Abstract
Parainfluenza virus type 5 (PIV5), formerly known as simian virus 5 (SV5), is a non-segmented negative strand RNA virus that offers several advantages as a vaccine vector. PIV5 infects many cell types causing little cytopathic effect, it replicates in the cytoplasm of infected cells, and does not have a DNA phase in its life cycle thus avoiding the possibility of introducing foreign genes into the host DNA genome. Importantly, PIV5 can infect humans but it is not associated with any known human illness. PIV5 grows well in tissue culture cells, including Vero cells, which have been approved for vaccine production, and the virus can be obtained easily from the media. To test the feasibility of using PIV5 as a live vaccine vector, the hemagglutinin (HA) gene from influenza A virus strain A/Udorn/72 (H3N2) was inserted into the PIV5 genome as an extra gene between the hemagglutinin-neuraminidase (HN) gene and the large (L) polymerase gene. Recombinant PIV5 containing the HA gene of Udorn (rPIV5-H3) was recovered and it replicated similarly to wild type PIV5, both in vitro and in vivo. The HA protein expressed by rPIV5-H3 infected cells was incorporated into the virions and addition of the HA gene did not increase virus virulence in mice. The efficacy of rPIV5-H3 as a live vaccine was examined in 6-week-old BALB/c mice. The results show that a single dose inoculation provides broad and considerable immunity against influenza A virus infection.
Introduction
PIV5, formerly known as simian virus 5 (SV5), is a non-segmented negative strand RNA virus in the paramyxovirus family. PIV5 contains an RNA genome of 15246 nucleotides that is surrounded by a nucleocapsid protein and the genome encodes eight known viral proteins (Lamb and Kolakofsky, 2001). Nucleocapsid protein (NP), phosphoprotein (P), V and large RNA polymerase (L) proteins are important for transcription and replication of the RNA genome. Several studies suggest that the V protein has a role in evasion of host immune responses as well as in regulating viral RNA synthesis (Didcock et al., 1999; He et al., 2002; Lin and Lamb, 2000; Lin et al., 2005; Sun et al., 2004). The fusion (F) glycoprotein mediates both virus to cell and cell to cell fusion in a pH independent manner. The hemagglutinin-neuraminidase (HN) glycoprotein, is the receptor binding protein and its neuraminidase activity is important for virus release from host cells (Schmitt, He, and Lamb, 1999; Yuan et al., 2005). The matrix (M) protein has an important role in the maturation of virus (Schmitt et al., 2005). The SH integral membrane protein may have an important role in inhibiting TNF-α-mediated apoptosis (Lin et al., 2003; Wilson et al., 2006).
Non-segmented negative strand RNA viruses (NNSVs) such as PIV5 are potential viral vector candidates for vaccine development. As compared to DNA viruses, the NNSVs are potentially safer because they do not have a DNA phase in their life cycles and they replicate in the cytoplasm, thus avoiding unintended consequences from genetic modifications of host cell DNA that may be associated with recombination or insertion. In addition, as compared to positive strand RNA viruses the genome of NNSVs are stable. These characteristics make NNSVs useful as potential vaccine vectors.
Despite the advantages of using NNSV as vaccine vectors, only in recent years has it been possible to manipulate their RNA genomes due to the development of methodologies for performing reverse genetics (Neumann, Whitt, and Kawaoka, 2002; Schnell, Mebatsion, and Conzelmann, 1994). This has allowed for successful generation of recombinant NNSV vectors that include vesicular stomatitis virus (VSV), human parainfluenza virus 3 (hPIV3), and Newcastle disease virus (NDV). VSV is a highly lytic NNSV which has been engineered to express a hemagglutinin (HA) gene of influenza A virus. The recombinant HA-VSV has been shown to provide a level of immunity in mice challenged with influenza A virus (Roberts et al., 1998). In addition, NDV has been used to express the HA gene of human (H1N1) influenza and the recombinant virus shown to provide immunity against influenza virus challenge in mice (Nakaya et al., 2001). Very recently, NDV was used to express the HA protein of avian (H5N1) influenza and this virus induces potent protection against both influenza and NDV infection in poultry (Park et al., 2006; Veits et al., 2006).
PIV5 infects a range of cell types including primary human cells (Arimilli, Alexander-Miller, and Parks, 2006). Indeed, there has been no report of a cell line that is resistant to PIV5 infection. Importantly, PIV5 causes very little cytopathic effect (CPE) in infected cells (Choppin, 1964; Zakstelskaya et al., 1976). PIV5 also infects most mammals including humans and is not associated with any clinical disease with the exception of canine kennel cough (Cohn et al., 1996; Cornwell et al., 1976; McCandlish et al., 1978). The ability of PIV5 to infect a large spectrum of cells with little cytopathic effect suggests this virus could be used for gene expression in a wide range of species to elicit a broad spectrum immune response without associated inflammation and pathogenicity. Moreover, the lack of CPE allows PIV5 grow in common cell lines, including MDBK cells, for more than 40 days while the virions are continually released into media at titers up to 8×108 PFU/ml (Choppin, 1964), providing an easy and cost effective means for mass production of the vaccine vector.
In previous studies, it was shown that PIV5 could be readily recovered from cloned DNA, and that a foreign gene encoding green fluorescent protein (GFP) could be inserted into the PIV5 genome (rPIV5-GFP) with GFP expressed cytoplasmically (He et al., 1997). Recombinant PIV5 containing GFP (rPIV5-GFP) replicates to similar titers as wild type (wt) PIV5 and insertion of the gene into the viral genome is stable. We tested the feasibility of utilizing recombinant PIV5 as a vaccine vector by expressing the HA gene of influenza A virus. We show that we can insert the HA gene from influenza A/Udorn/72 (H3N2 subtype) virus into the PIV5 genome at the gene junction between HN and L genes, recover infectious recombinant PIV5 containing the HA gene (rPIV5-H3), and that mice immunized with rPIV5-H3 are protected from challenge with influenza virus.
Results
Generation of rPIV5-H3
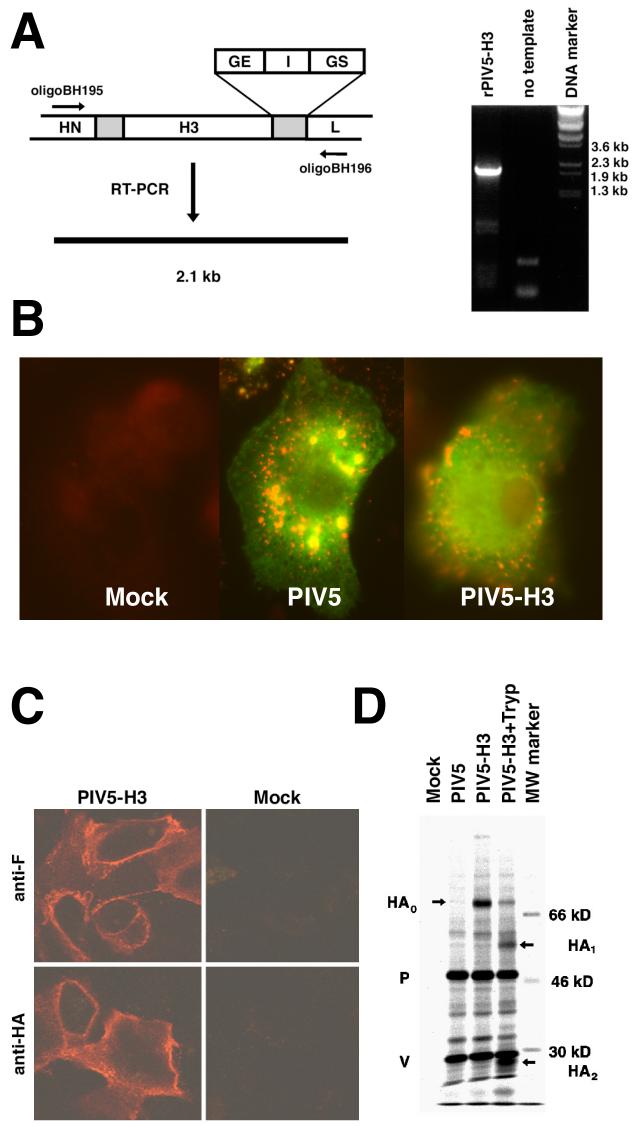
The HA gene of influenza A virus (A/Udorn/72, H3N2 subtype) was inserted between the HN and L genes of PIV5 in a plasmid (pBH311) containing the full length PIV5 genome as previously described for the GFP gene (Fig. 1A) (He et al., 1997). To ensure correct viral transcription initiation, termination and polyadenlyation of transcripts, the HA gene was flanked with gene start (GS), intergenic sequences (I) and gene end (GE) sequences from the junction region of the NP and V/P genes which gives high level transcription (He and Lamb, 1999). Recovery of rPIV5-HA was performed as described by He et al. (11) and recovery was as efficient as that of wild type PIV5. The authenticity of the rescued virus as rPIV5-H3 was confirmed by RT-PCR (Fig.1A). Insertion of the HA gene increases the expected size of RT-PCR fragment to 2.1 kb from 400 base pairs (bp) when using the specific primers that anneal to the HN and L genes (Fig. 1A). This 2.1 kb fragment was sequenced and found to match the input HA gene sequence (data not shown). It is known that PIV5 infects human cells (primary and established cell lines) (Arimilli, Alexander-Miller, and Parks, 2006). Thus to examine expression of HA in human primary cells, cells from human peripheral blood mononuclear cells (PBMC) were infected with PIV5 and PIV5-H3. Virus gene expression was detected in the human primary cells for both viruses (Fig.1B).
Figure 1. Generation of rPIV5-H3 and expression of HA in virus-infected cells.
(A). Generation of rPIV5-H3. HA gene was inserted between HN and L genes of PIV5. The shaded area represents regions between two genes. It contains GE (gene end), I (intergenic) and GS (gene start) sequences which are important for RNA transcription. A genomic nucleotide length divisible by six (the rule of six) was maintained. Oligomer BH195 that anneals to genome sense of HN gene was used for reverse-transcription; oligomer BH196 that anneals to mRNA sense of L gene and oligomer BH195 were used for PCR. The PCR product was resolved in 1% agarose gel. The DNA size markers are indicated. (B) Immunofluorescence of human primary cells infected with PIV5 and PIV5-H3. Cells obtained from primary human PBMC cells (mostly macrophages) were infected with PIV5, PIV5-H3 or mock infection. At 2 days p.i., the cells on coverslips were fixed and treated with anti-NP (red) and anti-V/P (green) antibodies. (C). Expression of HA in infected cells. CV-1 cells were infected by the rPIV5 or rPIV5-H3 and the cells were processed for immunofluorescence or immunoprecipitation at 20 h p.i.. CV-1 cells on coverslips were fixed and treated with anti-HA and anti-F antibodies as indicated. (D) Expression of HA on cell surface. Infected CV-1 cells were labeled with 35S-Met and 35S-Cys for 2 h and lysed directly or treated with trypsin and then lysed. The lysates were immunoprecipitated with antibodies against HA and the V and P proteins of PIV5. Sizes of molecular weight marker and positions of V, P, HA0, HA1 and HA2 are indicated.
Expression of HA on the surface of cells infected with rPIV5-H3
Expression of the HA gene in cells infected with rPIV5-H3 was examined by using both immunofluorescence and immunoprecipitation techniques. rPIV5-H3-infected CV-1 cells were stained with anti-HA antibody and anti-F antibody and surface expression of both HA and F could be detected (Fig. 1C). Cell surface-expressed F protein was also detected using an F specific mAb. Expression and cleavage of HA on the cell surface was confirmed by treatment of cell surfaces with trypsin. As shown in Fig. 1D, uncleaved HA0 could be cleaved by addition of exogenous trypsin to HA1 and HA2. Immunoprecipitation of the V/P proteins indicated that amounts of viral proteins expressed were similar between wild type PIV5 and rPIV5-H3 viruses.
Incorporation of HA into the rPIV5-H3 virion
To analyze the incorporation of HA into rPIV5-H3 virions, the virus was grown in MDBK cells, purified and polypeptides analyzed by SDS-PAGE. HA was observed in rPIV5-H3 virions by Commasie blue staining (Fig.2A) and its identity confirmed by immunoblotting (data not shown). Concomitantly, the amount of HN incorporated into rPIV5-H3 was decreased as compared to the amount of HN incorporated into wt PIV5. To eliminate the possibility that the HA protein observed was associated with contaminating vesicles and debris, purified rPIV5-H3 virions were immunogold labeled using HA antibody and examined by electron microscopy. HA labeling of virion was readily detected (Fig. 2B).
Figure 2. Incorporation of HA into purified virions.
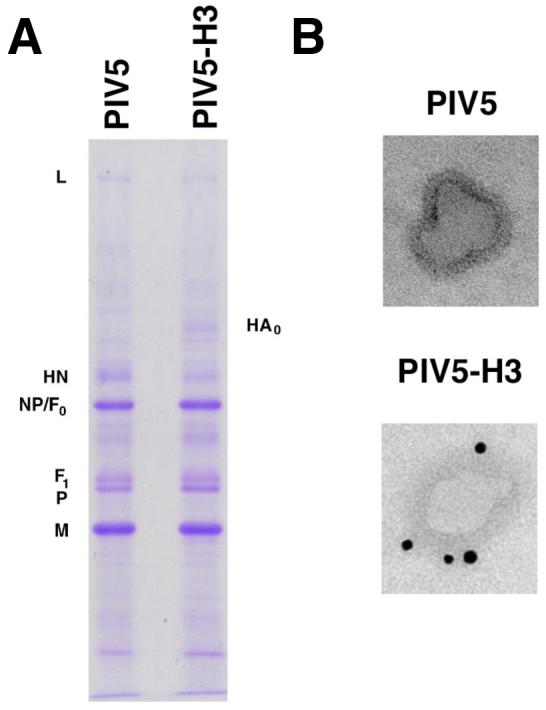
rPIV5-H3 was purified as described in the Material and Methods. (A) The purified virus polypeptides were resolved using a 15% SDS-PAGE gel and the gel was stained with Commasie blue. Positions of viral proteins are indicated. (B) The purified virions were treated with anti-HA mAb and gold particle-labeled secondary antibody and then examined by electron microscopy. Panels of different virions are indicated.
As PIV5 virions are pleomorphic with sizes ranging from 100 to 200 nm, we examined a large number of virions and found no change in virion morphology or size associated with the recombinant virus, indicating that the addition of 1700 nt into the PIV5 genome did not alter virion morphology (data not shown).
Growth of rPIV5-H3 in vitro and in vivo
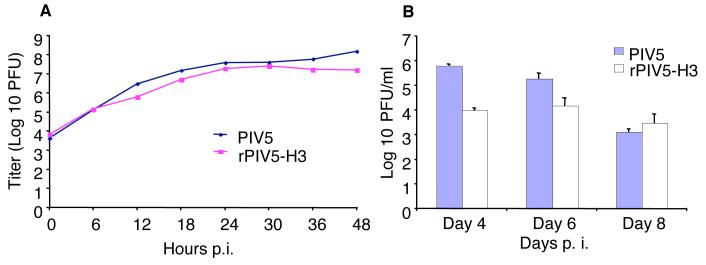
To determine if insertion of the HA gene had any detrimental effect on virus replication, single-step growth curves of wt PIV5 and rPIV5-H3 viruses were performed. MDBK cells were infected with 10 plaque-forming units (PFU) per cell of rPIV5 or rPIV5-H3 and supernatants were collected at 6 h intervals for up to 48 h and virus quantified by plaque assay (Fig. 3). The growth curve for rPIV5-H3 was very similar to that of wt virus. Both viruses had similar initial growth kinetics and reached a plateau at about 24 h post infection (p.i.). Additionally, the plaque sizes of the two viruses were comparable (data not shown).
Figure 3. Growth of rPIV5-H3 in vitro and in vivo.
(A). Growth curve in vitro. MDBK cells were infected at 10 PFU/cell. Media from the infected cells were collected at 6 h intervals up to 48 h p.i.. Titers of the media were examined using plaque assay in BHK-21F cells. (B). Growth in vivo. BALB/c mice (15 in a group) were infected with PIV5 or rPIV5-H3. Five mice per group were sacrificed on days 4, 6 and 8 p.i.. Titers of viruses in lung were determined by plaque assay.
To examine the growth of rPIV5-H3 in vivo, BALB/c mice were inoculated intranasally (i.n.) with 106 PFU of PIV5 or rPIV5-H3. Lungs of the infected animals were collected at 4, 6 and 8 days p.i., and virus titers in lungs were determined by using plaque assays as described previously (He et al., 2002). It was found that PIV5 replicated to higher titers than rPIV5-H3 at 4 days p.i. (Fig. 3B); however, the differences in virus titer diminished over time, e.g. at 8 days p.i. both viruses had approximately the same titers, i.e. ∼103 PFU.
Pathology of rPIV5-H3 in BALB/C mice
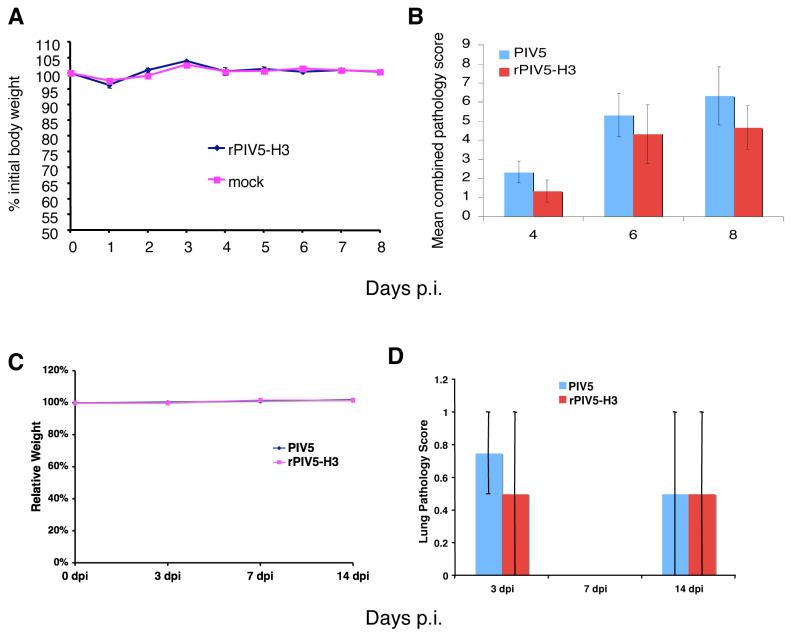
PIV5 infection does not cause known overt disease except kennel cough in canines (Chatziandreou et al., 2004). Whether PIV5 infection was pathogenic for mice and whether expression of the HA gene could affect the pathogenicity of the virus in mice have been determined. First, BALB/c mice were infected i.n. with 106 PFU of rPIV5-H3 and the body weights of the infected mice monitored daily. Infection of mice with rPIV5-H3 did not cause any detectable symptoms or weight loss compared with mock (PBS)-inoculated mice (Figure 4A). Second, to address other parameters of pathology that may be associated with PIV5 or rPIV5-H3 infection, BALB/c mice were inoculated i.n. with 106 PFU of PIV5 or rPIV5-H3. At 4, 6 and 8 days p.i., the animals were sacrificed and lung pathology examined by H&E staining. At each sampling period, histopathology changes were similar in character and magnitude in the lung lobes of individual animals between infected groups, i.e. PIV5 vs. rPIV5-H3. The lung histopathology generally was characterized by low-level perivascular, peribronchiolar, subpleural, and scattered intra-alveolar infiltrations that were predominantly lymphocytes. Infiltrates contained a few plasma cells by day 8 p.i. and intra-alveolar infiltrates were admixed with macrophages. Cellular infiltrations increased over the time-course of infection by both viruses, but were generally mild, and at each time period examined, there was no statistically significant difference (P≤0.05) between groups (Fig. 4B). However, as expected, the means for each group did increase with time. The mild cellular reaction suggested a progressive, localized immunologic response in the lung, apparently associated with PIV5 as addition of the HA gene did not exacerbate the reaction. To further confirm that rPIV5-H3 did not cause enhanced pathogenicity, the pathogenicity of the viruses in nude mice have been examined by injecting 107 PFU of each virus into the tail vein. Neither PIV5 nor PIV5-H3 caused signs of illness or weight loss at either early or late time-points post-infection, indicating the viruses are not pathologic even in immune deficient mice (Fig.4C). Examination of the organs from the mice infected with different viruses indicated that there was no difference in pathological scoring between PIV5 and rPIV5-H3-infected mice (Fig.4D). While signs of infection were observed as expected, there were no significant pathology detected in the spleen, heart, liver or kidney (data not shown) and only very mild patchy histeocytosis and very diffuse mixed inflammatory infiltrates, predominantly histocytes with a microgranuloma, were observed in lungs of some of the animals in both PIV5 and rPIV5-H3-infected mice.
Figure 4. Pathology of PIV5 and rPIV5-H3 in BALB/c mice.
(A). Weight loss. BALB/c mice (n=8) were infected with rPIV5-H3 (106 PFU in 50μl PBS) or mock-infected with PBS only. Body weights of the mice were recorded and graphed as percentage of the weight before inoculation. (B) Pathology of PIV5 in mice. BALB/c mice (n=9) were infected with rPIV5-H3 or rPIV5 (106 PFU in 50μl PBS). Animals were sacrificed on days 4, 6 and 8 p.i.and lungs removed and fixed. Lungs were sectioned, H&E stained and analyzed. For each animal, four major pulmonary changes (peribronchiolar infiltrations; perivascular infiltrations; parenchymal infiltrations; subpleural infiltrations) were subjectively scored 0-3 with 0=no change and 3=greatest change and a total score determined. Means of the total scores for each group are graphed and standard deviations are shown as error bars. Scores of the two groups (PIV5 vs. rPIV5-H3) were compared at each time point (day 4, 6 or 8) by Student's t-test and all p values are less than 0.05. No statistically significant differences (p<0.05) between the two viruses were observed. (C) Weights of nude mice injected with PIV5 or rPIV5-H3. Nude mice (14 week-old, n=10) were infected i.v. with PIV5 or PIV5-H3 (107 PFU) in 200μl volume. Body weights of the mice were recorded and graphed as percentage of the weight before inoculation. (B) Pathology scores of lung. The mice were sacrificed and organs (lung, heart, liver, kidney and spleen) were sectioned for H&E staining. Pathology scores of lung were shown. 1= mild pathology; 2 = intermediate pathology; 3 = server pathology. Bar is average score and the error bar represents the range. No statistically significant differences between two viruses were observed.
Immunity against influenza A virus infection in mice immunized with rPIV5-H3
To examine the efficacy of rPIV5-H3 as a live vaccine against influenza A virus infection, six-week-old BALB/c mice were inoculated i.n. using virus stock grown in MDBK cells. BALB/c mice were i.n. vaccinated with 0, 102, 104 or 106 PFU of rPIV5-H3 or rPIV5 viruses diluted in 50 μl PBS. Six weeks after vaccination, the mice were challenged i.n. with 106 PFU of influenza A virus strain WSN-H3. WSN-H3 is a 7+1 reassortant virus containing all of the genome segments from A/WSN/33 virus except the HA segment, which is from A/Udorn/72 (Jin et al., 1996). WSN-H3 was generated through recombination of genomes between A/WSN/33 (WSN), which is mouse-adapted and highly pathogenic, and Udorn, a human strain, which is not pathogenic in mice. WSN-H3 contains 7 genomic segments of WSN and the HA segment from Udorn (subtype H3). As noted, WSN-H3 is pathogenic in the mouse, although not as pathogenic as the parent strain, WSN. Following challenge, the mice were weighed daily. As expected, body weights of vaccinated and unvaccinated mice not challenged with WSN-H3 were similar. For the mice that were not vaccinated with rPIV5-H3, but challenged with the 106 PFU of WSN-H3, three out of four died by day 10 (Fig. 5A). The remaining mouse lost more than 30% of its body weight and had not recovered by day 12. A similar mortality rate was initially observed in the mouse group vaccinated with a low dose (102 PFU) of rPIV5-H3 and challenged with WSN-H3 in which four of the mice died at days 5, 8, 10 and 11 pi (Fig. 5B). However, the remaining 4 vaccinated mice recovered after 6 days after losing up to 20% to 30% of their body weight. By day 12, these mice regained their weight indicating that there was limited protection even for the mice inoculated with the low dosage of rPIV5-H3. The group of mice inoculated with 104 PFU of rPIV5-H3 suffered moderate weight loss (up to about 25%) and all of the mice survived. They also started to recover weight at day 6 p.i. (Fig. 5C). The group of mice vaccinated with 106 PFU of rPIV5-H3 had the lowest weight loss following challenge and all animals survived challenge (Fig. 5D). To confirm HA-specific immunity in mice vaccinated with rPIV5-H3, a group of mice were vaccinated i.n. with wild type rPIV5 alone and similarly challenged with WSN-H3 (Figure 5F). As expected, rPIV5 alone did not provide immunity against influenza A virus challenge. Two of the mice died at days 8 and 10, and the rest still were suffering from weight loss up to day 12.
Figure 5. Efficacy of rPIV5-H3 as a live vaccine vector.
Six-week-old BALB/c mice were immunized i.n. with PIV5-H3 or rPIV5. Six-weeks p.i., the mice were challenged i.n. with influenza A virus, WSN-H3. Weights of the mice were monitored and graphed as percentage of body weight of the mice. (A) Weights of individual mouse in the control group mice (n=4), i.e., weights of mock- (PBS) immunized mice challenged with WSN-H3. (B) to (D) Weights of individual immunized with 102 (B) (n=8), 104 (C) (n=8), or 106 PFU (D) (n=8) of rPIV5-H3 and then challenged WSN-H3. (E) Average weights of mice immunized with 104 and 106 PFU rPIV5-H3 and then challenged with WSN-H3. (F) Weights of individual PIV5-immunized mouse challenged with WSN-H3 (n=8).
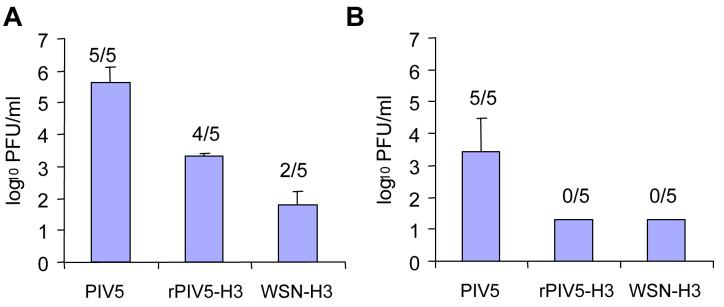
Influenza virus titers in the lungs of rPIV5-H3 inoculated mice
To further confirm protection associated with rPIV5-H3 vaccination, WSN-H3 titers in the lungs of infected mice were determined by plaque assay. Animals from each immunization group were sacrificed at days 4 and 7 post-challenge. The results show that vaccination with rPIV5-H3 provided protection from influenza A virus challenge, as indicated by the significant differences between PIV5 and rPIV5-H3 immunized groups (Fig. 6A, P<0.001, ANOVA). Similar to WSN-H3 immunized mice, rPIV5-H3 vaccinated mice showed reductions in lung virus titers and exhibited more rapid clearance of virus compared to rPIV5-vaccinated mice. While all of the PIV5-immunized mice still had some virus burden on day 7 p.i., the rPIV5-H3 mice had begun to clear the infection on day 4 (1/5 mice were virus-free) and all were clear of virus by day 7 (Fig. 6B). Thus, immunization with PIV5 virus expressing the influenza HA is sufficient to provide immunity that is comparable to that induced by influenza virus.
Figure 6. Reduction in influenza virus lung titer by rPIV5-H3 immunization.
BALB/c mice (n=10 per group) were immunized with rPIV5, rPIV5-H3, or WSN-H3 (sub-lethal dosage, 103 PFU) and 29 days later, challenged with 106 PFU WSN-H3 . Mice (n=4 or 5 per day per group) were sacrificed on day 4 (A) and day 7 (B) post-challenge and lung virus titers measured by plaque assay. Data is presented as average PFU/lung +/- SEM. The number in parenthesis indicates the number of samples with detectable virus. The limit of detection was 20 PFU/lung.
Anti-influenza virus antibody production in mice
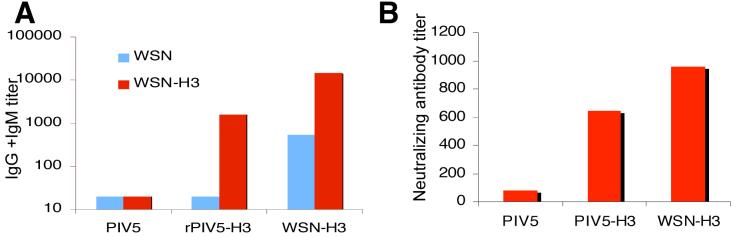
To investigate the level of the antibody responses in vaccinated mice, sera was collected from mice vaccinated with rPIV5-H3, PIV5, as well as from mice infected i.n. with a sub-lethal dose of WSN-H3-inoculated (Fig. 7). Total and neutralizing antibody levels against WSN (H1N1) and WSN-H3 (H3N1) were determined by ELISA against WSN. As predicted, mice vaccinated with rPIV5 inoculation did not produce any detectable amount of antibody against WSN since rPIV5 does not express any WSN genes. In contrast, mice vaccinated with rPIV5-H3 did produce high titers of antibody against WSN-H3, presumably specific for the H3 protein (Fig. 7A). The generation of neutralizing antibodies induced by rPIV5 or rPIV5-H3 vaccination was also examined. The results show that while PIV5 inoculation did not produce anti-WSN-H3 neutralizing antibodies, rPIV5-H3 inoculation produced a substantial neutralizing antibody response against WSN-H3 that was comparable to WSN-H3 infection (Fig. 7B). Higher antibody titers observed in sub-lethal WSN-H3 inoculated mice may be attributed to antibodies against other influenza virus proteins such as NA and M2. It is known that immunization with live influenza virus induces antibody and cellular immune responses to not only HA, but also other influenza antigens including neuraminidase, matrix, and nucleoprotein, which can contribute to protection and in some cases are sufficient to protect against virus challenge (reviewed in (Epstein, 2003)).
Figure 7. Antibodies in rPIV5-H3 immunized mice.
BALB/c mice (n=10 per group) were immunized with rPIV5, rPIV5-H3, or WSN-H3 and 19 days later, serum was collected for serum antibody analysis. (A). Influenza virus-specific antibody titers were measured in serum samples using an IgG+IgM-specific ELISA as described in the materials and methods; (B). Influenza A WSN-H3 neutralizing antibody titers were measured in serum samples by virus neutralizing assay described in the Materials and Methods.
Discussion
The development of reverse genetics technology for the recovery of NNSVs from their cDNAs, has permitted the expression of many foreign genes using these viruses (Bukreyev et al., 2005; Palese et al., 1996; Park et al., 2006; Pekosz, He, and Lamb, 1999; Roberts and Rose, 1998; Veits et al., 2006). Here we recovered PIV5-H3, which contains an HA gene from influenza A/Udorn/72 virus inserted between the HN and L genes of PIV5. rPIV5-H3 was viable and the insertion of the HA gene appeared to be stable and the virus grew well in tissue culture cells. A single dose inoculation of the rPIV5-H3 provided immunity to mice against influenza virus infection.
The HA protein was incorporated into PIV5 virions despite the absence of specific PIV5 sequences that may be important for incorporation into virions such as PIV5-specific cytoplasmic tails. The HA protein was detected in purified virions by both SDS-PAGE and electron microscopy (Fig. 2). Additionally, the amount of HN incorporated into rPIV5-H3 virions was reduced compared to rPIV5. It is perhaps surprising that HN was not preferentially incorporated into virions over HA. The efficient release of PIV5 from infected cells requires the cytoplasmic tail of HN (Schmitt, He, and Lamb, 1999) and it is thought that the cytoplasmic tail of HN is important for formation of budding virus and/or pulling viral components into virions. Despite having a cytoplasmic tail dissimilar to the HN, HA was incorporated into the PIV5 virions and presumably filled space at the expense of HN.
A single dose inoculation of rPIV5-H3 provided immunity to mice against influenza virus infection. The gradient of protection provided by the virus indicates effective immunity was dependent on the dosage of the virus used and the protection was provided by rPIV5-H3 inoculation but not by rPIV5. Although the effectiveness of rPIV5-H3 in preventing death by influenza A virus infection is evident, all mice lost body weight, indicating protection is not optimal. This is partially due to handling of mice as even mock inoculated and mock challenged mice suffered about 5% weight loss. To increase potency of rPIV5-H3 against influenza virus challenge, one possible remedy is to use multiple inoculations (i.e. prime and boost) instead of a single immunization prior to challenge. Alternatively, it may be possible to increase expression of HA by inserting the HA gene closer to the leader sequence of PIV5, the only de facto promoter of PIV5. Negative stranded RNA viruses such as PIV5 initiate transcription from the 3' end leader sequence, and transcription levels of the viral genes are affected by the distance of the gene from the leader sequence (Tokusumi et al., 2002). For example, the NP gene of PIV5, which is proximal to the leader sequence is the most abundantly transcribed, whereas the L gene, which is distal to the leader sequence is least transcribed. To increase the expression level of the HA gene, the HA gene can be inserted immediately downstream of the leader sequence and upstream of the NP gene.
Safety is a paramount concern for vaccine development. Expression of a foreign glycoprotein has the potential to expand the host range beyond the original virus, therefore creating a potential safety hazard. However, because PIV5 already infects all cell types that have been tested, it is less likely that adding an additional receptor-binding gene to its genome will change its tropism. PIV5 encodes two major glycoproteins F and HN that are important for PIV5 entry. The PIV5 F protein promotes virus to cell fusion at the plasma membrane independent of pH. In some cells, F alone is sufficient to promote cell-to-cell fusion without PIV5's receptor binding protein, HN (Paterson, Russell, and Lamb, 2000). The HN protein of PIV5 mediates virus attachment by binding to sialic acid, which is added to surface proteins post-translationally. Unlike influenza virus protein, the HN protein of PIV5 has no preference as to what kind of sialic acid it binds. For example, PIV5 may infect primary chicken cells (Peluso, Lamb, and Choppin, 1977), which has mainly N-acetylneuraminic acid α 2,3-galactose (NeuAcα2,3Gal) and may infect human cells (Lamb and Kolakofsky, 2001), which has mainly NeuAcα2,6Gal (Ito et al., 1997). Thus, it is unlikely that expressing influenza HA will expand the host range of PIV5. Nevertheless, to address the concern that enhanced pathogenicity may be caused by PIV5-H3, we examined the pathogenicity of the viruses in nude mice by injecting 107 PFU of each virus into the tail vein. Neither PIV5 nor PIV5-H3 caused signs of illness or weight loss at either early or late time-points post-infection, indicating the viruses are not pathological even in immune deficient mice (Fig.4C). Examination of organs of the mice indicated that there was no difference in pathological scoring between PIV5 and rPIV5-H3-infected mice (Fig.4D). Thus, as expected, insertion of the HA gene into PIV5 genome did not cause increased pathogenicity in mice. In the event that expressing a functional HA is a concern, a mutant HA without HA1/HA2 cleavage site can be used for influenza virus vaccine development.
Unlike icosahedral viruses, PIV5 virions have many forms and shapes. This pleomorphic virion structure provides flexibility to accommodate changes in sizes of PIV5's genome. This is an advantage for insertion of foreign genes as the changes to the viral genome will probably not be tightly restricted by the virion structure. While we do not know the upper limit for the size of insertion in PIV5, in our hands, insertion of 1700 nt, an increase of about 11% of genome size, does not affect virus growth or integrity of virions. We have now inserted and expressed a gene as large as 2.3 kb into PIV5 genome without difficulty (data not shown).
The origin and natural host of PIV5 is not clear. PIV5 was first isolated from monkey cells as a contaminant in 1956, hence the original name SV5 (Hull, Minner, and Smith, 1956). However, subsequent serological testing of monkeys in the wild indicated no exposure to this virus. In contrast, monkeys in captivity at an animal facility rapidly sero-converted, suggesting they contacted the virus in captivity (Atoynatan and Hsiung, 1969; Tribe, 1966). Thus, considerable evidence indicates that PIV5 is not a simian virus. PIV5 dose cause kennel cough in canines (Azetaka and Konishi, 1988; Binn et al., 1967; Cornwell et al., 1976; McCandlish et al., 1978; Rosenberg et al., 1971) and killed PIV5 is a component of the commercial vaccine “Vanguard” (Pfizer, Inc.) for dogs, which includes canine distemper virus vaccine. There is no convincing evidence that PIV5 causes diseases in humans, despite much speculation in the 1970's that PIV5 might be associated with a number of illnesses including multiple sclerosis (MS), subacute sclerosing panencepalitis (SSPE), Creutzfeldt-Jakob disease (CJD), pemphigus, athero-sclerosis, Paget's disease, hepatitis and the common cold. Subsequent studies have ruled out PIV5 as the etiological agent for these diseases (Chatziandreou et al., 2004; Hsiung, 1972; Vandvik and Norrby, 1989). In retrospect, two possible explanations exist for why PIV5 may have been linked to these diseases. One reason is based on the conditions used for virus isolation in the human studies, i.e. the labs used monkey cell lines which can be persistently infected with PIV5, and these cells often show no detectable cytopathic effects (Chatziandreou et al., 2004; Hsiung, 1972). Another reason is that there is known antigen cross-reactivity of PIV5 to ubiquitous paramyxoviruses such as human parainfluenza virus 2 and mumps virus, which are closely related to PIV5 and have almost 100 percent exposure in human population (Komada et al., 1991; Randall and Young, 1988; Tsurudome et al., 1989). Given the possibility of PIV5 antigen cross-reactivity, it may be a concern to use PIV5 as a vaccine vector. However there is no indication from studies in mice that antibodies can prevent PIV5 infection (Young et al., 1990). As shown in Fig.1B and others (Arimilli, Alexander-Miller, and Parks, 2006), PIV5 can infect human primary cells, suggesting it is likely that PIV5 can infect humans.
While live virus vectors hold great promise, there are many potential pitfalls, as has been demonstrated in studies using the two most studied virus vectors: adenovirus and VSV. For instance, adenovirus is a DNA virus and thus has the potential to integrate its genome. An additional problem is that there is a very high incidence of adenovirus sera- positive people. Also, a patient in a clinical trial died as a result of being treated with a high dose adenovirus-based gene vector. The other viral vector noted, VSV, is neurotrophic, thus its potential impact on immune comprised patients is of an important concern. Nonetheless, it is important to explore the potential use of these viral vectors due to the many advantages of live virus vectors over alternative vaccination strategies. As an RNA virus that infects respiratory epithelium, PIV5 offers an important alternative vector with properties that make it worthy of special consideration and further study, especially for developing an influenza virus vaccine. Thus, although several NNSV have been used previously to express foreign viral antigens and to protect mice for virus challenge, PIV5 offers an interesting alternative vector with properties that make it worthy of special consideration and further study.
Materials and Methods
Plasmid construction and RT-PCR
HA gene from pTF7.5-HA was amplified by PCR using oligomers BH291 (5'-ATTTCCTAGGTAATTTTTAAGAAAAAAACGATAGGACCGAACCTATCGATGCCGGC AAACATGAAGACTATCATTGCTTTG-3') and BH292 (5'-AAATTTGCATGCCATGGCCTAGGATATTCAAATGCAAATGTTGCACCT-3'). The PCR fragment was inserted into Hinc II site of pGEM2 (Promega) to generate pBH374. The coding region of HA gene in pBH374 was sequenced and found to be identical to the HA gene in pTF7.5-HA. The HA gene was excised out of the plasmid by digestion with Cla I and Nco I. The fragment was then filled-in by the Klenow fragment and inserted into filled-in Not I site of a plasmid (pBH311) (He et al., 1997) which contains a GFP gene between the HN and L genes in PIV5 genome to generate pBH412 which contains the PIV5 genome with the HA gene as an extra gene. Standard protocols of molecular cloning were followed. Oligomers were purchased from Macromolecular Resources (Colorado State University). All sequences of the plasmids are on file and available on request. Plasmids pTF7.5-HA, pT7-L, pT7-P and pT7-NP were described before (He et al., 1997; Murphy and Parks, 1997).
To sequence the viral RNA genome, total RNA from CV-1 cells infected by virus in 6 cm plate were purified using RNeasy kit (Qiagen®) according to manufacture's instruction. 19 μl out of 50 μl RNA were used with 20 ng of oligomer BH195 (5'-TTCAGATTGTCCCATTTATCCGTCAGG-3') for cDNA synthesis at 37°C for 1 hour in a total volume of 40 μl. 10 μl of cDNA were used in PCR with oligomers BH195 and BH196 (TGTAATGGACCTAAATCGTCAAGGTCG-3'). The PCR products were resolved in 1% agarose gel.
Viruses and cells
Virus recovery was carried out as described before (He et al., 1997). 5 μg of plasmid pBH412 which contains a PIV5 genome with HA gene insertion was transfected with 3μg of pT7-L, 0.2 μg of pT7-P and 2 μg of pT7-NP into A549 cells in 6-well plates which were infected with the modified vaccinia virus Ankara strain (MVA) containing a bacteriophage T7 RNA polymerase gene for one hour. Transfection media were changed to DMEM containing 2% fetal calf serum (FCS) 24 hours post transfection. Virus released into the media was amplified in CV-1 cells. Recovery of virus is indicated by syncytia formation in BHK 21F cells on incubation with the media from the CV-1 cells. The virus was then purified as a single plaque from BHK 21F cells. Human primary cells were obtained from PBMC. The human primary cells (mostly macrophages) from PBMCs were prepared as described (Lee, Zhou, and Henderson, 2001) and were seeded in 6-well plate with coverslips for 6 days before used for infection.
Viruses were grown in MDBK cells for 5 to 7 days using DMEM containing 2% FCS until their hema-absorption titers plateaued. Media were collected and pelleted at 3000 rpm using a Sorvall tabletop centrifuge for 10 minutes. BSA was added to the cleared supernatant to a final concentration of 1%. The virus stock was quickly frozen in dry ice and stored at −70°C. To purify the viruses, viruses in the cleared supernatant were pelleted in a Beckman ultracentrifuge Type 45Ti rotor at 19,000 rpm for 1 h. The pellet was then resuspended in NTE buffer and loaded onto 15% to 60% sucrose gradient and centrifuged in a SW 41Ti rotor for 2 h at 24,000 rpm. The virus band was collected and virus was pelleted in a Type 70Ti rotor for 1 h at 35,000 rpm (Paterson and Lamb, 1993). Virus titers were determined by plaque assay on BHK-21F cells as previously described (He et al., 1997).
MDBK cells, A549 cells and CV-1 cells were cultured in DMEM with 10% FCS. BHK-21F cells were grown in DMEM with 10% FCS and 10% tryptone phosphatase broth.
Immunoprecipitation of polypeptides
CV-1 cells in 6 cm plates were infected with rPIV5 and rPIV5-H3 at a m.o.i. of about 5 pfu/cell and labeled with 100 μCi/ml 35S-Met and 35S-Cys 20 hours p.i. (Paterson and Lamb, 1993). Half of the plates of CV-1 cells were treated with 0.01 mg/ml L-(tosylamido-2-phenyl) ethyl chloromethyl ketone (TPCK)-treated trypsin for one hour before the cells were lysed in RIPA buffer (Paterson and Lamb, 1993). Lysates were immunoprecipitated using mAb Pk specific for a shared region of the V and P proteins and rabbit polyclonal antibody specific for Udorn HA protein. Polypeptides were analyzed by 10% SDS-PAGE and their radioactivity was detected using a Fuji BioImager 1000.
Fluorescence and electron microscopy
CV-1 cells were grown on glass cover slips and infected with rPIV5 or rPIV5-H3 at a m.o.i. of about 5 PFU/cell. At 20 h p.i., the infected cells were washed with PBS and then fixed with 1% formaldehyde for 10 min at room temperature. The cells were washed three times with PBS and incubated for 30 minutes with 1:500 dilution of antibodies against PIV5 F or HA. The cells were washed extensively to remove unbound antibodies and secondary antibodies labeled FITC were added for 30 min. The cells were further washed and placed on slides. Fluorescence was examined using a Zeiss 410 confocal microscope.
To examine HA in virions, purified virions were absorbed onto parlodion-coated nickel grids for 30 seconds. Grids were floated on a drop of Tris-buffered saline (TBS), pH 7.4 for 5 minutes, after which they were incubated by floating on drops of 3% ovalbumin in TBS for 1 hour with HA specific mouse mAb ascites fluid 6D/1 diluted 1:300 in 1% ovalbumin in TBS. Following three successive washes with TBS for 10 minutes each, samples were incubated for 1 h with goat anti-mouse IgG coupled to 10 nm gold particles diluted 1:10 in 1% ovalbumin in TBS. Grids were washed in TBS as above and finally stained with 2% phosphotungsic acid, pH 6.6. Before viewing, a thin layer of carbon was evaporated over the grids for stability. The grids were then examined using JEOL 1230 transmission electron microscope (JEOL, Tokyo, Japan).
Virus titers in vivo and pathology of infection with PIV5 or rPIV5-H3
Six-week-old BALB/c mice were first anesthetized with Avertin i.p. and then infected intranasally (i.n.) with 106 PFU of PIV5 or rPIV5-H3 in 50 μl PBS. At various times post infection, mice were euthanized and lungs collected for either lung virus titer or histopathologic analysis. For virus titers, lungs were homogenized in 1 ml PBS and the homogenates cleared by centrifugation. Cleared homogenates were serially diluted and virus titers determined by plaque assay on BHK-21F cells (He et al., 1997). For histopathology, lungs were perfused with 10% buffered formalin via the trachea and then placed in formalin for fixation. Sections of all lung lobes were embedded in paraffin, sectioned at 3 μm, stained with hematoxylin and eosin, and examined by light microscopy. For each animal, four major pulmonary changes (peribronchiolar infiltrations; perivascular infiltrations; parenchymal infiltrations; subpleural infiltrations) were subjectively scored 0-3 with 0=no change and 3=greatest change and a total score determined. For pathology studies in nude mice, 107 PFU of each virus in a 200 μl volume was injected into the tail veins of nude mice. The mice were weighed and sacrificed at 3, 7 and 14 days post injection. The organs of the mice (lung, heart, liver, kidney and spleen) were sectioned and processed for H&E staining.
Influenza A virus challenge experiments
Six-week-old BALB/c mice were first anesthetized and then inoculated intranasally by dropping 50 μl PBS containing 0, 102, 104 or 10 6 PFU of viruses. Six weeks later, the mice were inoculated intranasally with 50μl PBS containing 106 PFU of an influenza virus 7+1 reassortant WSN-H3 that contains all segments from WSN strain except the HA segment which is derived from A/Udorn/72. Body weights of the mice were recorded everyday thereafter.
Antibody measurement
Influenza virus-specific serum antibody titers were measured using an IgG+IgM ELISA. Enzyme immunoassay plates (Corning Costar, Inc.) were coated with 50 μl/well of WSN or WSN-H3 containing approximately 105 PFU of virus. Viruses were UV inactivated and the plates washed. The wells were blocked with 200 μl/well Starting Block Buffer (Pierce, Inc.) twice for 1 minute. After washing, dilutions of sera were added (50 μl/well) and incubated at room temperature for two hours. Plates were washed and 100 μl/well of a 1:1,000 dilution of alkaline phosphate-labeled rat goat-mouse IgG+IgM, IgG2a, IgG2b and/or IgA (KPL, Inc.) was added. After incubation for one hour at room temperature, the plates were washed, 100 μl/well pNPP phosphatase substrate (KPL, Inc.) added, and the enzymatic reaction was allowed to develop at room temperature. O.D. was measured at 405 nm on a BioTek plate reader. The ELISA titer was the lowest serum dilution with an OD 2 standard deviations above the OD of the same dilution of naïve serum.
Influenza neutralizing antibody titers were measured in serum samples by virus neutralizing assay. Sera were serially diluted in 50μl DMEM+5% FBS. 2000 TCID50 of WSN-H3 virus was added to diluted sera and incubated for 60 minutes at 37°C. Serum and virus were added to 96-well microtiter plates containing 80-90% confluent MDCK cells and incubated overnight at 37°C. The wells were washed with PBS and the medium replaced with 0.2mL of MEM+0.25μg/ml TPCK-treated trypsin (Worthington) and incubated for 4 days at 37°C. Individual wells were scored for CPE and supernatants assayed for virus by hemagglutination of chicken red blood cells (cRBCs).
In brief, fresh cRBCs were washed in Alsevier's solution, counted, and resuspended in PBS at a final concentration of 0.5%. In a round-bottom, 96-well microtiter plate 50μl of 0.5% cRBCs were added to 50 μl of culture supernatant from the virus neutralization, MDCK plate. Plates were tapped to gently mix the cells and supernatants. The plates were incubated at 4°C for 1 h and scored for hemagglutination. The absence of hemagglutination indicated a lack of infectious virus in the neutralization assay, indicating the presence of influenza-neutralizing antibodies in the serum samples. The neutralizing titer was the lowest serum dilution neutralizing 2000 TCID50 of virus.
Statistical analysis
Lung virus titers were compared using one-way ANOVA statistical analysis on log-transformed data, followed by pairwise multiple comparison using the Holm-Sidak method. Lung histology scores were compared by Student's t-test. All statistical analysis was done with SigmaStat Software v3.11 (Systat Software, Point Richmond, CA).
Acknowledgment
We thank Amy Strasner and Dr. Andrew Henderson for providing human primary cells. We appreciate other members of Biao He's lab for discussion and technical help. This work was supported by research grant AI-23173 (R.A.L.) and AI51372 (B.H.) from the National Institute of Allergy and Infectious Diseases and a grant from the Pennsylvania Department of Agriculture (B.H.). R.A.L. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arimilli S, Alexander-Miller MA, Parks GD. A simian virus 5 (SV5) P/V mutant is less cytopathic than wild-type SV5 in human dendritic cells and is a more effective activator of dendritic cell maturation and function. J Virol. 2006;80:3416–3427. doi: 10.1128/JVI.80.7.3416-3427.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atoynatan T, Hsiung GD. Epidemiologic studies of latent virus infections in captive monkeys and baboons. II. Serologic evidence of myxovirus infections with special reference to SV5. Am J Epidemiol. 1969;89:472–479. doi: 10.1093/oxfordjournals.aje.a120959. [DOI] [PubMed] [Google Scholar]
- Azetaka M, Konishi S. Kennel cough complex: confirmation and analysis of the outbreak in Japan. Nippon Juigaku Zasshi. 1988;50:851–858. doi: 10.1292/jvms1939.50.851. [DOI] [PubMed] [Google Scholar]
- Binn LN, Eddy GA, Lazar EC, Helms J, Murnane T. Viruses recovered from laboratory dogs with respiratory disease. Proc Soc Exp Biol Med. 1967;126:140–145. doi: 10.3181/00379727-126-32386. [DOI] [PubMed] [Google Scholar]
- Bukreyev A, Huang Z, Yang L, Elankumaran S, Claire M, Murphy BR, Samal SK, Collins PL. Recombinant Newcastle Disease Virus Expressing a Foreign Viral Antigen Is Attenuated and Highly Immunogenic in Primates. J. Virol. 2005;79:13275–13284. doi: 10.1128/JVI.79.21.13275-13284.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatziandreou N, Stock N, Young D, Andrejeva J, Hagmaier K, McGeoch DJ, Randall RE. Relationships and host range of human, canine, simian and porcine isolates of simian virus 5 (parainfluenza virus 5) J Gen Virol. 2004;85:3007–3016. doi: 10.1099/vir.0.80200-0. [DOI] [PubMed] [Google Scholar]
- Choppin PW. Multiplication of a myxovirus (SV5) with minimal cytopathic effects and without interference. Virology. 1964;23:224–233. doi: 10.1016/0042-6822(64)90286-7. [DOI] [PubMed] [Google Scholar]
- Cohn ML, Robinson ED, Thomas D, Faerber M, Carey S, Sawyer R, Goswami KK, Johnson AH, Richert JR. T cell responses to the paramyxovirus simian virus 5: studies in multiple sclerosis and normal populations. Pathobiology. 1996;64:131–135. doi: 10.1159/000164026. [DOI] [PubMed] [Google Scholar]
- Cornwell HJ, McCandlish IA, Thompson H, Laird HM, Wright NG. Isolation of parainfluenza virus SV5 from dogs with respiratory disease. Vet Rec. 1976;98:301–302. doi: 10.1136/vr.98.15.301. [DOI] [PubMed] [Google Scholar]
- Didcock L, Young DF, Goodbourn S, Randall RE. The V protein of simian virus 5 inhibits interferon signalling by targeting STAT1 for proteasome-mediated degradation. J. Virol. 1999;73:9928–9933. doi: 10.1128/jvi.73.12.9928-9933.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein SL. Control of influenza virus infection by immunity to conserved viral features. Expert Rev Anti Infect Ther. 2003;1:627–638. doi: 10.1586/14787210.1.4.627. [DOI] [PubMed] [Google Scholar]
- He B, Lamb RA. Effect of inserting paramyxovirus simian virus 5 gene junctions at the HN/L gene junction: analysis of accumulation of mRNAs transcribed from rescued viable viruses. J. Virol. 1999;73:6228–6234. doi: 10.1128/jvi.73.8.6228-6234.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Paterson RG, Stock N, Durbin JE, Durbin RK, Goodbourn S, Randall RE, Lamb RA. Recovery of paramyxovirus simian virus 5 with a V protein lacking the conserved cysteine-rich domain: the multifunctional V protein blocks both interferon-beta induction and interferon signaling. Virology. 2002;303:15–32. doi: 10.1006/viro.2002.1738. [DOI] [PubMed] [Google Scholar]
- He B, Paterson RG, Ward CD, Lamb RA. Recovery of infectious SV5 from cloned DNA and expression of a foreign gene. Virology. 1997;237:249–260. doi: 10.1006/viro.1997.8801. [DOI] [PubMed] [Google Scholar]
- Hsiung GD. Parainfluenza-5 virus. Infection of man and animal. Prog Med Virol. 1972;14:241–74. [PubMed] [Google Scholar]
- Hull RN, Minner JR, Smith JW. New viral agents recovered from tissue cultures of monkey kidney cells. 1. Origin and properties of cytopathic agents S.V.1, S.V.2, S.V.4, S.V.5, S.V.6, S.V.11, S.V.12, and S.V.15. Am. J. Hyg. 1956;63:204–215. doi: 10.1093/oxfordjournals.aje.a119804. [DOI] [PubMed] [Google Scholar]
- Ito T, Suzuki Y, Mitnaul L, Vines A, Kida H, Kawaoka Y. Receptor specificity of influenza A viruses correlates with the agglutination of erythrocytes from different animal species. Virology. 1997;227:493–499. doi: 10.1006/viro.1996.8323. [DOI] [PubMed] [Google Scholar]
- Jin H, Subbarao K, Bagai S, Leser GP, Murphy BR, Lamb RA. Palmitylation of the influenza virus hemagglutinin (H3) is not essential for virus assembly or infectivity. J. Virol. 1996;70:1406–1414. doi: 10.1128/jvi.70.3.1406-1414.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komada H, Klippmark E, Orvell C, Randall RE, Ito Y, Norrby E. Immunological relationships between parainfluenza virus type 4 and other paramyxoviruses studied by use of monoclonal antibodies. Arch Virol. 1991;116:277–283. doi: 10.1007/BF01319249. [DOI] [PubMed] [Google Scholar]
- Lamb RA, Kolakofsky D. Paramyxoviridae: The viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology (Fourth Edition) Lippincott, Williams and Wilkins; Philadelphia: 2001. [Google Scholar]
- Lee ES, Zhou H, Henderson AJ. Endothelial cells enhance human immunodeficiency virus type 1 replication in macrophages through a C/EBP-dependent mechanism. J Virol. 2001;75:9703–9712. doi: 10.1128/JVI.75.20.9703-9712.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin GY, Lamb RA. The paramyxovirus simian virus 5 V protein slows progression of the cell cycle. J Virol. 2000;74:9152–9166. doi: 10.1128/jvi.74.19.9152-9166.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Bright AC, Rothermel TA, He B. Induction of apoptosis by paramyxovirus simian virus 5 lacking a small hydrophobic gene. J Virol. 2003;77:3371–3383. doi: 10.1128/JVI.77.6.3371-3383.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Horvath F, Aligo JA, Wilson R, He B. The role of simian virus 5 V protein on viral RNA synthesis. Virology. 2005;338:270–280. doi: 10.1016/j.virol.2005.05.014. [DOI] [PubMed] [Google Scholar]
- McCandlish IA, Thompson H, Cornwell HJ, Wright NG. A study of dogs with kennel cough. Vet. Rec. 1978;102:293–301. doi: 10.1136/vr.102.14.293. [DOI] [PubMed] [Google Scholar]
- Murphy SK, Parks GD. Genome nucleotide lengths that are divisible by six are not essential but enhance replication of defective interfering RNAs of the paramyxovirus simian virus 5. Virology. 1997;232:145–157. doi: 10.1006/viro.1997.8530. [DOI] [PubMed] [Google Scholar]
- Nakaya T, Cros J, Park MS, Nakaya Y, Zheng H, Sagrera A, Villar E, Garcia-Sastre A, Palese P. Recombinant Newcastle disease virus as a vaccine vector. J Virol. 2001;75:11868–11873. doi: 10.1128/JVI.75.23.11868-11873.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann G, Whitt MA, Kawaoka Y. A decade after the generation of a negative-sense RNA virus from cloned cDNA - what have we learned? J Gen Virol. 2002;83:2635–2362. doi: 10.1099/0022-1317-83-11-2635. [DOI] [PubMed] [Google Scholar]
- Palese P, Zheng H, Engelhardt OG, Pleschka S, Garcia-Sastre A. Negative-strand RNA viruses: genetic engineering and applications. Proc. Natl. Acad. Sci. USA. 1996;93:11354–11358. doi: 10.1073/pnas.93.21.11354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MS, Steel J, Garcia-Sastre A, Swayne D, Palese P. From the Cover: Engineered viral vaccine constructs with dual specificity: Avian influenza and Newcastle disease. Proc Natl Acad Sci U S A. 2006;103:8203–8208. doi: 10.1073/pnas.0602566103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson RG, Lamb RA. The molecular biology of influenza viruses and paramyxoviruses. In: Davidson A, Elliott RM, editors. Molecular Virology: A Practical Approach. IRL Oxford University Press; Oxford: 1993. pp. 35–73. [Google Scholar]
- Paterson RG, Russell CJ, Lamb RA. Fusion protein of the paramyxovirus SV5: destabilizing and stabilizing mutants of fusion activation. Virology. 2000;270:17–30. doi: 10.1006/viro.2000.0267. [DOI] [PubMed] [Google Scholar]
- Pekosz A, He B, Lamb RA. Reverse genetics of negative-strand RNA viruses: closing the circle. Proc. Natl. Acad. Sci. USA. 1999;96:8804–8806. doi: 10.1073/pnas.96.16.8804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peluso RW, Lamb RA, Choppin PW. Polypeptide synthesis in simian virus 5-infected cells. J. Virol. 1977;23:177–187. doi: 10.1128/jvi.23.1.177-187.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall RE, Young DF. Comparison between parainfluenza virus type 2 and simian virus 5: monoclonal antibodies reveal major antigenic differences. J Gen Virol. 1988;69:2051–2060. doi: 10.1099/0022-1317-69-8-2051. [DOI] [PubMed] [Google Scholar]
- Roberts A, Kretzschmar E, Perkins AS, Forman J, Price R, Buonocore L, Kawaoka Y, Rose JK. Vaccination with a recombinant vesicular stomatitis virus expressing an influenza virus hemagglutinin provides complete protection from influenza virus challenge. J. Virol. 1998;72:4704–4711. doi: 10.1128/jvi.72.6.4704-4711.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, Rose JK. Recovery of negative-strand RNA viruses from plasmid DNAs: a positive approach revitalizes a negative field. Virology. 1998;247:1–6. doi: 10.1006/viro.1998.9250. [DOI] [PubMed] [Google Scholar]
- Rosenberg FJ, Lief FS, Todd JD, Reif JS. Studies of canine respiratory viruses. I. Experimental infection of dogs with an SV5-like canine parainfluenza agent. Am J Epidemiol. 1971;94:147–165. doi: 10.1093/oxfordjournals.aje.a121307. [DOI] [PubMed] [Google Scholar]
- Schmitt AP, He B, Lamb RA. Involvement of the cytoplasmic domain of the hemagglutinin-neuraminidase protein in assembly of the paramyxovirus simian virus 5. J. Virol. 1999;73:8703–8712. doi: 10.1128/jvi.73.10.8703-8712.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt AP, Leser GP, Morita E, Sundquist WI, Lamb RA. Evidence for a New Viral Late-Domain Core Sequence, FPIV, Necessary for Budding of a Paramyxovirus. J. Virol. 2005;79(5):2988–2997. doi: 10.1128/JVI.79.5.2988-2997.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell MJ, Mebatsion T, Conzelmann K-K. Infectious rabies viruses from cloned cDNA. EMBO J. 1994;13:4195–4203. doi: 10.1002/j.1460-2075.1994.tb06739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Rothermel TA, Shuman L, Aligo JA, Xu S, Lin Y, Lamb RA, He B. Conserved cysteine-rich domain of paramyxovirus simian virus 5 V protein plays an important role in blocking apoptosis. J Virol. 2004;78:5068–5078. doi: 10.1128/JVI.78.10.5068-5078.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokusumi T, Iida A, Hirata T, Kato A, Nagai Y, Hasegawa M. Recombinant Sendai viruses expressing different levels of a foreign reporter gene. Virus Res. 2002;86:33–38. doi: 10.1016/s0168-1702(02)00047-3. [DOI] [PubMed] [Google Scholar]
- Tribe GW. An investigation of the incidence, epidemiology and control of Simian virus 5. Br J Exp Pathol. 1966;47:472–479. [PMC free article] [PubMed] [Google Scholar]
- Tsurudome M, Nishio M, Komada H, Bando H, Ito Y. Extensive antigenic diversity among human parainfluenza type 2 virus isolates and immunological relationships among paramyxoviruses revealed by monoclonal antibodies. Virology. 1989;171:38–48. doi: 10.1016/0042-6822(89)90508-4. [DOI] [PubMed] [Google Scholar]
- Vandvik B, Norrby E. Paramyxovirus SV5 and multiple sclerosis. Nature. 1989;338:769–771. doi: 10.1038/338769a0. [DOI] [PubMed] [Google Scholar]
- Veits J, Wiesner D, Fuchs W, Hoffmann B, Granzow H, Starick E, Mundt E, Schirrmeier H, Mebatsion T, Mettenleiter TC, Romer-Oberdorfer A. Newcastle disease virus expressing H5 hemagglutinin gene protects chickens against Newcastle disease and avian influenza. Proc Natl Acad Sci U S A. 2006;103:8197–8202. doi: 10.1073/pnas.0602461103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RL, Fuentes SM, Wang P, Taddeo EC, Klatt A, Henderson AJ, He B. Function of Small Hydrophobic Proteins of Paramyxovirus. J Virol. 2006;80:1700–1709. doi: 10.1128/JVI.80.4.1700-1709.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young DF, Randall RE, Hoyle JA, Souberbielle BE. Clearance of a persistent paramyxovirus infection is mediated by cellular immune responses but not by serum-neutralizing antibody. J Virol. 1990;64:5403–5411. doi: 10.1128/jvi.64.11.5403-5411.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan P, Thompson TB, Wurzburg BA, Paterson RG, Lamb RA, Jardetzky TS. Structural Studies of the Parainfluenza Virus 5 Hemagglutinin-Neuraminidase Tetramer in Complex with Its Receptor, Sialyllactose. Structure. 2005;13:803–815. doi: 10.1016/j.str.2005.02.019. [DOI] [PubMed] [Google Scholar]
- Zakstelskaya LY, Zhdanov VM, Yakhno MA, Gushchin BV, Klimenko SM, Demidova SA, Konovalova NG, Gushchina EA. Persistent SV5 virus infection in continuous cell cultures. Acta Virol. 1976;20:506–511. [PubMed] [Google Scholar]