Abstract
Rabex-5 is a guanine nucleotide exchange factor (GEF) for Rab5. Here, we report the identification of a novel functional domain of Rabex-5 that is essential for its membrane targeting and Rab5 GEF activity in vivo. The data show that full-length Rabex-5 efficiently activates Rab5 in the cell. However, the GEF domain itself (residues 135–399) is inactive in this respect, despite its activity in vitro. Generation and characterization of a series of Rabex-5 constructs reveal that the GEF domain is unable to target to early endosomes and that a sequence N-terminal to the GEF domain can restore its early endosomal targeting and its ability to activate Rab5 in the cell. This region (residues 81–135) is termed membrane-binding motif, which together with the downstream helical bundle domain (residues 135–230) forms an early endosomal targeting (EET) domain necessary and sufficient for association with early endosomes. Furthermore, several active Rabex-5 constructs do not contain the Rabaptin-5-binding domain in the C-terminal region. Thus, Rabex-5 can target to early endosomes via the EET domain and activate Rab5 in a Rabaptin-5–independent manner in vivo. We discuss a model to reconcile these in vivo data with previous in vitro results on Rabex-5 function and its interaction with Rabaptin-5.
INTRODUCTION
Rabex-5 is a guanine nucleotide exchange factor (GEF) for Rab5 (Horiuchi et al., 1997), a small GTPase regulating early endosome fusion and endocytosis (Gorvel et al., 1991; Bucci et al., 1992; Li et al., 1994; Hoffenberg et al., 1995; Li and Liang, 2001). Interestingly, Rabex-5 was originally purified as a soluble complex with Rabaptin-5, and immunodepletion of this complex can reduce early endosome fusion in vitro (Horiuchi et al., 1997). Rabex-5 itself shows very little GEF activity in vitro, and it requires interaction with Rabaptin-5 to gain activity (Esters et al., 2001; Lippe et al., 2001). Furthermore, the core GEF domain (Vps9 domain plus the upstream helical bundle and the downstream α-helix, residues 132–391) of Rabex-5 is also much more active than the full-length protein in in vitro biochemical assays (Delprato et al., 2004). It is unclear why the full-length Rabex-5 has such low GEF activity in vitro and how interaction with Rabaptin-5 can stimulate this activity. This could be due to a folding or conformational problem of the full-length Rabex-5 in vitro or it could reflect a physiologically relevant regulatory mechanism in the cell.
Rabex-5 contains multiple functional domains (Delprato et al., 2004). A coiled-coil domain downstream of the GEF domain may mediate the binding to Rabaptin-5, as shown in a yeast two-hybrid assay (Mattera et al., 2006), whereas a Zn2+ finger (ZnF) domain at the N terminus is shown to bind ubiquitin (Lee et al., 2006; Mattera et al., 2006; Penengo et al., 2006). Recently, Rabex-5 (also called RabGEF1) knockout mice have been generated, and they die early and develop severe skin inflammation (Tam et al., 2004). Mast cells isolated from these Rabex-5–deficient mice show enhanced stem cell factor/c-Kit–mediated signal transduction and biological responses (Kalesnikoff et al., 2006). It is not yet clear whether these effects are related to the Rabex-5 GEF activity for Rab5. In addition to Rabex-5, there are other Vps9 domain-containing proteins, e.g., the RIN proteins (Tall et al., 2001; Saito et al., 2002), which may contain signal transduction-activated Rab5 GEF activity (Carney et al., 2006).
In this study, we investigate Rabex-5 function in vivo, and we specifically address the following questions: 1) Is the full-length Rabex-5 protein itself active as a Rab5 GEF in the cell? 2) Is the GEF domain itself active as a Rab5 GEF in the cell? 3) What is the role of Rabaptin-5 in Rabex-5 GEF activity? and 4) Can Rabex-5 target to early endosomes independently of Rabaptin-5 in the cell? If so, which domain is responsible? Our data identify a novel mechanism in the membrane targeting and function of Rabex-5 in the cell, and they clarify unresolved issues by reconciliation with in vitro studies.
MATERIALS AND METHODS
Plasmids
pGEX-6p-1, pGEX-4T-2, and pGEX-3X were purchased from GE Healthcare (Little Chalfont, Buckinghamshire, United Kingdom). pET-11a, pET-15b, and pET28a were from Novagen (Madison, WI). pMAL-2c was from New England Biolabs (Natick, MA). pBI was from BD Biosciences (San Jose, CA).
Protein Expression and Purification
Rabex-5 cDNA (Bos taurus, NCBI accession no. NM_174591) and truncated fragments were cloned into the pGEX and pMAL vectors for expression as glutathione S-transferase (GST) and maltose-binding protein (MBP) fusion proteins or into the pET-28a vector for expression as His-tagged proteins. Rabaptin-5 cDNAs were cloned into pET-15b for expression as His-tagged proteins or into pET-11a vectors for expression as free proteins (Zhu et al., 2004b). The plasmid constructs were transformed into the Escherichia coli strain BL21(DE3), and the expressed proteins were purified as soluble proteins by following the procedure described previously (Zhu et al., 2004a). To purify Rabex-5–Rabaptin-5 complexes, pET-28a/Rabex-5(135-480) and pET-11a/Rabaptin-5(572-641) were cotransformed into BL21(DE3), and the transformed bacteria were grown in LB medium containing both ampicillin (60 mg/l) and kanamycin (30 mg/l). Protein expression was induced by adding 0.1 mM isopropyl β-d-thiogalactoside at OD600 of 0.6, and the bacterial cultures were allowed to grow for another 15 h at 16°C, before being harvested and lysed with lysozyme. Recombinant proteins were purified from the supernatants of cell lysates by affinity His-Select resin (Sigma-Aldrich, St. Louis, MO). The eluted complex was cleaved by thrombin to remove the His-tag and further purified with Resource Q ion-exchange chromatography (GE Healthcare). The purified complex was analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) (20% gel) and visualized by Coomassie brilliant blue staining.
GST and MBP Pull-Down Assays
Recombinant GST- and MBP–Rabex-5 fusion proteins were immobilized on glutathione Sepharose-4B (GE Healthcare) and Amylose (New England Biolabs) resins, respectively. Five micrograms of recombinant Rabaptin-5 proteins were incubated with the resin containing 5 μg of GST- or MBP-Rabex-5 as indicated for 30 min in the binding buffer (200 μl) according to the manufacturer's instructions. The final concentration of each protein was <2 μM for detection of specific binding. The resins were subsequently washed three times with phosphate-buffered saline (PBS) and resuspended in 20 μl of SDS sample buffer. The samples were subjected to 20% SDS-PAGE analysis, and the proteins were visualized by Coomassie brilliant blue staining.
Guanosine 5′-O-(3-thio)triphosphate (GTPγS) Loading Assay
GST-Rab5 was purified with glutathione Sepharose-4B resin in PBS containing 10 mM MgCl2 and 1 mM GDP to keep GDP on Rab5. Rab5-GDP was then separated from GST and released into the supernatant by thrombin, which was later removed by benzamidine-Sepharose (GE Healthcare). The protein was dialyzed in the binding buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 5 mM MgCl2, 1 mM EDTA, and 1 mM dithiothreitol). A modified version of the filter binding assay (Lippe et al., 2001) was used to determine the [35S]GTPγS binding rate of Rab5-GDP, which reflected its nucleotide exchange rate. Briefly 1 μM Rab5-GDP was incubated with 2 μM [35S]GTPγS (GE Healthcare) in 100 μl of the binding buffer in the absence or presence of 0.1 μM of various Rabex-5 fragments or Rabex-5–Rabaptin-5 complexes. At indicated times, samples were taken and filtered through a hemagglutinin-type nitrocellulose membrane (0.45 μm; Millipore, Billerica, MA) by using a vacuum manifold. After washing with 2 ml of the binding buffer, the membrane was dried and [35S]GTPγS retained on the membrane was quantified with a liquid scintillation counter.
Mammalian Cell Cultures and Transfection
Baby hamster kidney (BHK)-21 cell monolayers were grown in 35-mm culture dishes with 3 ml of α-minimal essential medium containing 10% fetal bovine serum (Invitrogen, Carlsbad, CA). Cells were transfected with the plasmid constructs capable of expressing Rabex-5, Rabaptin-5, or Rab5 proteins as indicated via the Lipofectamine 2000-mediated procedure (Invitrogen), and they were incubated at 37°C for 24 h. The expression plasmids used included pcDNA3 (Invitrogen) and pBI (BD Biosciences). The pBI vector requires cotransfection with pTet-Off, and they can express two cloned proteins simultaneously. Protein expression was confirmed by immunoblot analysis and intracellular localization and endosomal morphology were determined by confocal fluorescence microscopy (see below).
Immunoblot Analysis
Cells were lysed in 1% SDS (200 μl/dish), and the lysates were sheared to reduce the stickiness by passing through a 26-gauge needle five times with a 1-ml syringe, followed by SDS-PAGE (12% gel) and immunoblot assay by using the enhanced chemiluminescence reagents (GE Healthcare). The primary antibodies used in these assays included anti-MYC monoclonal antibody (mAb) (Sigma-Aldrich), anti-FLAG mAb (Sigma-Aldrich), and anti-Rabaptin-5 antibody (BD Biosciences) as indicated.
Confocal Fluorescence Microscopy
We used a Leica confocal laser scanning microscope with Ar-488 and Kr-568 laser excitation in the Flow and Image Lab on campus (University of Oklahoma, Oklahoma City, OK) and followed a procedure described previously (Li and Liang, 2001). Briefly BHK-21 cells were grown on coverslips and transfected with pBI and/or pcDNA3 constructs expressing various Rabex-5, Rabaptin-5, or Rab5 proteins as indicated. At 24 h after transfection, the cells were processed for microscopy. Some of the proteins were expressed as green fluorescent protein (GFP) (enhanced green fluorescent protein; BD Biosciences) or red fluorescent protein (RFP) (ds-Red monomer; BD Biosciences) fusion proteins. For these fluorescent protein-tagged proteins, cells were rinsed three times with PBS and fixed for 20 min with 4% paraformaldehyde (wt/v in PBS) at room temperature. The coverslips were then mounted in PBS on glass slides and viewed with the microscope. For Myc-tagged proteins, indirect immunofluorescence microscopy was performed to identify the proteins. In this case, after fixation (see above), the cells were permeabilized with 0.05% saponin (in PBS) for 15 min and incubated with the anti-Myc antibody (Sigma-Aldrich) for 60 min in PBS containing 1% bovine serum albumin. Cells were rinsed three times with PBS to remove unbound primary antibody, followed by incubation with the secondary antibody (goat anti-mouse immunoglobulin G conjugated with Alexa568; Invitrogen) for 60 min. The coverslips were rinsed, mounted, and viewed as described above.
Subcellular Fractionation
BHK-21 cell monolayers in 35-mm dishes were grown and transfected as described above. At 24 h after transfection, the cells were rinsed with ice-cold PBS and scraped into 250 μl of 100 mM Tris-HCl, pH 7.4, containing the protease inhibitor cocktail (Sigma-Aldrich) with a cell scraper (Fisher Scientific, Pittsburgh, PA). The cells were then homogenized by passing through a 26-gauge needle attached to a 1-ml syringe 20 times. Cell homogenates were centrifuged at 850g for 5 min to remove nuclei and cell debris, and postnuclear supernatants were then subjected to ultraspeed centrifugation at 200,000 × g for 7 min in a TL-100 centrifuge (Beckman Coulter, Fullerton, CA) to separate the membrane fraction (pellet) from the cytosol fraction (supernatant). The membrane pellet was resuspended in the same volume of 100 mM Tris-HCl buffer as the cytosol fraction, and SDS was added to both fractions at a final concentration of 1% (wt/v). Proteins in each fraction (10 μl) were analyzed by SDS-PAGE and immunoblot assay as described above.
RESULTS
Biochemical Characterization of the GEF Domain and the Rabaptin-5-binding Domain in Rabex-5
Purified recombinant Rabex-5 showed only weak GEF activity for Rab5 in vitro (kcat = 0.007 s−1), but preformed Rabex-5-Rabaptin-5 complex was much more active (Esters et al., 2001; Lippe et al., 2001). In addition, the core GEF domain (residues 132–391) was also highly active (kcat >0.1 s−1) (Delprato et al., 2004). These observations might reflect an “autoinhibition” mechanism in which other domain(s) in Rabex-5 blocks the GEF domain activity, and Rabaptin-5 interaction may relieve this inhibition. Alternatively the purified recombinant Rabex-5 used in these in vitro studies might have some folding/conformation problems, and Rabaptin-5 interaction could help regain proper conformation and activity. Our results described in this and following sections support the second possibility, and they indicate that Rabex-5 activity in vivo in intact cells can bypass the requirement for Rabaptin-5.
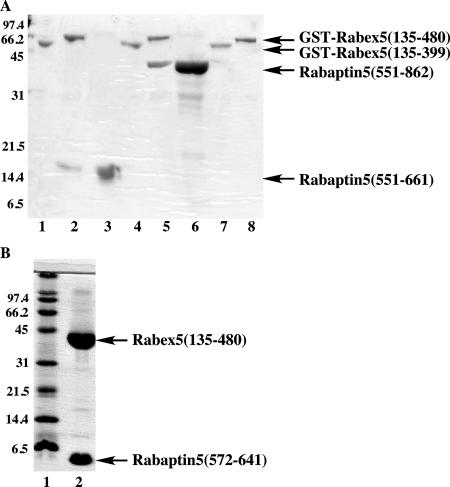
We first dissected the domains involved in the interaction between Rabex-5 and Rabaptin-5, and we found that the Rabaptin-5-binding domain of Rabex-5 blocked its GEF activity in vitro. To identify the interacting domains in Rabex-5 and Rabaptin-5, we made a number of GST or MBP fusion proteins of Rabex-5, and we used them to pull-down 6-His–tagged Rabaptin-5 proteins. Our results showed the interaction between the coiled-coil domain (residues 401–480) of Rabex-5 and the four-helical bundle region (residues 572-641) of Rabaptin-5 (Figure 1). Rabex-5(135-480), which includes the GEF domain and the downstream coiled-coil domain, formed a complex with Rabaptin-5(551-661) in the pull-down assay (Figure 1A, lane 2), but Rabex-5(135-399), the GEF domain itself, failed to bind Rabaptin-5(551-661) (Figure 1A, lane 1). A longer fragment Rabaptin-5(551-862), which contains all the C-terminal sequence, showed the same binding characteristics (Figure 1A, lanes 4–6). The GST Rabex-5(135-480) and Rabex-5(135-399) resins alone contained no contamination of any Rabaptin-5 proteins, which served as negative controls (Figure 1A, lanes 8 and 7). Furthermore, MBP-Rabex-5(401-480), the coiled-coil domain alone, was sufficient to bind a 70-amino acid Rabaptin-5(572-641) fragment in the pull-down assay (data not shown). Finally, we coexpressed Rabex-5(135-480) and Rabaptin-5(572-641) in E. coli, and we demonstrated that they were copurified as a complex (Figure 1B, lane 2). These biochemical results are consistent with a recent report showing similar domain interactions in a yeast two-hybrid assay (Mattera et al., 2006).
Figure 1.
Domains involved in Rabex-5 and Rabaptin-5 interaction. (A) The GST-Rabex-5 proteins (5 μg) were used to pull-down purified Rabaptin-5(551-661) (5 μg) in a 200-μl reaction. The bound proteins were analyzed by SDS-PAGE and visualized by Coomassie blue staining. Lane 1, GST-Rabex-5(135-399) incubation with 6-His-Rabaptin-5(551-661); lane 2, GST-Rabex-5(135-480) incubation with 6-His-Rabaptin-5(551-661); lane 3, 6-His-Rabaptin-5(551-661) directly loaded as a control; lane 4, GST-Rabex-5(135-399) incubation with 6-His-Rabaptin-5(551-862); lane 5, GST-Rabex-5(135-480) incubation with 6-His-Rabaptin-5(551-862); lane 6, 6-His-Rabaptin-5(551-862) directly loaded as a control; lane 7, GST-Rabex-5(135-399) directly loaded as a control; lane 8, GST-Rabex-5(135-480) directly loaded as a control. Molecular mass standards (in kilodaltons) are indicated on the left of the panel. (B) 6-His-tagged Rabex-5(135-480) was coexpressed with Rabaptin-5(572-641) in E. coli BL21 (DE3) and affinity purified with the His-Select resin (Sigma-Aldrich), followed by thrombin cleavage and further purification with Resource Q ion-exchange chromatography (GE Healthcare). Lane 1, molecular mass standards (in kilodaltons); lane 2, purified Rabex-5(135-480) and Rabaptin-5(572-641) complex.
We next examined the GEF activity of these Rabex-5 domains by determining their effect on [35S]GTPγS loading onto Rab5-GDP. The intrinsic nucleotide exchange rate, as reflected by the GTPγS loading rate, was extremely low in the presence of 5 mM Mg2+ and served as a negative control in these experiments (Figure 2A). The GEF domain alone (residues 135–399) strongly stimulated the GTPγS loading of Rab5-GDP (Figure 2A), consistent with a previous report (Delprato et al., 2004). In this regard, full-length Rabex-5 had little exchange activity (Esters et al., 2001; Lippe et al., 2001). To determine which region in Rabex-5 was responsible for blocking the high activity of the GEF domain, we made a series of N- and C-terminal extensions to the GEF domain. Rabex-5(1-399), which contained the entire sequence N terminal to the GEF domain, showed similar high exchange activity as the GEF domain itself (Figure 2A). In contrast, Rabex-5(135-480), which contained the coiled-coil domain C terminal to the GEF domain, showed much reduced exchange activity (Figure 2A), indicating that the coiled-coil domain (i.e., the Rabaptin-5-binding domain) blocked the GEF domain activity. However, the purified Rabex-5(135-480)–Rabaptin-5(572-641) complex (Figure 1B) exhibited full exchange activity similar to the GEF domain itself (Figure 2A), indicating that the 70-amino acid Rabaptin-5(572-641) fragment was sufficient to bind the coiled-coil domain (residues 401–480) of Rabex-5 and to overcome its inhibitory effect on the GEF domain.
Figure 2.
GEF activity of Rabex-5 and mutants in vitro and in vivo. (A) Purified Rabex-5(1-399), Rabex-5(135-399), Rabex-5(135-480), and Rabex-5(135-480)/Rabaptin-5(572-641) complex were examined for their ability to stimulate the loading of [35S]GTPγS onto Rab5-GDP. The reaction without any of the Rabex-5 constructs (Rab5 alone) served as a negative control. Samples were taken at the indicated times, and the amount of [35S]GTPγS bound to Rab5 in each case was determined by the filter binding assay. The results were reproducible in two independent experiments. (B) FLAG-tagged Rab5 was coexpressed in BHK cells with either the pBI vector control or pBI constructs expressing Rabex-5(1-135), Rabex-5, Rabex-5(135-399), Rabex-5(135-480), Rabex-5(135-480)/Rabaptin-5, Rabex-5(81-399) as indicated. Top, amount of Rab5-GTP in each case as determined by GST-R5BD pull-down assay, followed by immunoblot analysis with the anti-FLAG antibody. Middle, total amount of Rab5 in each cell lysate used for the pull-down assay as determined by immunoblot analysis of the lysate directly (1% of the amount for the pull-down assay) with the anti-FLAG antibody. Bottom, expression of the indicated Rabex-5 constructs (Myc-tagged) in the cell as determined by immunoblot analysis with the anti-Myc antibody. Molecular mass standards (in kilodaltons) are indicated on the left. The results were reproducible in three experiments. (C) Ratio of Rab5-GTP over total Rab5 quantified by densitometry of the immunoblots in B. Error bars indicate SEM in three experiments.
Rabex-5 Activates Rab5 in a Rabaptin-5–independent Manner In Vivo
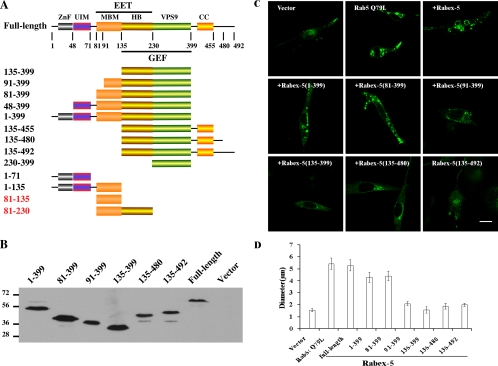
Strikingly, when we expressed Rabex-5 and the truncation mutants in mammalian cell cultures and examined their GEF activity in vivo, full-length Rabex-5 showed full activity, whereas Rabex-5(135-399), the GEF domain, showed reduced activity (Figure 2B), which was in contrast to the results obtained in vitro (Figure 2A). In this case, we coexpressed Rab5 and Rabex-5 or various Rabex-5 truncation mutants in BHK cells, and the amount of activated GTP-bound Rab5 in cell lysates was determined by pull-down assays by using GST-R5BD (the Rab5-binding domain of Rabaptin-5). Rabex-5 strongly stimulated GTP loading on Rab5, and it increased the Rab5-GTP level in the cell, in comparison with control cells without coexpression of Rabex-5 (Figure 2, B and C). In contrast, the GEF domain alone [Rabex-5(135-399)] was much less active than the full-length Rabex-5 (Figure 2, B and C), after standardizing Rab5-GTP level with total Rab5 level in each sample. Interestingly, Rabex-5(81-399), which contains an additional sequence N terminal to the GEF domain, restored the Rab5 GEF activity to a level similar to full-length Rabex-5 (Figure 2, B and C). Rabex-5(135-480), like Rabex-5(135-399), showed low GEF activity in vivo (Figure 2, B and C). However, Rabaptin-5, coexpressed from the same vector, significantly increased the activity of Rabex5(135-480) (Figure 2, B and C). The Rabex-5(1-135) fragment without the GEF domain showed no activity (Figure 2, B and C).
The data indicate the differences in the ways Rabex-5 activates Rab5 in solution and in the cell where Rab5 is mostly on the endosomal membrane, and these differences can be reconciled if in the cell there is a membrane-targeting step by Rabex-5 before it can act on its substrate, i.e., membrane-associated Rab5-GDP. In addition to the Rabaptin-5–mediated membrane targeting, Rabex-5 may also directly target to early endosomes and activate Rab5. In this regard, the low activity of Rabex-5(135-399) and Rabex-5(135-480) is probably due to defective endosomal targeting, and addition of the endosomal targeting domain, as in Rabex-5(81-399), restores full activity. This direct endosomal targeting may not require Rabaptin-5, because Rabex-5(81-399) does not contain the downstream Rabaptin-5-binding domain. Furthermore, the activity of full-length Rabex-5 may also result from direct membrane targeting rather than from interaction with endogenous Rabaptin-5, which is apparently insufficient to interact with and support Rabex-5(135-480). The residual activity of Rabex-5(135-399) and Rabex-5(135-480) may reflect their activation of the cytosolic fraction of Rab5, because the pull-down assay does not distinguish cytosolic and membrane-bound Rab5-GTP signals. These concepts are further investigated and confirmed by the following microscopy experiments that focus on the early endosome-associated Rab5 activation in the cell.
We expressed GFP-Rab5 to label early endosomes and coexpressed Rabex-5 or various Rabex-5 truncation mutants (Figure 3, A and B) to determine whether these Rabex-5 proteins can activate Rab5 and consequently enlarge the early endosomes in BHK cells. This assay was based on previous observations that Rab5 activity in these cells is rate limiting (Bucci et al., 1992; Li and Stahl, 1993). Indeed, the full-length Rabex-5, which has little activity in vitro (Esters et al., 2001; Lippe et al., 2001), exhibited high activity in the cell and led to great enlargement of the Rab5-positive early endosomes, similar to the effect of constitutive active Rab5:Q79L mutant (Figure 3, C and 3D). In contrast, Rabex-5(135-399), which is the GEF domain and highly active in vitro (Figure 2), was inactive in the cell, and it failed to enlarge the early endosomes (Figure 3, C and D), indicating that the GEF domain alone cannot activate early endosome-associated Rab5 in the cell. We examined >100 Rabex-5(135-399)-transfected cells, and none contained the large endosomes seen in Rabex-5–transfected cells, i.e., endosomes with diameter >4 μm. Corroborating the results with the GFP-Rab5–labeled early endosomes, GFP-EEA1–labeled early endosomes were also enlarged by full-length Rabex-5 but not by Rabex-5(135-399) (Supplemental Figure 1S). The data indicate that the Rabaptin-5-binding domain does not block the GEF activity of Rabex-5 in vivo, in contrast to its negative effect in vitro. Furthermore, endogenous Rabaptin-5 is insufficient to account for the observed Rabex-5 activity in the cell (Figure 3C; see below).
Figure 3.
Expression and activity of Rabex-5 and various truncation mutants in BHK cells. (A) Schematic illustration of the domain structures of full-length Rabex-5 and the truncation mutants used in this and the following experiments. Vps9, Vps9 domain; CC, coiled-coil domain. The arrows and numbers indicate the positions and amino acid residue numbers where the truncations were made. (B) Immunoblot showing the expression of some of the above Rabex-5 constructs with the GEF domain and N-terminal Myc-tag in BHK cells as identified with the anti-Myc antibody. Molecular mass standards (in kilodaltons) are indicated on the left. (C) Confocal fluorescence microscopy images showing the morphological changes of GFP-Rab5–labeled early endosomes in BHK cells coexpressing the indicated Rabex-5 constructs. GFP-Rab5 and GFP-Rab5:Q79L alone (cotransfection with the empty vector) serve as negative (vector) and positive (Rab5:Q79L) controls. Bar, 16 μm. (D) The graph quantifies the experiments described in C, and it shows the different sizes of GFP-Rab5–labeled early endosomes in control cells and cells expressing the indicated Rabex-5 constructs. The diameters of 90 of the largest GFP-Rab5–labeled endosomes in 30 cells were measured in each case, and the graph shows the mean and calculated SEM. All cells measured coexpressed the indicated Rabex-5 constructs as evidenced by immunofluorescence microscopy with the anti-Myc antibody as shown in Figure 4.
The data also indicate that in addition to the core GEF domain, one or more other domains are necessary for Rabex-5 to activate Rab5 in vivo. We made several Rabex-5 constructs with various combinations of domains (Figure 3A) to identify regions essential for the GEF activity (Figure 3C) and the endosomal targeting (Figure 4) of Rabex-5. To determine whether sequences N terminal to the GEF domain can confer Rab5 GEF activity to Rabex-5(135-399) in the cell, Rabex-5(1-399), Rabex-5(81-399), and Rabex-5(91-399) were expressed in BHK cells (Figure 3B) to determine their ability in the enlargement of early endosomes. Although Rabex-5(91-399) remained inactive, the two longer N-terminal extension constructs Rabex-5(81-399) and Rabex-5(1-399) were both able to enlarge early endosomes (Figure 3, C and D, and Supplemental Figure 1S), albeit to a lesser extent than full-length Rabex-5. Another N-terminal extension construct, Rabex-5(48-399), also colocalizes with Rab5 on early endosomes, and it was active in enlarging the early endosomes (Supplemental Figure 2; data not shown). Importantly, these active Rabex-5 constructs do not contain the downstream coiled-coil domain involved in Rabaptin-5 binding (residues 401–455), indicating that interaction with Rabaptin-5 is not necessary for Rabex-5 to exhibit Rab5 GEF activity in vivo.
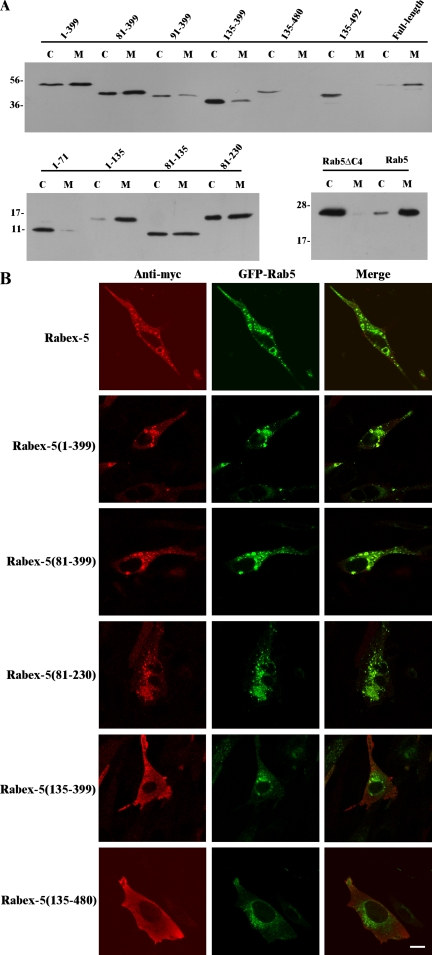
Figure 4.
The EET domain necessary and sufficient for early endosomal targeting by Rabex-5. (A) Immunoblot showing the membrane and cytosol distribution of the indicated Rabex-5 constructs, which are schematically illustrated in Figure 3A. M, membrane; C, cytosol. Molecular mass standards (in kilodaltons) are indicated on the left. (B) Confocal fluorescence microscopy images showing intracellular localization of the indicated Rabex-5 constructs and coexpressed GFP-Rab5 in BHK cells. The Myc-tagged Rabex-5 constructs were identified by indirect immunofluorescence microscopy with the anti-Myc antibody. Bar, 16 μm.
To determine whether sequences C-terminal to the GEF domain, such as the Rabaptin-5-binding domain, may restore the Rab5 GEF activity of Rabex-5(135-399) in vivo, we made Rabex-5(135-455), Rabex-5(135-480), and Rabex-5(135-492), all of which contained the Rabaptin-5-binding domain with the last one extending all the way to the C terminus. However, like Rabex-5(135-399), Rabex-5(135-480) and Rabex-5(135-492) were inactive in terms of enlarging the early endosomes in BHK cells (Figure 3, C and D, and Supplemental Figure 1S), as was Rabex-5(135-455) (Supplemental Figure 2S; data not shown), indicating that endogenous Rabaptin-5 is insufficient to confer activity to these C-terminal extension constructs. Along this line, endogenous Rabaptin-5 is unlikely to contribute to the full-length Rabex-5 activity observed in the cell. Together, the data suggest that Rabex-5 can function independently of Rabaptin-5 in the cell.
A Novel Early Endosomal Targeting Domain in Rabex-5 Is Essential for Its Rab5 GEF Activity In Vivo
To further investigate why the sequence N terminal to the GEF domain is critical for its activity in vivo, we determined the membrane-targeting properties of the aforementioned and additional Rabex-5 constructs, and we found that the region (residues 81–135) immediately upstream of the GEF domain represents a novel membrane-binding motif (MBM), which together with the downstream helical bundle (HB) domain (residues 135–230) forms a novel early endosomal targeting (EET) domain for Rabex-5 (Figures 3A and 4). In the initial experiments, Myc-tagged Rabex-5 fragments were expressed in BHK cells, and cell homogenates were subjected to centrifugation to separate membrane and cytosol fractions. The Rabex-5 proteins in each fraction were identified by immunoblot analysis with an anti-Myc antibody. Full-length Rabex-5 was mostly membrane associated (Figure 4A). However, Rabex-5(135-399), i.e., the GEF domain, was mostly cytosolic (Figure 4A), suggesting that its inability to activate Rab5 in vivo may be due to defective membrane targeting. In this regard, all Rabex-5 constructs that showed Rab5 GEF activity in vivo, including Rabex-5(1-399) and Rabex-5(81-399), were also significantly membrane associated, whereas all inactive Rabex-5 constructs, including Rabex-5(135-480), Rabex-5(135-492), and Rabex-5(91-399), shifted toward a mostly cytosolic distribution (Figure 4A). Thus, there is a general correlation between efficient membrane association of a Rabex-5 construct and its Rab5 GEF activity in vivo. Examination of additional Rabex-5 constructs were consistent with this observation, including the active Rabex-5(48-399) and inactive Rabex-5(135-455) and Rabex-5(230-399), which is the Vps9 domain itself (Supplemental Figure 2S). Although Rabex-5(135-399) might also interact directly with membrane-bound Rab5, which could account for the residual membrane-bound fraction (∼20%), this direct interaction is apparently too inefficient to activate Rab5 sufficiently and enlarge endosomes in the cell. We also examined the membrane/cytosol distribution of additional Rabex-5 constructs lacking the Vps9 domain, including Rabex-5(1-71) Rabex-5(1-135), Rabex-5(81-135), and Rabex-5(81-230). Rabex-5(1-71), which encompasses the ZnF and UIM domains, was in the cytosol, but Rabex-5(1-135), Rabex-5(81-135), and Rabex-5(81-230) were able to associate with the membrane (Figure 4A), suggesting that the region encompassing residues 81–135 represents a novel MBM. Rab5 and the Rab5ΔC4 mutant that lacks C-terminal prenylation served as membrane and cytosol controls, respectively (Figure 4A).
To address more specifically whether the Rabex-5 proteins indeed targeted to Rab5-containing early endosomes, we coexpressed the Myc-tagged Rabex-5 constructs with GFP-Rab5 in BHK cells, and we determined whether they colocalize with GFP-Rab5 by confocal immunofluorescence microscopy with the anti-Myc antibody. GFP-Rab5–labeled early endosomes, which exhibited a punctate pattern in the cell (Figure 3C). All Rabex-5 constructs that were able to activate Rab5 in vivo, such as full-length Rabex-5, Rabex-5(1-399), Rabex-5(81-399), and Rabex-5(48-399), were targeted and colocalized to the GFP-Rab5–labeled early endosomes, which as a result were generally larger (Figure 4B and Supplemental Figure 2S). In contrast, the Rabex-5 constructs that failed to activate Rab5 in vivo, such as the core GEF domain Rabex-5(135-399) and Rabex-5(135-480), showed no detectable colocalization with GFP-Rab5 on the early endosomes (Figure 4B), neither did Rabex-5(135-455) and Rabex-5(135-492) (Supplemental Figure 2S). Instead, these inactive Rabex-5 proteins exhibited a diffused cytosolic staining pattern throughout the entire cell (Figure 4B and Supplemental Figure 2S).
Importantly, Rabex-5(81-230), which contains the newly identified MBM and HB domains, was sufficient to target to early endosomes and colocalize with Rab5 (Figure 4B), although the MBM alone [Rabex-5(81-135)] occurred mostly on the plasma membrane (Supplemental Figure 2S). Rabex-5(1-71) containing the ZnF and UIM domains showed cytosolic distribution (Supplemental Figure 2S). Interestingly, Rabex-5(1-135) were associated with vesicle-like structures, but these structures were distinct from Rab5-containing endosomes, even though they were occasionally found adjacent to each other (Supplemental Figure 2S, arrow and arrowhead). Further investigation revealed that Rabex-5(1-135) colocalized with Rab7 on the late endosomes (Supplemental Figure 3S). Thus, MBM (residues 81–135) represents a novel membrane-binding motif, and they can associate with different membranes in different sequence contexts. Importantly, MBM and the downstream HB domain (residues 135–230) together represent a novel EET domain, which is critical for Rabex-5 to associate with early endosomes and to activate Rab5 in vivo. Because the EET domain lacks the Vps9 domain necessary for interacting with Rab5, it must associate with early endosomes through other early endosomal protein(s) or lipid(s). Phosphatidylinositol 3-phosphate is an important phospholipid involved in recruiting early endosomal proteins, but it seems to not be required for the association of EET domain or Rabex-5 with early endosomes, as evidenced by its insensitivity to wortmannin, a phosphatidylinositol 3-kinase inhibitor (data not shown).
Rabex-5 Can Also Target to Early Endosomes in a Rabaptin-5-dependent Manner In Vivo
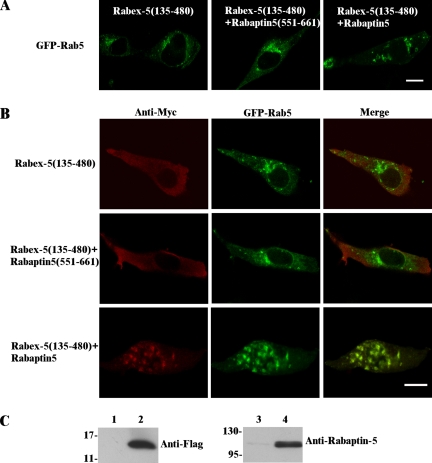
The expression of Rabex-5(135-480) and Rabex-5(135-492) constructs both of which contained the Rabaptin-5-binding domain (residues 400–480) did not show any enlargement of early endosomes in the cell (Figure 3C). However, it was possible that endogenous Rabaptin-5 was limiting. Thus, we coexpressed Rabex-5(135-480) with either full-length Rabaptin-5 or Rabaptin-5(551-661) with the bidirectional vector pBI, which can simultaneously express two proteins on a single plasmid. Indeed, the full-length Rabaptin-5 was able to rescue Rabex-5(135-480) activity in terms of enlargement of GFP-Rab5–labeled early endosomes (Figure 5A). In contrast, the Rabaptin-5(551-661) fragment was unable to do so (Figure 5A), even though it formed an active complex with Rabex-5(135-480) in vitro (Figures 1 and 2). Consistent with the recovery of GEF activity, full-length Rabaptin-5 but not Rabaptin-5(551-661) helped Rabex-5(135-480) localize to the Rab5-positive endosomes (Figure 5B). The expression of Rabaptin-5 and Rabaptin-5(551-661) was further confirmed by immunoblot analysis (Figure 5C). Because the full-length Rabaptin-5 contains the Rab5-binding domain at the C terminus, the results are consistent with previous reports that Rabex-5–Rabaptin-5 complex can target to early endosomes via Rabaptin-5 binding to Rab5-GTP (Lippe et al., 2001; Zhu et al., 2004b).
Figure 5.
Rabaptin-5–mediated Rabex-5 targeting to early endosomes in BHK cells. (A) Confocal fluorescence microscopy images showing the morphology of GFP-Rab5–labeled early endosomes in BHK cells coexpressing Rabex-5(135-480), Rabex-5(135-480)/Rabaptin-5(551-661), or Rabex-5(135-480)/Rabaptin-5, as indicated. Bar, 16 μm. (B) Confocal fluorescence microscopy images showing intracellular localization of GFP-Rab5 and coexpressed Rabex-5(135-480) when expressed alone or together with Rabaptin-5 or Rabaptin-5(551-661), as indicated. The Myc-tagged Rabex-5(135-480) was identified by indirect immunofluorescence microscopy with the anti-Myc antibody. Bar, 16 μm. (C) Immunoblot confirming the coexpression of Rabaptin-5(551-661) that contains a FLAG tag (lane 2) and Rabaptin-5 (lane 4) with the anti-FLAG and anti-Rabaptin-5 antibodies, respectively. Either of the Rabaptin-5 constructs is on the same pBI vector with Myc-Rabex-5(135-480); thus, they are coexpressed with Myc-Rabex-5(135-480) in the same cells as shown in B. The pBI vector that contains only Myc-Rabex-5(135-480) shows no expression of either FLAG-Rabaptin-5(551-661) (lane 1) or Rabaptin-5 (lane 3). Molecular mass standards (in kilodaltons) are indicated on the left of each panel.
Rabex-5 Can Rescue Rab5:S34N-mediated Inhibition of Early Endosome Fusion
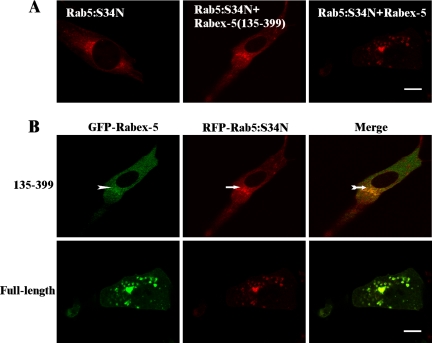
Dominant-negative mutants, such as Rab5:S34N, were suggested to inhibit endogenous Rab5 by sequestration of a Rab5 GEF (Li and Stahl, 1993; Stenmark et al., 1994), but this contention was not formally demonstrated. If endogenous Rabex-5 is the target of sequestration by Rab5:S34N, then overexpression of Rabex-5 should be able to overcome Rab5:S34N-mediated inhibition of early endosome fusion. We coexpressed GFP-Rabex-5 with RFP-Rab5:S34N in BHK cells, and then we examined the morphology of RFP-Rab5:S34N-labeled early endosomes by confocal fluorescence microscopy. Indeed, GFP-Rabex-5 targeted to RFP-Rab5:S34N–labeled early endosomes, and it restored their fusion, as evidenced by the enlargement of RFP-Rab5:S34N–labeled early endosomes in these cells (Figure 6, A and B). In control cells without Rabex-5 overexpression, the RFP-Rab5:S34N–labeled endosomes were much smaller, and they accumulated at the perinuclear region (Figure 6A), due to the inhibition of endosome fusion (Li et al., 1994; Stenmark et al., 1994). Expression of GFP-Rabex-5(135-399), i.e., the GEF domain, did not show any activity to enlarge the Rab5:S34N-labeled endosomes (Figure 6A). Although GFP-Rabex-5(135-399) exhibited a mostly diffused cytosolic pattern, a portion of the protein was consistently found on the Rab5:S34N-labeled endosomes (Figure 6B, arrows). Because Rabex-5(135-399) lacks the EET domain and it is not found on normal early endosomes (Figure 4B), its partial localization to Rab5:S34N-labeled endosomes is likely due to direct interaction with Rab5:S34N, consistent with the contention that the dominant-negative Rab mutant has higher affinity for and thus can sequester the GEF. The sequestered Rabex-5(135-399) on the Rab5:S34N-labeled endosomes is apparently inactive, because it cannot activate endogenous Rab5 to enlarge these endosomes (Figure 6). In this regard, full-length Rabex-5 contains the EET domain to mediate its targeting to Rab5:S34N-containing endosomes; thus, it can bypass Rab5:S34N sequestration and retain the ability to activate endogenous Rab5 and enlarge these endosomes (Figure 6), even though a fraction of Rabex-5 molecules may still bind Rab5:S34N and become sequestered and inactive. That the sequestration occurs on the membrane is consistent with previous findings that the dominant-negative phenotype of Rab5:S34N is dependent on its membrane association and that it can be abolished by truncation of its C-terminal isoprenylation motif (Li et al., 1994).
Figure 6.
Rescue of Rab5:S34N-blocked endosome fusion by overexpression of Rabex-5. (A) Confocal fluorescence microscopy images showing the morphology of RFP-Rab5:S34N-labeled early endosomes in BHK cells with or without coexpression of GFP-Rabex-5 or GFP-Rabex-5(135-399) as indicated. Bar, 16 μm. (B) Confocal fluorescence microscopy images showing localization of GFP-Rabex-5 or GFP-Rabex-5(135-399) in the above-mentioned cells expressing RFP-Rab5:S34N. Arrows indicate partial localization of GFP-Rabex-5(135-399) on RFP-Rab5:S34N endosomes. Bar, 16 μm.
DISCUSSION
This study investigates Rabex-5 function in vivo and identifies a direct, Rabaptin-5–independent targeting pathway to early endosomes by Rabex5. Rabex-5 needs to associate with early endosomes first before it can interact effectively with Rab5, and this two-dimensional interaction in the endosomal membrane is not reflected by in vitro nucleotide exchange reactions in solution. For example, the soluble GEF domain itself (residues 135–399) is a potent Rab5 GEF in in vitro nucleotide exchange reactions, but it is inactive in the cell in terms of activating Rab5 on the early endosomes. These data strongly suggest that the soluble GEF domain cannot directly act on membrane-bound Rab5-GDP or at least that this interaction is very inefficient, unless it contains additional early endosomal targeting information. The EET domain itself (residues 81–230) is sufficient to target to early endosomes, and it contains a novel MBM (residues 81–135) and the HB domain (residues 135–230). The MBM is not hydrophobic, but it is rich in positively charged residues (10 Lys and 3 Arg), and it is likely to form an amphipathic helix for binding to the membrane, whereas the HB domain may provide specificity via interaction with an early endosome-specific protein or lipid whose nature remains to be investigated. The GEF domain lacks the MBM, and it cannot target to early endosomes efficiently. The two-step mechanism for Rab activation, i.e., GEF targeting to the membrane followed by GEF–Rab interaction, seems conserved in the Rab GTPase family and another well-characterized Rab GEF, Sec2p (the GEF for Sec4p), also contains a membrane-targeting domain necessary for its in vivo function (Elkind et al., 2000).
Rabex-5 efficiently targets to early endosomes and activates Rab5 in the cell, as evidenced by the Rab5-GTP pull-down assay and enlargement of early endosomes. This process does not require interaction with Rabaptin-5, because Rabex-5 truncation mutants lacking the Rabaptin-5-binding domain can target to early endosomes and activate Rab5 in the same manner. Increased Rabex-5 expression can increase Rab5 activity and endosome fusion, suggesting that Rabex-5 level is limiting in these cells. The increased Rabex-5 activity is unlikely to be mediated by Rabaptin-5, because endogenous Rabaptin-5 level is too low and insufficient to form new complexes with the newly expressed Rabex-5 (Figure 5; see below). In this context, it is necessary to reconcile with previous in vitro data, which show that full-length Rabex-5 has little Rab5 GEF activity in in vitro biochemical reactions (Lippe et al., 2001). Our results described in Figure 2 extend this observation and they reveal that the low activity of Rabex-5 in vitro is due to its Rabaptin-5-binding domain. The data are most consistent with the interpretation that the purified Rabex-5 may have a folding/conformational problem in vitro with its active site blocked by the Rabaptin-5-binding domain. Truncation of the Rabaptin-5-binding domain, like binding to Rabaptin-5, can greatly enhance the Rabex-5 GEF activity in vitro. However, in the cell, Rabex-5 has no such folding/conformational problem and it is fully active without Rabaptin-5, although the Rabex-5–Rabaptin-5 complex may play a role in establishing a positive feedback loop to increase rapidly the number of Rab5-GTP molecules in the formation of functional Rab5 domains in the endosomal membranes (Zerial and McBride, 2001; Grosshans et al., 2006).
Rabex-5 specifically targets to early endosomes in the cell, and it is not detected in other intracellular membranes. Rabex-5 can also associate with early endosome preparations in vitro in a Rab5-independent manner, although the in vitro targeting process is rather inefficient in comparison with the Rab5-dependent recruitment of Rabex-5–Rabaptin-5 complex (Lippe et al., 2001). The aforementioned conformational problem may contribute to the inefficient membrane targeting of Rabex-5 in vitro. However, in the cell, Rabex-5 is fully active, and there is no further increase of Rabex-5 activity upon coexpression of Rabaptin-5 (data not shown), suggesting that there is no additive or synergistic effect between the EET and Rabaptin-5-bindng domains. Nonetheless, coexpression of Rabaptin-5 can rescue the activity of Rabex-5(135-480), which itself cannot target to early endosomes because of the truncation of the EET domain, indicating that the Rabaptin-5–mediated Rabex-5 membrane targeting pathway identified in vitro (Lippe et al., 2001) also functions in the cell. In addition, these data suggest that endogenous Rabaptin-5 is already in complexes with endogenous Rabex-5 and/or other proteins and that it is unavailable to form new complexes.
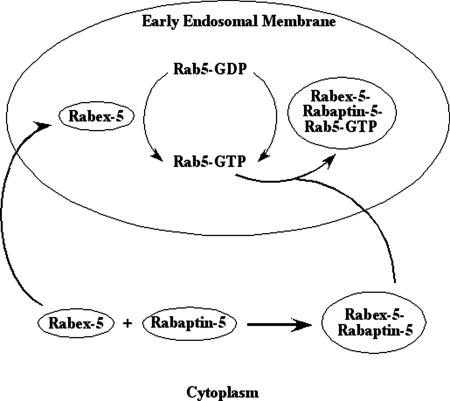
Thus, there are two parallel pathways for Rabex-5 to associate with early endosomes: direct targeting via the EET domain and indirect targeting via Rabaptin-5 binding to Rab5-GTP (Figure 7). Direct targeting may account for most of the membrane-associated pool of Rabex-5, whereas Rabaptin-5 determines the cytosolic pool of Rabex-5, taken into consideration that Rabex-5 was originally isolated as a soluble Rabex-5–Rabaptin-5 complex (Horiuchi et al., 1997), and there is little free Rabex-5 in the cytosol (Lippe et al., 2001). Interestingly, Rabaptin-5 binding to Rabex-5 seems to block the direct membrane targeting pathway, possibly by masking the EET domain, because the Rabex-5–Rabaptin-5 complex either remains in the cytosol or targets to early endosomes in a Rab5-dependent manner via Rabaptin-5 binding to Rab5-GTP (Lippe et al., 2001; Zhu et al., 2004b). Because the intrinsic exchange rate from Rab5-GDP to Rab5-GTP is extremely low, an apparent advantage of direct targeting is to provide a basal level of Rabex-5 on early endosomal membranes, which in turn produces a basal level of Rab5-GTP. In this context, the cytosolic Rabex-5–Rabaptin-5 complex can function via binding to Rab5-GTP and targeting to early endosomes, which forms a positive feedback loop to produce more Rab5-GTP and consequently establish a functional Rab5 domain in the endosomal membrane (Zerial and McBride, 2001; Grosshans et al., 2006).
Figure 7.
Model on direct and indirect membrane targeting and Rab5 activation by Rabex-5. Rabex-5 targets to the early endosomal membrane in two parallel pathways: direct targeting via the EET domain identified in this report and indirect targeting via Rabaptin-5–mediated binding to Rab5-GTP described previously (Lippe et al., 2001). Direct targeting of Rabex-5 is necessary to promote the production of a basal level of Rab5-GTP, which in turn recruits Rabaptin-5–Rabex-5 complexes to the endosomal membrane to convert more Rab5-GDP to Rab5-GTP. This effectively creates a positive feedback loop to accumulate Rab5-GTP molecules on the membrane, leading to the establishment of functional Rab5 domains.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Harald Stenmark (Norwegian Radium Hospital) and Marino Zerial (Max Planck Institute of Molecular Cell Biology and Genetics) for kindly providing the cDNAs of EEA1 and Rabex-5, respectively. We thank Jim Henthorn for expert assistance with the confocal fluorescence microscopy. This work was supported in part by the NIH grant GM074692 (to G.L.).
Abbreviations used:
- BHK
baby hamster kidney
- EET
early endosomal targeting
- GEF
guanine nucleotide exchange factor
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- HB
helical bundle
- MBM
membrane-binding motif
- MBP
maltose-binding protein
- PBS
phosphate-buffered saline
- R5BD
Rab5-binding domain
- RFP
red fluorescent protein
- ZnF
Zn2+ finger
- UIM
ubiquitin-interacting motif.
Note Added in Proof.
While this article was in revision, a new paper was published and showed that a region downstream of the Vps9 domain exerted “autoinhibition” on the GEF activity of Rabex-5 and that this inhibition could be partially relieved by Rabaptin-5 in vitro (Delprato and Lambright, 2007). These results are consistent with our data in Figure 2.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-02-0100) on August 15, 2007.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Bucci C., Parton R. G., Mather I. M., Stunnenberg H., Simons K., Hoflack B., Zerial M. The small GTPase Rab5 functions as a regulatory factor in the early endocytic pathway. Cell. 1992;70:715–728. doi: 10.1016/0092-8674(92)90306-w. [DOI] [PubMed] [Google Scholar]
- Carney D. S., Davies B. A., Horazdovsky B. F. Vps9 domain-containing proteins: activators of Rab5 GTPases from yeast to neurons. Trends Cell Biol. 2006;16:27–35. doi: 10.1016/j.tcb.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Delprato A., Lambright D. G. Structural basis for Rab GTPaes activation by VPS9 domain exchange factors. Nat. Struct. Mol. Biol. 2007;14:406–412. doi: 10.1038/nsmb1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delprato A., Merithew E., Lambright D. G. Structure, exchange determinants, and family-wide rab specificity of the tandem helical bundle and Vps9 domains of Rabex-5. Cell. 2004;118:607–617. doi: 10.1016/j.cell.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Elkind N. B., Walch-Solimena C., Novick P. J. The role of the COOH terminus of Sec2p in the transport of post-Golgi vesicles. J. Cell Biol. 2000;149:95–110. doi: 10.1083/jcb.149.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esters H., Alexandrov K., Iakovenko A., Ivanova T., Thoma N., Rybin V., Zerial M., Scheidig A. J., Goody R. S. Vps9, Rabex-5 and DSS 4, proteins with weak but distinct nucleotide-exchange activities for Rab proteins. J. Mol. Biol. 2001;310:141–156. doi: 10.1006/jmbi.2001.4735. [DOI] [PubMed] [Google Scholar]
- Gorvel J.-P., Chavrier P., Zerial M., Gruenberg J. rab5 controls early endosome fusion in vitro. Cell. 1991;64:915–925. doi: 10.1016/0092-8674(91)90316-q. [DOI] [PubMed] [Google Scholar]
- Grosshans B. L., Ortiz D., Novick P. Rabs and their effectors: achieving specificity in membrane traffic. Proc. Natl. Acad. Sci. USA. 2006;103:11821–11827. doi: 10.1073/pnas.0601617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffenberg S., Sanford J. C., Liu S., Daniel D. S., Tuvin M., Knoll B. J., Wessling-Resnick M., Dickey B. F. Biochemical and functional characterization of a recombinant GTPase, Rab5, and two of its mutants. J. Biol. Chem. 1995;270:5048–5056. doi: 10.1074/jbc.270.10.5048. [DOI] [PubMed] [Google Scholar]
- Horiuchi H., et al. A novel Rab5 GDP/GTP exchange factor complexed to Rabaptin-5 links nucleotide exchange to effector recruitment and function. Cell. 1997;90:1149–1159. doi: 10.1016/s0092-8674(00)80380-3. [DOI] [PubMed] [Google Scholar]
- Kalesnikoff J., Rios E. J., Chen C. C., Nakae S., Zabel B. A., Butcher E. C., Tsai M., Tam S. Y., Galli S. J. RabGEF1 regulates stem cell factor/c-Kit-mediated signaling events and biological responses in mast cells. Proc. Natl. Acad. Sci. USA. 2006;103:2659–2664. doi: 10.1073/pnas.0511191103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Tsai Y. C., Mattera R., Smith W. J., Kostelansky M. S., Weissman A. M., Bonifacino J. S., Hurley J. H. Structural basis for ubiquitin recognition and autoubiquitination by Rabex-5. Nat. Struct. Mol. Biol. 2006;13:264–271. doi: 10.1038/nsmb1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Barbieri M. A., Colombo M. I., Stahl P. D. Structural features of the GTP-binding defective Rab5 mutants required for their inhibitory activity on endocytosis. J. Biol. Chem. 1994;269:14631–14635. [PubMed] [Google Scholar]
- Li G., Liang Z. Phosphate-binding loop and Rab GTPase function: mutations at Ser29 and Ala30 of Rab5 lead to loss-of-function as well as gain-of-function phenotype. Biochem. J. 2001;355:681–689. doi: 10.1042/bj3550681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Stahl P. D. Structure-function relationship of the small GTPase Rab5. J. Biol. Chem. 1993;268:24475–24480. [PubMed] [Google Scholar]
- Lippe R., Miaczynska M., Rybin V., Runge A., Zerial M. Functional synergy between Rab5 effector Rabaptin-5 and exchange factor Rabex-5 when physically associated in a complex. Mol. Biol. Cell. 2001;12:2219–2228. doi: 10.1091/mbc.12.7.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattera R., Tsai Y. C., Weissman A. M., Bonifacino J. S. The Rab5 guanine nucleotide exchange factor Rabex-5 binds ubiquitin (Ub) and functions as a Ub ligase through an atypical Ub-interacting motif and a zinc finger domain. J. Biol. Chem. 2006;281:6874–6883. doi: 10.1074/jbc.M509939200. [DOI] [PubMed] [Google Scholar]
- Penengo L., Mapelli M., Murachelli A. G., Confalonieri S., Magri L., Musacchio A., Di Fiore P. P., Polo S., Schneider T. R. Crystal structure of the ubiquitin binding domains of rabex-5 reveals two modes of interaction with ubiquitin. Cell. 2006;124:1183–1195. doi: 10.1016/j.cell.2006.02.020. [DOI] [PubMed] [Google Scholar]
- Saito K., Murai J., Kajiho H., Kontani K., Kurosu H., Katada T. A novel binding protein composed of homophilic tetramer exhibits unique properties for the small GTPase Rab5. J. Biol. Chem. 2002;277:3412–3418. doi: 10.1074/jbc.M106276200. [DOI] [PubMed] [Google Scholar]
- Stenmark H., Parton R. G., Steele-Mortimer O., Lutcke A., Gruenberg J., Zerial M. Inhibition of rab5 GTPase activity stimulates membrane fusion in endocytosis. EMBO J. 1994;13:1287–1296. doi: 10.1002/j.1460-2075.1994.tb06381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tall G. G., Barbieri M. A., Stahl P. D., Horazdovsky B. F. Ras-activated endocytosis is mediated by the Rab5 guanine nucleotide exchange activity of RIN1. Dev Cell. 2001;1:73–82. doi: 10.1016/s1534-5807(01)00008-9. [DOI] [PubMed] [Google Scholar]
- Tam S. Y., Tsai M., Snouwaert J. N., Kalesnikoff J., Scherrer D., Nakae S., Chatterjea D., Bouley D. M., Galli S. J. RabGEF1 is a negative regulator of mast cell activation and skin inflammation. Nat. Immunol. 2004;5:844–852. doi: 10.1038/ni1093. [DOI] [PubMed] [Google Scholar]
- Zerial M., McBride H. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- Zhu G., Zhai P., He X., Wakeham N., Rodgers K., Li G., Tang J., Zhang X. C. Crystal structure of human GGA1 GAT domain complexed with the GAT-binding domain of Rabaptin5. EMBO J. 2004a;23:3909–3917. doi: 10.1038/sj.emboj.7600411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G., Zhai P., Liu J., Terzyan S., Li G., Zhang X. C. Structural basis of Rab5-Rabaptin5 interaction in endocytosis. Nat. Struct. Mol. Biol. 2004b;11:975–983. doi: 10.1038/nsmb832. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.