Abstract
Osteoporosis is a common disease with a strong genetic component. We previously described a polymorphic Sp1 binding site in the COL1A1 gene that has been associated with osteoporosis in several populations. Here we explore the molecular mechanisms underlying this association. A meta-analysis showed significant associations between COL1A1 “s” alleles and bone mineral density (BMD), body mass index (BMI), and osteoporotic fractures. The association with fracture was stronger than expected on the basis of the observed differences in BMD and BMI, suggesting an additional effect on bone strength. Gel shift assays showed increased binding affinity of the “s” allele for Sp1 protein, and primary RNA transcripts derived from the “s” allele were approximately three times more abundant than “S” allele–derived transcripts in “Ss” heterozygotes. Collagen produced from osteoblasts cultured from “Ss” heterozygotes had an increased ratio of α1(I) protein relative to α2(I), and this was accompanied by an increased ratio of COL1A1 mRNA relative to COL1A2. Finally, the yield strength of bone derived from “Ss” individuals was reduced when compared with bone derived from “SS” subjects. We conclude that the COL1A1 Sp1 polymorphism is a functional genetic variant that predisposes to osteoporosis by complex mechanisms involving changes in bone mass and bone quality.
Introduction
Osteoporosis is a common disease characterized by reduced bone mass, microarchitectural deterioration of bone tissue, and an increased risk of fragility fractures (1). Genetic factors play an important role in the pathogenesis of osteoporosis by complex mechanisms involving variation in several genes that regulate bone mineral density (BMD) and bone geometry and quality (2). One of the most important candidate genes for predisposition to osteoporosis is the COL1A1 gene, which encodes the α1(I) protein chain of type I collagen, the major protein of bone. We previously identified a single nucleotide G→T polymorphism affecting a binding site for the transcription factor Sp1 in the COL1A1 gene (3). This polymorphism has been associated with low BMD and an increased risk of osteoporotic fracture in several studies (3–13). Haplotype analysis has demonstrated that the association with fracture is primarily driven by allelic variation at the Sp1 site, rather than other polymorphic sites in the COL1A1 gene (12). Although COL1A1 alleles have been found by some workers to predict fractures after correcting for BMD (6, 8, 9), other investigators have found no association between COL1A1 genotypes and BMD or osteoporotic fractures (14–17). The polymorphic Sp1 site lies within the first intron of the COL1A1 gene in a region that has previously been shown to be important for regulation of collagen transcription (18). However, the effects of the polymorphism on COL1A1 gene regulation and protein production remain unclear. In this study we have explored the mechanisms that underlie the association between COL1A1 alleles and osteoporotic fractures by a meta-analysis of published work and functional studies in which we analyzed the relationship between COL1A1 genotype and DNA-protein binding, allele-specific transcription, collagen mRNA and protein production, and the biomechanical properties of bone.
Methods
Meta-analysis of clinical studies.
Clinical studies in which the COL1A1 Sp1 polymorphism had been related to BMD or osteoporotic fracture in adults were identified by electronic searches of MEDLINE between October 1996 and October 2000. Sixteen eligible studies were identified (3–17, 19). For quantitative variables, standardized mean differences between genotypes (equivalent to SD units or Z-scores) were calculated from source data using a fixed effects model. Peto odds ratios were similarly calculated for categorical variables by calculating the number of events in each genotype group. When relevant data were unavailable in the source publications, they was obtained from the corresponding author. Funnel plots (20) were used to detect evidence of possible bias resulting from selective publication of positive studies.
Patient samples.
Functional studies were performed using samples of bone tissue obtained from patients undergoing routine orthopedic surgical procedures. Studies of allele-specific transcription were performed using RNA extracted from bone samples obtained at transiliac biopsy (n = 6) or from osteoblasts cultured from femoral heads obtained during routine orthopedic surgical procedures (n = 4) in 7 female and 3 male patients of mean ± SD age of 68 ± 11.2 years. Studies of collagen protein production and RNA expression were performed on primary osteoblasts cultured from samples of trabecular bone obtained from the femoral heads of 17 female and 11 male patients undergoing surgery for hip fracture (n = 13) or osteoarthritis (n = 15). The mean ± SD age of these patients was 79 ± 12.2 years. Biomechanical studies were performed on trabecular bone samples from femoral heads obtained from 17 females and 6 males of mean ± SD age 76.3 ± 8.4 years. In 17 cases, bone samples were obtained from femoral heads removed from patients undergoing hip replacement surgery as a result of osteoporotic fracture. In the remaining six cases, samples were obtained at post-mortem from individuals with no known history of bone disease who had died suddenly. None of the patients included in any of these studies had received medication known to affect calcium metabolism such as corticosteroids, bisphosphonates, calcitonin or active metabolites of vitamin D. The study was approved by the local hospital ethical committee, and all patients or their relatives gave informed consent to tissue samples being included.
Electrophoretic mobility shift assays.
Sense and antisense oligonucleotides corresponding to the “S” allele (AGGGAATGGGGGCGGGATGAGGGCCT) and the “s” allele (AGGGAATGTGGGCGGGATGAGGGCCT) (sense strand shown, Sp1 binding site underlined, and polymorphic base in boldface type) were synthesized, annealed, and end-labeled with γ32P-ATP (Amersham Pharmacia Biotech UK Ltd., Buckinghamshire, United Kingdom) using T4 Polynucleotide Kinase (Promega Corp., Madison, Wisconsin, USA). The labeled oligonucleotides (0.04 μM) were incubated at 30°C for 30 minutes with human recombinant Sp1 (hrSp1; 10 ng; Promega Corp.); BSA (20 μg; Promega Corp.); and varying concentrations of either “S” or “s” unlabeled competitor oligonucleotides. In some experiments, anti Sp1 antibody (2 μg; Santa Cruz Biotechnology, Santa Cruz, California, USA) was used to confirm specificity of binding. The samples were analyzed on a 4% polyacrylamide gel and visualized by Bio-Rad Personal Molecular Imager, and band intensity was quantitated by Quantity One software (both, Bio-Rad Laboratories Inc., Hercules, California, USA).
Allele-specific transcription.
Allele specific transcription was assessed by a semi-nested RT-PCR assay designed to detect relative abundance of unspliced nuclear RNA derived from each allele, based on analysis of an RFLP created by the polymorphic Sp1 site in intron 1 (Figure 3a). Total RNA was extracted from transiliac bone samples or primary human osteoblasts obtained from patients who were heterozygous for the polymorphism using an acid phenol-guanidinium thiocyanate–based method as described previously (21). Complementary DNA (cDNA) was prepared by reverse-transcribing 5 μg total RNA from each patient using SuperScript RNaseH̄ RT (200 U; Life Technologies Ltd., Paisley, United Kingdom) with random hexamer primers according to the manufacturers protocol. The first round of the semi-nested PCR was carried out with the following primer set: 5′- TAACTTCTGGACTATTTGCGGACTTTTTGG –3′ (p1) and 5′– GGGCGAGGGAGGAGAGAA –3′ (p3), which amplifies a 283-bp region surrounding the polymorphic site in the first intron of COL1A1. The thermal cycling protocol was: 95°C for 2 minutes, 58°C for 1 minute, 72°C for 1 minute (one cycle), 95°C for 50 seconds, 58°C for 1 minute, 72°C for 1 minute (13 cycles); 95°C for 50 seconds, 58°C for 1 minute, 72°C for 3 minutes (one cycle). Undiluted products from this reaction were used as template in a second-round PCR using a fluorescently labeled forward primer of the same sequence as in the first round and the following reverse primer: 5′– GTCCAGCCCTCATCCTGGCC – 3′ (p2), which introduces a restriction site for the enzyme MscI in products derived from the “s” allele (3). The second-round PCR used the following thermal cycling protocol: 94°C for 3 minutes, 62°C for 10 seconds, ramping at 1°C per 10 seconds to 72°C, 72°C for 15 seconds (one cycle); 94°C for 50 seconds, 62°C for 10 seconds, ramping 1°C per 10 seconds to 72°C, 72°C for 15 seconds (26 cycles); 94°C for 50 seconds, 62°C for 10 seconds, ramping 1°C per 10 seconds to 72°C, 72°C for 5 minutes (one cycle). Allele-specific transcripts were detected by digesting the products of the second-round PCR with the restriction enzyme MscI and quantitated by electrophoresis using an ABI 377 sequencer and GeneScan software (Applied Biosystems, Warrington, United Kingdom). Results were expressed as the ratio of “s” allele product abundance to “S” allele product abundance. In each assay, a sample of genomic DNA and non–reverse-transcribed RNA from the same patient was amplified along with the sample of cDNA to control for possible allele-specific differences in the efficiency of the PCR and contamination of cDNA with genomic DNA. Extensive experiments were conducted to validate the assay, including spiking experiments in which samples of target DNA corresponding to each allele were mixed in known amounts and product abundance assessed, and varying cycle number in the second round of the PCR to ensure that product abundance lay within the linear phase of PCR amplification under the conditions chosen. These experiments showed that the ratio of allele-specific products accurately reflected the relative abundance of target molecules under the conditions chosen (data not shown).
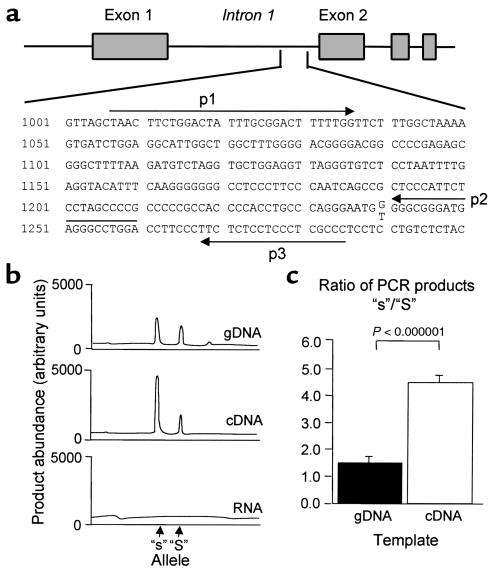
Figure 3.
Effect of the COL1A1 polymorphism on allele-specific transcription in heterozygotes. (a) Position of the primers designed to detect the polymorphic site at position 1240 in the first intron of the human COL1A1 gene (Sequence Accession number M20789). (b) Representative electropherograms from GeneScan analysis of RFLP-PCR assays from one patient. Equivalent amounts of genomic DNA (gDNA; top), cDNA (middle), and non–reverse-transcribed RNA (bottom) were used as PCR template. The increased abundance of “s” allele–derived transcripts is reflected by the greater area under the curve of the “s”-derived PCR products on RFLP-PCR analysis of cDNA. (c) Mean ± SEM ratios of product abundance derived from the “s” and “S” alleles for gDNA. Ratios were measured in duplicate samples of gDNA and cDNA from ten patients.
Collagen protein production.
Collagen protein production was assessed using primary cultures of human osteoblasts obtained from patients undergoing joint replacement surgery as described previously (22). The cells were grown to confluence and pulse labeled for 4 hours with 14C-proline (Amersham Pharmacia Biotech UK Ltd.) (2 μCi/ml) in the presence of 50 μg/ml ascorbic acid. After 24 hours, the conditioned medium was removed, protease inhibitors added (100 μM PMSF, 5 mM EGTA, 2 μg/ml leupeptin) and proteins were precipitated by the addition of ammonium sulfate (176 mg/ml) with slow stirring for 16 hours at 4°C. Cell layer proteins were extracted into 0.5 M acetic acid (pH 2.0). Proteins derived from the cell layer and conditioned medium were combined and digested with pepsin 1 mg/ml overnight at 4°C with stirring to degrade noncollagenous proteins and remove nonhelical portions of collagen. The reaction was terminated by neutralization of samples to pH 7.0, and samples were then extensively dialyzed against 0.1 M acetic acid before lyophilization. Collagen samples from each culture were resolved on SDS-PAGE in triplicate using 4% stacking gel and 6% separating gel and α1(I) and α2(I) chains quantified using Packard Instant Phosphorimaging system (Packard Instrument Co., Meriden, Connecticut, USA).
RNase protection assay.
Steady-state mRNA levels were assessed by RNase protection assay (RPA) using the RPA III kit (Ambion Inc., Austin, Texas, USA). For these experiments, a 267-bp fragment of human COL1A1 cDNA-spanning exons 1 and 2 (Sequence Accession number Z74615 bases 83–350) and a 495-bp fragment of human COL1A2 cDNA spanning exons 25-32 (Sequence Accession number J03464 bases 2001–2496) were cloned into pGEM T-easy vectors (Promega Corp.). As an internal control, we used a 170-bp fragment of the human β-actin cDNA spanning exon 1 (Sequence Accession number X63432 bases 545–715) cloned into the same vector. Antisense transcripts labeled with [α-32P]UTP were generated from each clone with SP6 RNA polymerase using Riboprobe in vitro transcription system (Promega Corp.). Patient samples (10 μg RNA) were hybridized in a single tube with COL1A1, COL1A2, and β-actin probes. Protected fragments were separated on a 5% denaturing polyacrylamide gel and quantified using by a Packard Instant Phosphorimager. The ratio of COL1A1/COL1A2 transcripts was then calculated after correcting for β-actin expression.
Bone composition and biomechanical studies.
Cylindrical cores of bone were removed from the superior, posterior, medial, and central sites in the femoral head using a 9-mm-diameter hollow drill bit as described previously (23). These sites were chosen to represent regions subjected to a range of loading conditions in vivo (24). Articular cartilage and the subchondral bone plate were removed from the bone cores, and both ends were trimmed parallel. The resulting cores of trabecular bone, having a mean height 7.7 ± 1.6 mm, were subjected to an unconstrained compression test using an Instron 5564 materials testing machine (Instron, High Wycombe, United Kingdom). The test was performed at a strain rate of 20% per minute (0.0033%/second). The apparent density of washed and defatted bone samples was calculated by dividing the total mass by the sample volume, calculated from caliper measurements of diameter and length. The stiffness, yield stress, and energy to yield were determined from the characteristics of the stress-strain curve using standard techniques described previously (23). Bone composition was determined by dehydrating the samples at 105°C for 48 hours followed by ashing at 600°C for 24 hours. Water content was defined as the difference between wet and dry masses; mineral content by the final mass after ashing, and organic content by the difference between the dry mass and the ash mass. In our laboratory, the precision of bone composition measurements is less than 0.1% (25).
Statistical analysis.
Data from the meta-analysis were analyzed using the RevMan 4.1 software package obtained from the Cochrane Collaboration (The Nordic Cochrane Centre, Rigshospitalet, Copenhagen, Denmark; http://www.cochrane.dk). Odds ratios and standardized mean differences were calculated under fixed effects and random effects models, with essentially similar results; because of this, data are presented for the fixed effects models only. Funnel plots (20) were made to check for evidence of publication bias. Statistical analysis of EMSA data was carried out using GraphPad Prism software (GraphPad Software Inc., San Diego, California, USA), assuming a one-site competitor binding model and nonlinear regression analysis. Statistical analysis of all other data was carried out using Minitab version 12 (Minitab Inc., State College, Pennsylvania, USA). Between-genotype differences in allele-specific transcription, protein levels, mRNA levels, and bone composition were assessed by Student’s t test. Analysis of genotype-specific differences in yield strength was by general linear model (GLM) ANOVA using sampling site and genotype as grouping variables and sample density as a covariate. The influences of age, sex, diagnosis, and genotype on other data resulting from in vitro experiments was assessed by GLM ANOVA by entering sex and diagnosis as grouping variables and age as a covariate.
Results
Meta-analysis of clinical studies.
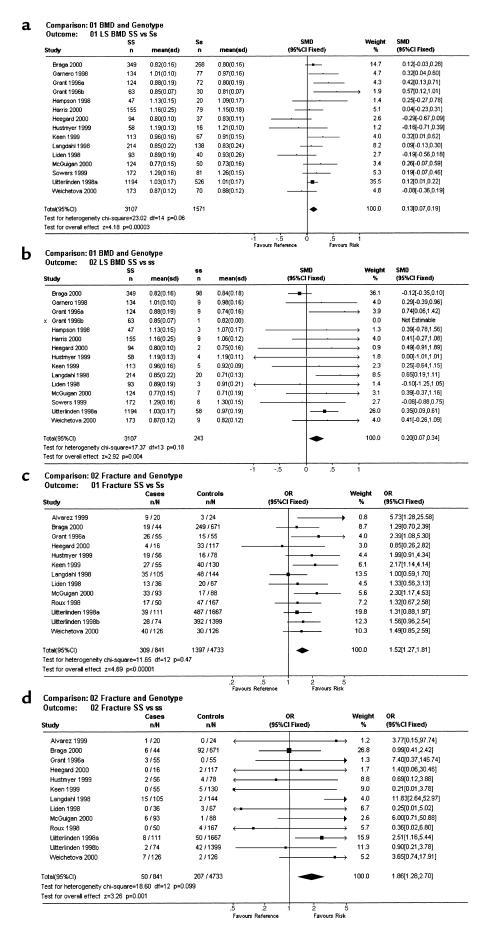
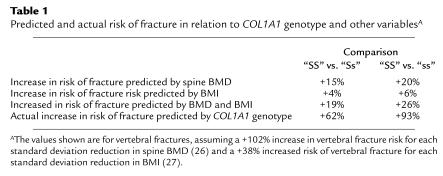
The meta-analysis was conducted using data from 16 studies published between October 1996 and October 2000 (3–17, 19). At the lumbar spine, “Ss” heterozygotes (n = 1,571) had BMD values of 0.13 (95% confidence interval, 0.07–0.19) Z-score units lower than did “SS” individuals (n = 3,107) (P = 0.0003) (Figure 1a). The differences were greater still for the “SS” (n = 3,107) versus “ss” (n = 243) comparison: 0.20 (0.07–0.34) (P = 0.004) (Figure 1b). Analysis of femoral neck BMD values in relation to COL1A1 genotype (data from individual studies not shown) showed similar results with values of 0.10 (0.04–0.17) for the “SS” versus “Ss” comparison (P = 0.0008) and 0.27 (0.13–0.40) for the “SS” versus “ss” comparison (P = 0.0001). In five of the studies, data were available for a total of 3,312 patients (3, 7, 8, 13, 19) on BMD values that had been adjusted for age, body mass index (BMI), and other confounding variables in the source publications. A meta-analysis of adjusted BMD values from these studies showed an effect size very similar to that of the unadjusted BMD data. Thus, for spine BMD values, the differences were “SS” versus “Ss” = 0.14 (0.06–0.21) (P = 0.003), “SS” versus “ss” = 0.19 (0.04–0.35) (P = 0.02); and for femoral neck BMD the differences were “SS” versus “Ss” = 0.12 (0.04–0.19) (P = 0.002) and “SS” versus “ss” = 0.23 (0.08–0.39) (P = 0.003). Analysis of 12 studies that had included information on fractures showed a highly significant association between COL1A1 genotype and fracture, with an odds ratios of 1.52 (1.27–1.81) for the “SS” versus “Ss” comparison (Figure 1c) (P < 0.0001) and 1.86 (1.28–2.70) for the “SS” versus “ss” comparison (Figure 1d) (P = 0.001). Analysis of vertebral fractures only (ten studies including 560 fractures and 2,269 controls) showed an odds ratio of 1.62 (1.30–2.02) for the “SS” versus “Ss” comparison (P = 0.00002) and 1.93 (1.14–3.26) for the “SS” versus “ss” comparison (P = 0.01). A further meta-analysis was conducted for confounding variables known to influence bone density and susceptibility to osteoporotic fracture, including smoking, dietary calcium intake, age, age at menopause, and BMI. This only difference observed was for BMI, which was 0.10 (0.03–0.16) SDs lower in “Ss” individuals when compared with “SS” (P = 0.004) and 0.17 (0.01–0.32) SDs lower in “ss” individuals (P = 0.03). Funnel plots (20) of the SE of individuals studies versus effect size (standardized mean difference for BMD and odds ratio for fracture) were symmetrical, which is evidence against publication bias as an explanation for the findings observed (data not shown). To assess whether the genotype-specific reduction in BMD and BMI was sufficient to explain the increase in fracture risk associated with the COL1A1 polymorphism, we compared the actual increase in fracture risk in the “Ss” and “ss” genotype groups with that which would have been predicted on the basis of the differences in BMD and BMI. The calculations were done for spine BMD in predicting vertebral fractures, as these were by far the most common fractures represented in the meta-analysis. Previous work has shown that the odds ratio vertebral fracture is 2.02 for each SD reduction in spine BMD, representing an increase in risk of 102% per SD (26) and 1.38 for each reduction in BMI (27), representing a 38% increase per SD. Table 1 shows that the predicted increase in risk of vertebral fracture associated with reduced BMD and BMI ranged from 19% with the “Ss” genotype to 26% with the “ss” genotype. The predicted increase in fracture risk associated with these difference in BMD and BMI was insufficient to explain the observed increase in fracture risk associated with COL1A1 alleles, as patients with the “Ss” genotype had a 62% increased incidence of vertebral fractures when compared with “SS” subjects, and those with the “ss” genotype had a 93% increase.
Figure 1.
Meta-analysis of COL1A1 polymorphisms in relation to BMD and osteoporotic fractures. (a) Standardized mean difference (SMD) and 95% confidence intervals between the “SS” and “Ss” genotype groups for lumbar spine bone density (LS BMD). (b) Corresponding data for the “SS” versus “ss” genotype comparison. Also shown are the number of individuals studied, the mean and SD BMD values in each genotype group, and the weight and confidence intervals for individual studies. The results of heterogeneity testing and tests for overall effect are shown at the bottom left of each panel. Note that data from both populations studied by Grant et al. (3) are shown separately as Grant 1996a and Grant 1996b. (c) Odds ratios for fracture for the “SS” versus “Ss” genotype comparison. (d) Corresponding data for the “SS” versus “ss” comparison. Numbers in the cases and controls columns refer to the number of individuals with the risk genotype (“Ss” or “ss”) in relation to the total number of individuals studied. In the study of Uitterlinden et al. (8), data on both vertebral and nonvertebral fractures were available, and these are shown respectively as Uitterlinden 1998a and Uitterlinden 1998b.
Table 1.
Predicted and actual risk of fracture in relation to COL1A1 genotype and other variablesA
Effects of Sp1 polymorphism on DNA-protein binding affinity.
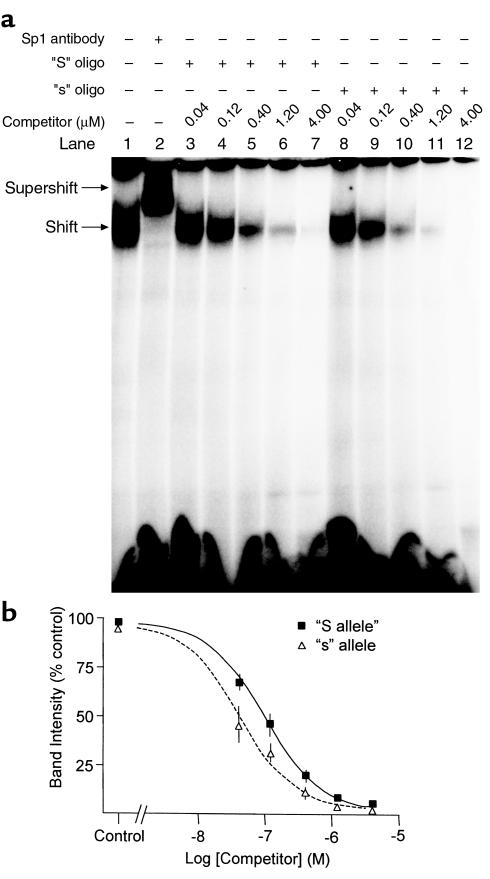
Binding affinity of the Sp1 protein for the polymorphic DNA recognition site in COL1A1 was analyzed using gel shift assays (Figure 2). The unlabeled “s” competitor bound Sp1 with greater affinity than did the unlabeled “S” competitor as demonstrated by the reduced intensity of the radiolabeled Sp1-DNA complex at equivalent competitor concentrations. The concentrations at which 50% inhibition of band intensity occurred (pIC50) were significantly different for the “S” (pIC50 = 0.095 μM) and the “s” (pIC50 = 0.039 μM) oligonucleotides (P < 0.001). Similar differences in DNA-protein binding affinity were found when the radiolabeled “s” allele was used as the probe instead of the “S” allele and when nuclear protein extracts derived from MG63 osteoblast-like cells were used in place of hrSp1 (data not shown).
Figure 2.
(a) Typical gel shift assay with 32P-labeled “S” oligonucleotide and hrSp1 protein (all lanes), illustrating the DNA-Sp1 complex (shift) and the supershifted complex (supershift) created by incubating the hrSp1 with Sp1 antibody (lane 2). The shift is attenuated more rapidly with increasing concentrations of the unlabeled “s” oligonucleotide (lanes 8–12) compared with the unlabeled “S” oligonucleotide (lanes 3–7). (b) Analysis of Sp1 binding affinity. Band intensity of the shift in lanes containing competitor oligonucleotides was expressed as a percentage of the band intensity in the absence of competitor (control, lane 1) and analyzed by GraphPad Prism. Data points are means ± SEM from three experiments.
Abundance of allele-specific transcripts in heterozygotes.
To examine whether the differences seen in the DNA-protein binding assays were accompanied by alterations in COL1A1 transcription, we studied transcript abundance in “Ss” heterozygotes using the polymorphic Sp1 site to detect allele-specific transcripts in unspliced RNA by a semi-nested RFLP-PCR assay (Figure 3a). The abundance of product derived from the “s” allele was significantly greater than that derived from the “S” allele–derived product for ten heterozygous patients in whom it was studied (representative electropherogram, Figure 3b, middle). Analysis of genomic DNA from the same patients showed that the “s”-derived product was also slightly more abundant than the “S”-derived product probably because of allele-specific differences in PCR amplification efficiency (representative electropherogram Figure 3b, top). Even after correcting for the slight allele-specific differences observed in genomic DNA, there remained a highly significant difference in product abundance between the “s” and “S” alleles in cDNA (P < 0.000001) (Figure 3c). There was no significant difference in allele-specific product abundance in samples derived from men or women, or in samples derived from transiliac biopsies compared with those derived from osteoblast cultures (data not shown). These data indicate that increased binding affinity of the “s” allele for Sp1 protein is accompanied by a relative increase in abundance of “s”-derived allele-specific transcripts in heterozygotes.
Effects of Sp1 polymorphism on collagen mRNA and protein production.
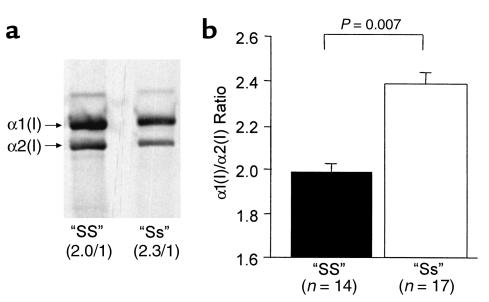
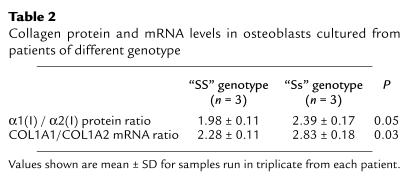
The effect of the polymorphism on collagen protein production was studied using cultured primary human osteoblasts from patients of different genotype (Figure 4). Quantitative analyses of type I collagen produced by osteoblasts derived from “SS” homozygotes showed values close to the expected 2:1 ratio of α1(I) to α2(I) collagen protein chains (1.99 ± 0.07 to 1). Osteoblasts derived from “Ss” heterozygotes produced an increased ratio of α1(I) to α2(I) chains (2.36 ± 0.10 to 1), and this was significantly different from the ratio in “SS” homozygotes (P = 0.007). These data indicate that type I collagen produced by osteoblasts derived from “Ss” heterozygotes contains an increased amount of the collagen α1(I) chain relative to the α2(I) chain. Given that the osteoblast cultures were derived from patients of differing age, sex, and underlying diagnosis, we performed GLM ANOVA to determine whether any of these confounding factors may have influenced the results. This showed that of all these factors, only genotype was a significant predictor of α1(I) to α2(I) ratio (P = 0.016). The increased ratio of α1(I) to α2(I) raises the possibility that some of the collagen may have been present in the form of homotrimers composed of three collagen α1(I) chains ([α1(I)3]) (28), as the method of protein preparation means that only trimeric collagen molecules are loaded onto the gel. We attempted to detect intact [α1(I)3] in these samples by various techniques including differential salt precipitation (29), ion-exchange chromatography (28), and thermal stability (30); however, the results were inconclusive, possibly because there were limited amounts of material available for analysis, and only a small proportion of the collagen, calculated to be approximately 13%, was likely to be present in the form of [α1(I)3]. To determine whether the altered ratio of α1(I) to α2(I) collagen protein chains in “Ss” heterozygotes was accompanied by differences in mRNA abundance, we performed RNase protection assays in six samples in which both RNA and protein were available for analysis. The results of these experiments are summarized in Table 2, which shows that the increase in relative abundance of collagen α1(I) to α2(I) protein chains noted in “Ss” heterozygotes was accompanied by an increase in the relative abundance of COL1A1 to COL1A2 mRNAs.
Figure 4.
Effect of the COL1A1 polymorphism on type I collagen protein production by primary human osteoblasts. (a) Representative samples derived from two patients showing the expected 2:1 ratio of α(I)1/α(I)2 chains in the “SS” homozygote and the increased relative abundance (2.3:1) of collagen α1(I)/α2(I) chains in the “Ss” heterozygote. (b) Cumulative data (mean ± SEM) from 14 “SS” homozygotes and 17 “Ss” heterozygotes.
Table 2.
Collagen protein and mRNA levels in osteoblasts cultured from patients of different genotype
Effects of polymorphism on biomechanical and material properties of bone.
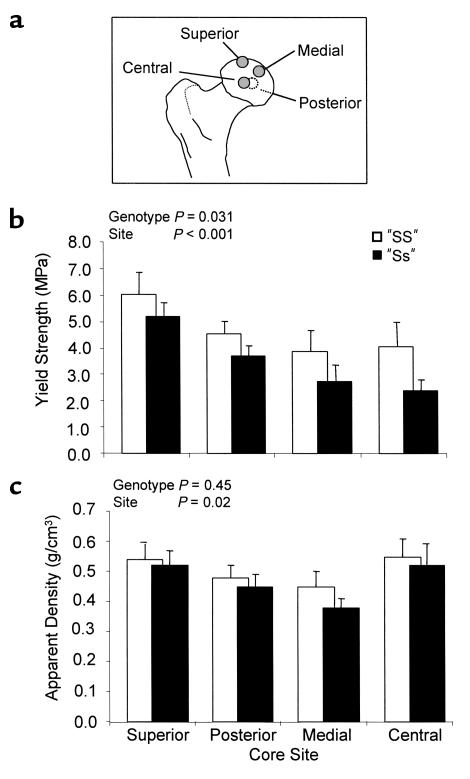
We studied the biomechanical and material properties of bone from patients of different genotype, using trabecular bone cores derived from four sites in the femoral head (Figure 5a). Analysis of the data using GLM ANOVA entering diagnosis, genotype, sex, density of the bone core, and sample site into the model showed significant site-to-site variation in yield strength (P < 0.001); significantly reduced yield strength of “Ss” bone cores compared with “SS” cores (mean ± SD) = 3.74 ± 0.30 vs. 4.53 ± 0.27, P = 0.031 (Figure 5b); and a significant effect of apparent density on yield strength (P < 0.0001; data not shown). There was no significant difference between apparent density of the bone cores according to genotype (“SS” = 0.55 ± 0.02 vs. “Ss” = 0.53 ± 0.02, P = 0.45) (Figure 5c), between water content and genotype (“SS” = 17.1 ± 0.5% vs. “Ss” = 18.1 ± 0.6%, P = 0.14), or between stiffness and genotype (“SS” = 300.1 ± 13.4 vs. “Ss” = 278.3 ± 15.2, P = 0.22). Cores derived from “Ss” heterozygotes did, however, have a significantly reduced inorganic content (“Ss” = 51.7 ± 0.6% vs. “SS” = 54.3 ± 0.6%, P = 0.001) and increased organic content (“Ss” = 30.1 ± 0.4% vs. “SS” = 28.5 ± 0.5%, P = 0.001) when compared with cores from “SS” individuals.
Figure 5.
Relationship between COL1A1 alleles and material properties of bone. Bone samples from various sites in the femoral head (a) derived from “Ss” heterozygotes had significantly reduced yield strength when compared with samples derived from “SS” homozygotes (b), but there was no significant difference in density of the bone cores according to genotype (c). Values are expressed as mean ± SD. MPa, megapascals.
Discussion
In this study, we have tried to address the molecular mechanisms that underlie the associations that have previously been reported between COL1A1 alleles, BMD, and osteoporotic fracture. A meta-analysis of data from 16 published studies including a total of 4,965 individuals showed a significant association between carriage of the COL1A1 “s” allele and low BMD. This was apparent at both the lumbar spine and femoral neck with evidence of an allele-dose effect. The “s” allele was also associated with reduced BMI, but the association between genotype and BMD was still highly significant in a subgroup of five studies in which BMD data had been adjusted for BMI and other confounding factors. This indicates that the association between COL1A1 alleles and BMD is direct and not simply the result of reduced BMI. Publication bias also needs to be considered as a possible explanation for the findings observed. Although it is difficult to absolutely exclude this possibility, funnel plots of BMD values were symmetrical and, as such, did not suggest the presence of publication bias (20).
Osteoporotic fractures were also strongly associated with the COL1A1 “s” allele in the meta-analysis, and the magnitude of risk was too great to be accounted for by the genotype-specific differences in BMD and BMI that we observed. These data support previous work which has shown COL1A1 genotype predicts osteoporotic fractures after correcting for important confounding factors such BMD, BMI, and age (6, 8, 9) and raises the possibility that COL1A1 alleles act as a marker for reduced bone quality. Other variables such as ethnic background and sex may also influence fracture risk, but neither of these can readily be invoked as an explanation for the findings observed. Ethnic differences were excluded by the fact that all studies included in the meta-analysis were conducted in white Caucasian subjects and sex differences were excluded by the fact that only one study included men and, in this study, the male cases were matched with male controls.
Studies of DNA-binding, collagen gene regulation, and the material properties of bone showed clear evidence of functional differences between COL1A1 Sp1 alleles. Gel shift assays showed that the COL1A1 “s” allele had increased affinity for Sp1 protein binding in vitro when compared with the “S” allele, and this was accompanied by increased abundance of “s”-derived transcripts in unspliced RNA extracted from bone samples in individuals who were heterozygous for the polymorphism. These observations suggest either that allele-specific transcription is increased in relation to the osteoporosis-associated “s” allele or that transcripts from the “s” allele are spliced or processed differently within the nucleus. Further experiments will be required to determine which of these mechanisms is responsible for the findings observed.
Analysis of collagen protein production by metabolic labeling showed an altered production of the collagen α1(I) chain relative to the α2(I) chain in osteoblasts derived from “Ss” heterozygotes. The ratio of collagen α1(I) to α2(I) was 2.3 to 1 in “Ss” heterozygotes compared with the expected value of 2 to 1 that was observed in “SS” homozygotes. These differences in ratio of α1(I) to α2(I) protein chains were accompanied by relative overexpression of the COL1A1 mRNA relative to the COL1A2 mRNA in osteoblasts cultured in vitro derived from a subgroup of individuals in which samples of both RNA and protein were available for analysis.
Since only intact trimeric collagen molecules are analyzed by the PAGE technique used in this study, the above observation is consistent with the hypothesis that some of the collagen produced by “Ss” heterozygous individuals is composed of [α1(I)3], although we were unable to formally demonstrate the presence of [α1(I)3], owing to the limited amounts of patient material available (31). Increased ratios of α1(I) relative to α2(I) collagen consistent with [α1(I)3] production have previously been found by other workers in fetal tissues and material derived from tumors and chronic fibrotic conditions (32, 33). The abnormal ratio of collagen α1(I) relative to α2(I) reported here is potentially relevant for the pathogenesis of osteoporotic fractures in the light of previous studies which have shown [α1(I)3] is associated with impaired mechanical strength of bone. The mechanism by which [α1(I)3] impairs bone strength is unclear, but cases of severe osteogenesis imperfecta have been reported in association with inactivating mutations in the COL1A2 gene, which results in production of type I collagen that comprises solely [α1(I)3] (28). A similar phenotype has been observed in the oim/oim mouse, which has a null mutation of the COL1A2 gene and type I collagen composed entirely of [α1(I)3] (32). Mice that are heterozygous for the oim mutation have a milder increase in bone fragility, however (33, 34); and here, the bone collagen comprises a mixture of [α1(I)3] and normal collagen α1(I)2α2(I). Although we have been unable to demonstrate the presence of [α1(I)3] in bone derived from patients who carry the “s” allele, the biomechanical data support the hypothesis that individuals who carry the “s” allele have reduced bone strength independent of differences in BMD. Analysis of bone cores from four separate sites in the femoral head of patients with the “Ss” genotype showed evidence of a modest reduction in yield strength when compared with similar cores derived from “SS” heterozygotes, after correcting for differences in the density of the cores and site specific differences in bone strength. Although we cannot absolutely exclude the possibility that these differences were due to differences in bone trabecular structure, this seems unlikely given the consistency of the observations over four separate sites. We also observed slight differences between genotypes in composition of bone such that “Ss”-derived cores had a reduced inorganic content (mainly reflecting mineral content) and an increased organic content when compared with “SS”-derived cores. Although this may reflect subtle abnormalities of mineralization in the “Ss” genotype, further work with more sensitive techniques such as backscatter electron imaging (35) will be required to confirm this observation.
In summary, the evidence presented here indicates that the COL1A1 Sp1 binding site polymorphism has functional effects on collagen gene regulation that leads to abnormal production of the α1(I) collagen chain relative to α2(I) and reduced bone strength by mechanisms that are partly independent of bone mass. Our studies therefore provide evidence to suggest that the Sp1 polymorphism is a functional genetic variant that predisposes to osteoporotic fractures by mechanisms that involve a reduction in bone quality and quantity.
Acknowledgments
We thank colleagues from the department of Orthopaedic Surgery for assistance with collection of tissue samples; Dan McBride for supplying oim mouse bone as an internal control in assays for collagen [α1(I)3] homotrimer; and G. Hampson, A. Uitterlinden, G. Liden, G. Lucotte, H. Jorgensen, and J. Stepan for providing unpublished data used in the meta-analysis. These studies were supported by grants from the Arthritis Research Council, the Medical Research Council, and the Wellcome Trust. E.E. Hobson is recipient of a Medical Research Council Clinical Fellowship and R.M. Aspden of a Medical Research Council Senior Fellowship.
Footnotes
Val Mann and Emma E. Hobson contributed equally to this work.
References
- 1.Kanis JA, Melton LJ, Christiansen C, Johnston CC, Khaltaev N. The diagnosis of osteoporosis. J Bone Miner Res. 1994;9:1137–1141. doi: 10.1002/jbmr.5650090802. [DOI] [PubMed] [Google Scholar]
- 2.Stewart TL, Ralston SH. Role of genetic factors in the pathogenesis of osteoporosis. J Endocrinol. 2000;166:235–245. doi: 10.1677/joe.0.1660235. [DOI] [PubMed] [Google Scholar]
- 3.Grant SFA, et al. Reduced bone density and osteoporosis associated with a polymorphic Sp1 site in the collagen type I alpha 1 gene. Nat Genet. 1996;14:203–205. doi: 10.1038/ng1096-203. [DOI] [PubMed] [Google Scholar]
- 4.Hampson G, et al. Bone mineral density, collagen type 1 alpha 1 genotypes and bone turnover in premenopausal women with diabetes mellitus. Diabetologia. 1998;41:1314–1320. doi: 10.1007/s001250051071. [DOI] [PubMed] [Google Scholar]
- 5.Roux C, Dougados M, Abel L, Mercier G, Lucotte G. Association of a polymorphism in the collagen I α 1 gene with osteoporosis in French women. Arthritis Rheum. 1998;41:187–188. doi: 10.1002/1529-0131(199801)41:1<187::AID-ART32>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 6.Langdahl BL, Ralston SH, Grant SFA, Eriksen EF. An Sp1 binding site polymorphism in the COLIA1 gene predicts osteoporotic fractures in men and women. J Bone Miner Res. 1998;13:1384–1389. doi: 10.1359/jbmr.1998.13.9.1384. [DOI] [PubMed] [Google Scholar]
- 7.Garnero P, Borel O, Grant SFA, Ralston SH, Delmas PD. Collagen I α 1 polymorphism, bone mass and bone turnover in healthy French pre-menopausal women: the OFELY study. J Bone Miner Res. 1998;13:813–818. doi: 10.1359/jbmr.1998.13.5.813. [DOI] [PubMed] [Google Scholar]
- 8.Uitterlinden AG, et al. Relation of alleles of the collagen type I α 1 gene to bone density and risk of osteoporotic fractures in postmenopausal women. N Engl J Med. 1998;338:1016–1022. doi: 10.1056/NEJM199804093381502. [DOI] [PubMed] [Google Scholar]
- 9.Keen RW, et al. Polymorphism at the type I collagen (COLIA1) locus is associated with reduced bone mineral density, increased fracture risk and increased collagen turnover. Arthritis Rheum. 1999;42:285–290. doi: 10.1002/1529-0131(199902)42:2<285::AID-ANR10>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 10.Alvarez L, et al. Collagen type I alpha1 gene Sp1 polymorphism in premenopausal women with primary osteoporosis: improved detection of Sp1 binding site polymorphism in the collagen type 1 gene. Clin Chem. 1999;45:904–906. [PubMed] [Google Scholar]
- 11.Weichetova M, et al. COLIA1 polymorphism contributes to bone mineral density to assess prevalent wrist fractures. Bone. 2000;26:287–290. doi: 10.1016/s8756-3282(99)00280-x. [DOI] [PubMed] [Google Scholar]
- 12.McGuigan FEA, Reid DM, Ralston SH. Susceptibility to osteoporotic fracture is determined by allelic variation at the Sp1 site, rather than other polymorphic sites, at the COLIA1 locus. Osteoporos Int. 2000;11:338–343. doi: 10.1007/s001980070123. [DOI] [PubMed] [Google Scholar]
- 13.Braga V, et al. Association of CTR and COLIA1 alleles with BMD values in peri- and postmenopausal women. Calcif Tissue Int. 2000;67:361–366. doi: 10.1007/s002230001160. [DOI] [PubMed] [Google Scholar]
- 14.Heegaard A, Jorgensen HL, Vestergaard AW, Hassager C, Ralston SH. Lack of influence of collagen type Ialpha1 Sp1 binding site polymorphism on the rate of bone loss in a cohort of postmenopausal Danish women followed for 18 years. Calcif Tissue Int. 2000;66:409–413. doi: 10.1007/s002230010083. [DOI] [PubMed] [Google Scholar]
- 15.Hustmyer FG, Lui G, Johnston CC, Christian J, Peacock M. Polymorphism at an Sp1 binding site of COLIA1 and bone mineral density in pre-menopausal female twins and elderly fracture patients. Osteoporos Int. 1999;9:346–350. doi: 10.1007/s001980050157. [DOI] [PubMed] [Google Scholar]
- 16.Sowers M, et al. Genetic markers, bone mineral density and serum osteocalcin levels. J Bone Miner Res. 1999;14:1411–1419. doi: 10.1359/jbmr.1999.14.8.1411. [DOI] [PubMed] [Google Scholar]
- 17.Liden M, Wilen B, Ljunghall S, Melhus H. Polymorphism at the Sp 1 binding site in the collagen type I alpha 1 gene does not predict bone mineral density in postmenopausal women in sweden. Calcif Tissue Int. 1998;63:293–295. doi: 10.1007/s002239900529. [DOI] [PubMed] [Google Scholar]
- 18.Bornstein P, McKay J, Morishima JK, Devarayalu S, Gelinas RE. Regulatory elements in the first intron contribute to transcriptional control of the human collagen alpha 1 (I) collagen gene. Proc Natl Acad Sci USA. 1987;84:8869–8873. doi: 10.1073/pnas.84.24.8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harris SS, Patel MS, Cole DE, Dawson-Hughes B. Associations of the collagen type I alpha1 Sp1 polymorphism with five-year rates of bone loss in older adults. Calcif Tissue Int. 2000;66:268–271. doi: 10.1007/pl00005842. [DOI] [PubMed] [Google Scholar]
- 20.Egger M, Davey SG, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. Br Med J. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ralston SH. Analysis of gene expression in human bone biopsies by polymerase chain reaction: evidence for enhanced cytokine expression in postmenopausal osteoporosis. J Bone Miner Res. 1994;9:883–890. doi: 10.1002/jbmr.5650090614. [DOI] [PubMed] [Google Scholar]
- 22.Ralston SH, Todd D, Helfrich M, Benjamin N, Grabowski PS. Human osteoblast-like cells produce nitric oxide and express inducible nitric oxide synthase. Endocrinology. 1994;135:330–336. doi: 10.1210/endo.135.1.7516867. [DOI] [PubMed] [Google Scholar]
- 23.Li B, Aspden RM. Composition and mechanical properties of cancellous bone from the femoral head of patients with osteoporosis or osteoarthritis. J Bone Miner Res. 1997;12:641–651. doi: 10.1359/jbmr.1997.12.4.641. [DOI] [PubMed] [Google Scholar]
- 24.Hodge WA, et al. Contact pressures in the human hip joint measured in vivo. Proc Natl Acad Sci USA. 1986;83:2879–2883. doi: 10.1073/pnas.83.9.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aspden RM, Li B. Reproducibility of techniques using Archimedes’ principle in measuring cancellous bone volume by L. Zou, R. D. Bloebaum and K. N. Bachus. Med Eng Phys. 1998;20:393–395. doi: 10.1016/s1350-4533(98)00057-5. [DOI] [PubMed] [Google Scholar]
- 26.Marshall D, Johnell O, Wedel H. Meta-analysis of how well measures of bone mineral density predict occurrence of osteoporotic fractures. Br Med J. 1996;312:1254–1259. doi: 10.1136/bmj.312.7041.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lunt M, et al. Bone density variation and its effects on risk of vertebral deformity in men and women studied in thirteen European centers: the EVOS Study. J Bone Miner Res. 1997;12:1883–1894. doi: 10.1359/jbmr.1997.12.11.1883. [DOI] [PubMed] [Google Scholar]
- 28.Deak SB, van der Rest M, Prockop DJ. Altered helical structure of a homotrimer of alpha 1(I)chains synthesized by fibroblasts from a variant of osteogenesis imperfecta. Coll Relat Res. 1985;5:305–313. doi: 10.1016/s0174-173x(85)80020-0. [DOI] [PubMed] [Google Scholar]
- 29.Jimenez SA, Bashey RI, Benditt M, Yankowski R. Identification of collagen alpha1(I) trimer in embryonic chick tendons and calvaria. Biochem Biophys Res Commun. 1977;78:1354–1361. doi: 10.1016/0006-291x(77)91441-3. [DOI] [PubMed] [Google Scholar]
- 30.McBride DJ, Jr, Choe V, Shapiro JR, Brodsky B. Altered collagen structure in mouse tail tendon lacking the alpha 2(I) chain. J Mol Biol. 1997;270:275–284. doi: 10.1006/jmbi.1997.1106. [DOI] [PubMed] [Google Scholar]
- 31.Kuo MY, Chen HM, Hahn LJ, Hsieh CC, Chiang CP. Collagen biosynthesis in human oral submucous fibrosis fibroblast cultures. J Dent Res. 1995;74:1783–1788. doi: 10.1177/00220345950740111101. [DOI] [PubMed] [Google Scholar]
- 32.Chipman SD, et al. Defective pro alpha 2(I) collagen synthesis in a recessive mutation in mice: a model of human osteogenesis imperfecta. Proc Natl Acad Sci USA. 1993;90:1701–1705. doi: 10.1073/pnas.90.5.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McBride DJ, Jr, Shapiro JR, Dunn MG. Bone geometry and strength measurements in aging mice with the oim mutation. Calcif Tissue Int. 1998;62:172–176. doi: 10.1007/s002239900412. [DOI] [PubMed] [Google Scholar]
- 34.Saban J, et al. Heterozygous oim mice exhibit a mild form of osteogenesis imperfecta. Bone. 1996;19:575–579. doi: 10.1016/s8756-3282(96)00305-5. [DOI] [PubMed] [Google Scholar]
- 35.Roschger P, Fratzl P, Eschberger J, Klaushofer K. Validation of quantitative backscattered electron imaging for the measurement of mineral density distribution in human bone biopsies. Bone. 1998;23:319–326. doi: 10.1016/s8756-3282(98)00112-4. [DOI] [PubMed] [Google Scholar]