifluoromethyl)phenyl]ethane-1,2-dione (30)
1,2-bis[4-(trifluoromethyl)phenyl]ethane-1,2-dione was synthesized from 15 to give yellow crystals in 97% yield.
4.3.15. 1,2-bis[3-(trifluoromethy)phenyl]ethane-1,2-dione (31)
1,2-bis[3-(trifluoromethy)phenyl]ethane-1,2-dione was synthesized from 16 to give yellow crystals in 90% yield.
4.4. Synthesis of 1,2-bis[2,4-bis(trifluoromethyl)phenyl]ethane-1,2-dione (32), 1,2-bis[3,5-bis(trifluoromethyl)phenyl]ethane-1,2-dione (33) and 1,2-bis[2,5-bis(trifluoromethyl)phenyl]ethane-1,2-dione (34)
Synthesis of the above compounds was achieved by direct condensation of the substituted benzaldehyde using the method described for the benzoins (see above). Under these conditions, the benzil analog was produced, presumably via immediate oxidation of the benzoin intermediate.
4.4.1. 1,2-bis[2,4-bis(trifluoromethyl)phenyl]ethane-1,2-dione (32)
1,2-bis[2,4-bis(trifluoromethyl)phenyl]ethane-1,2-dione was synthesized from bis(2,4-trifluoromethyl)benzaldehyde to give yellow crystals in 76% yield.
4.4.2. 1,2-bis[3,5-bis(trifluoromethyl)phenyl]ethane-1,2-dione (33)
1,2-bis[3,5-bis(trifluoromethyl)phenyl]ethane-1,2-dione was synthesized from bis(3,5-trifluoromethyl)benzaldehyde to give yellow crystals in 76% yield.
4.4.3. 1,2-bis[2,5-bis(trifluoromethyl)phenyl]ethane-1,2-dione (34)
1,2-bis[2,5-bis(trifluoromethyl)phenyl]ethane-1,2-dione was synthesized from bis(2,5-trifluoromethyl)benzaldehyde to give yellow crystals in 76% yield.
4.5. Enzymes
Pure rCE and hCE1 were prepared as described previously17. hiCE was prepared by concentration of baculovirus media from Sf9 cells expressing a secreted form of the protein. While not homogeneous, the preparation was at least 60% pure. Since no CE activity is expressed or secreted from uninfected Sf9 cells, the only CE present in the culture media was the recombinant hiCE protein. The Genbank accession numbers of the cDNAs used to generate the enzymes for this study were as follows: hiCE, Y0961618; hCE1, M7349919; rCE, AF03693020.
Human acetylcholinesterase (AChE) and butyrylcholinesterase (BChE) were obtained from Sigma Aldrich.
4.6. Inhibition of carboxylesterases
CE inhibition was assessed using a spectrophotometric multiwell plate assay using 3mM o-nitrophenyl acetate (o-NPA) as a substrate6,11. Briefly, the test compound and substrate (o-NPA) were aliquoted into duplicate wells of a 96-well plate and enzyme was added using a multiwell pipettor. The rate of change in absorbance at 420 nm was measured at 15 s intervals for 5 min and compared to wells containing no inhibitor. Routinely, inhibitor concentrations ranged from 1nM to 100μM. All assays were performed in duplicate and included both positive (50μM bis(4-nitrophenyl)phosphate) and negative controls (DMSO, no enzyme).
4.7. Inhibition of acetylcholinesterase and butyrylcholinesterase
The ability of compounds to inhibit AChE and BChE was performed as previously described using either 1mM acetylthiocholine (AcTCh) or butyrylthiocholine (BuTCh), respectively, as substrates21, 22.
4.8. Determination of Ki values
Data obtained from the above assays were fitted to the following equation23 to determine the inhibition constant (Ki):
where i is the fractional inhibition, [I] is the inhibitor concentration, [s] is the substrate concentration, α is the change in affinity of substrate for enzyme, β is the change in the rate of enzyme substrate complex decomposition, Ks is the dissociation constant for the enzyme substrate complex, and Ki is the inhibitor constant. Curve fits were generated (where α ranged from 0 to ∞ and β ranged from 0 to 1) using GraphPad Prism software (San Diego, CA) and those generating the highest r2 values were further analyzed using Akaike's information criteria24, 25. After determination of the best fit for the experimental data, Ki values were then calculated using Prism.
4.9. Computational chemistry
All calculations were carried out using the Gaussian 03 software package (Gaussian, Wallingford, CT). Each compound was constructed using Gauss-View and geometry optimizations were performed at the B3LYP/6-31G(p,d) level of theory26,27. Mulliken atomic charges for atoms within the molecules were calculated from these datasets. pKa values were predicted using ChemSilico Predict v2.0 software (ChemSilico LLC, Tewksbury, MA) and Hammett substituent constants were obtained from previously published reports28.
4.10. Linear regression and Spearman correlation analyses
Datasets were analyzed using GraphPad Prism software. This allowed for simultaneous calculation of both linear regression correlates (r2) and Spearman r coefficients. For the latter analyses, Spearman r values close to 1 or −1 indicate good correlations, whereas values closer to 0 indicate a lack of statistical correlation.
4.11. 3D-QSAR analysis
3D-QSAR analysis was performed as previously described.6, 10 Briefly, compounds were initially constructed using Chem3D and atom types were determined using the antechamber module of AMBER7 (University of California, San Francisco, CA). After assignment of partial atomic charges using the bond charge correction approach29 compounds were analyzed using Quasar 4.0 software29-31. This program generates a 3D-receptor-surface model that contains the molecular properties of both the receptor site and the ligand that will be docked into this domain. Typically, 200 independent models are generated for each data set and these are evaluated to yield 7000 pseudoreceptor site models. Model evaluation was then performed until the cross correlation coefficients (q2) exceed 0.7 for the observed versus the predicted Ki values. Routinely this produced correlation coefficients (r2) of >0.9.
Acknowledgments
This work was supported in part by NIH Grants CA76202, CA79763, CA98468, CA108775, DA18116, a Cancer Center Core Grant P30 CA 21765 and by the American Lebanese Syrian Associated Charities.
The abbreviations used are
- AChE
acetylcholinesterase
- AcTCh
acetylthiocholine
- BChE
butyrylcholinesterase
- BuTCh
butyrylthiocholine
- CE
carboxylesterase
- CPT-11
irinotecan, 7-ethyl-10-[4-(1-piperidino)-1-piperidino]carbonyloxycamptothecin
- EDG
electron donating group
- EWG
electron withdrawing group
- hCE1
human carboxylesterase 1
- hiCE
human intestinal carboxylesterase
- Ki
inhibition constant
- o-NPA
o-nitrophenyl acetate
- q2
cross validation coefficient
- QSAR
quantitative structure activity relationship
- rCE
rabbit liver carboxylesterase
- TRAP
Target Related Affinity Profiling
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES AND NOTES
- 1.Cashman J, Perroti B, Berkman C, Lin J. Environ. Health Perspect. 1996;104:23. doi: 10.1289/ehp.96104s123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khanna R, Morton CL, Danks MK, Potter PM. Cancer Res. 2000;60:4725. [PubMed] [Google Scholar]
- 3.Redinbo MR, Bencharit S, Potter PM. Biochem. Soc. Trans. 2003;31:620. doi: 10.1042/bst0310620. [DOI] [PubMed] [Google Scholar]
- 4.Redinbo MR, Potter PM. Drug Discov. Today. 2005;10:313–325. doi: 10.1016/S1359-6446(05)03383-0. [DOI] [PubMed] [Google Scholar]
- 5.Molinoff PB, Ruddon RW. In: Goodman & Gilman's the pharmacological basis of therapeutics. Hardman JG, Limbird LE, editors. McGraw-Hill; New York: 1996. p. 1905. [Google Scholar]
- 6.Wadkins RM, Hyatt JL, Yoon KJ, Morton CL, Lee RE, Damodaran K, Beroza P, Danks MK, Potter PM. Mol. Pharmacol. 2004;65:1336. doi: 10.1124/mol.65.6.1336. [DOI] [PubMed] [Google Scholar]
- 7.Beroza P, Villar HO, Wick MM, Martin GR. Drug Discov. Today. 2002;7:807. doi: 10.1016/s1359-6446(02)02371-1. [DOI] [PubMed] [Google Scholar]
- 8.Dixon SL, Villar HO. J. Chem. Inf. Comput. Sci. 1998;38:1192. doi: 10.1021/ci980105+. [DOI] [PubMed] [Google Scholar]
- 9.Kauvar LM, Higgins DL, Villar HO, Sportsman JR, Engqvist_Goldstein A, Bukar R, Bauer KE, Dilley H, Rocke DM. Chem. & Biol. 1995;2:107. doi: 10.1016/1074-5521(95)90283-x. [DOI] [PubMed] [Google Scholar]
- 10.Wadkins RM, Hyatt JL, Wei X, Yoon KJ, Wierdl M, Edwards CC, Morton CL, Obenauer JC, Damodaran K, Beroza P, Danks MK, Potter PM. J. Med. Chem. 2005;48:2905. doi: 10.1021/jm049011j. [DOI] [PubMed] [Google Scholar]
- 11.Hyatt JL, Stacy V, Wadkins RM, Yoon KJ, Wierdl M, Edwards CC, Zeller M, Hunter AD, Danks MK, Crundwell G, Potter PM. J. Med. Chem. 2005;48:5543. doi: 10.1021/jm0504196. [DOI] [PubMed] [Google Scholar]
- 12.Wadkins RM, Morton CL, Weeks JK, Oliver L, Wierdl M, Danks MK, Potter PM. Mol. Pharmacol. 2001;60:355. doi: 10.1124/mol.60.2.355. [DOI] [PubMed] [Google Scholar]
- 13.Lundstedt T, Seifert E, Abramo L, Thelin B, Nystrom A, Pettersen J, Bergman B. Chemom. Intell. Lab. Syst. 1998;42:3. [Google Scholar]
- 14.Mohrig JR, Hammond CN, Schatz PF, Morrill TC. Modern projects and experiments in organic chemistry: Miniscale and Williamson microscale. W.H. Freeman & Co.; New York: 2003. pp. 367–368. [Google Scholar]
- 15.Demir AS, Sesenoglu O, Eren E, Hosrik B, Pohl M, Janzen E, Kolter D, Feldmann R, Dunkelmann P, Muller M. Adv. Synth. Catal. 2002;344:96. [Google Scholar]
- 16.Weiss M, Appel M. J. Am. Chem. Soc. 1948;70:3666. doi: 10.1021/ja01191a036. [DOI] [PubMed] [Google Scholar]
- 17.Morton CL, Potter PM. Mol. Biotechnol. 2000;16:193. doi: 10.1385/MB:16:3:193. [DOI] [PubMed] [Google Scholar]
- 18.Schwer H, Langmann T, Daig R, Becker A, Aslanidis C, Schmitz G. Biochem. Biophys. Res. Comm. 1997;233:117. doi: 10.1006/bbrc.1997.6413. [DOI] [PubMed] [Google Scholar]
- 19.Munger JS, Shi GP, Mark EA, Chin DT, Gerard C, Chapman HA. J. Biol. Chem. 1991;266:18832. [PubMed] [Google Scholar]
- 20.Potter PM, Pawlik CA, Morton CL, Naeve CW, Danks MK. Cancer Res. 1998;52:2646. [PubMed] [Google Scholar]
- 21.Doctor BP, Toker L, Roth E, Silman I. Anal. Biochem. 1987;166:399. doi: 10.1016/0003-2697(87)90590-2. [DOI] [PubMed] [Google Scholar]
- 22.Ellman GL, Courtney KD, Anders V, Featherstone RM. Biochem. Pharmacol. 1961;7:88. doi: 10.1016/0006-2952(61)90145-9. [DOI] [PubMed] [Google Scholar]
- 23.Webb JL. Enzyme and Metabolic Inhibitors. Volume 1. General Principles of Inhibition. Academic Press Inc.; New York: 1963. [Google Scholar]
- 24.Akaike H. In: Petrov BN, Csaki F, editors. Information theory and an extension of the maximum likelihood principle; Second International Symposium on Information Theory; Budapest. 1973; Budapest: Akademiai Kiado; 1973. pp. 267–281. [Google Scholar]
- 25.Akaike H. IEEE Trans. Automatic Control. 1974;AC-19:716. [Google Scholar]
- 26.Stephens PJ, Devlin FJ, Chabalowski CF, Frisch MJ. J. Phys. Chem. 1994;98:11623. [Google Scholar]
- 27.Petersson GA, Bennett A, Tensfeldt TG, Al-Laham MA, Shirley WA, Mantzaris J. J. Chem. Phys. 1988;89:2193. [Google Scholar]
- 28.Buckley A, Chapman NB, Schorter J. J. Chem. Soc. B-Phys. Org. 1969;2:195. [Google Scholar]
- 29.Jakalian A, Jack DB, Bayly CI. J. Med. Chem. 2002;23:1623. doi: 10.1002/jcc.10128. [DOI] [PubMed] [Google Scholar]
- 30.Vedani A, Dobler M. J. Med. Chem. 2002;45:2139. doi: 10.1021/jm011005p. [DOI] [PubMed] [Google Scholar]
- 31.Vedani A, Dobler M. Quant. Struct.-Act. Relat. 2002;21:382. [Google Scholar]
- 32.Kraulis PJ. J. Appl. Cryst. 1991;24:946. [Google Scholar]