Abstract
Kinetochore–passenger complexes in metazoans have been proposed to coordinate the segregation of chromosomes in anaphase with the induction of cytokinesis. Passenger protein homologues in the budding yeast Saccharomyces cerevisiae play a critical role early in mitosis, ensuring proper biorientation of kinetochore–microtubule attachments. Our recent work has implicated the passenger protein Bir1p (Survivin) and the inner kinetochore complex centromere binding factor 3 (CBF3) in the regulation of septin dynamics during anaphase. Here, we present data that is consistent with there being multiple passenger protein complexes. Our data show that Bir1p links together a large passenger complex containing Ndc10p, Sli15p (INCENP), and Ipl1p (Aurora B) and that the interaction between Bir1p and Sli15p is specifically involved in regulating septin dynamics during anaphase. Neither conditional alleles nor mutants of BIR1 that disrupt the interaction between Bir1p and Sli15p resulted in mono-attached kinetochores, suggesting that the Bir1p–Sli15p complex functions in anaphase and independently from Sli15p–Ipl1p complexes. We present a model for how discrete passenger complexes coordinate distinct aspects of mitosis.
INTRODUCTION
To ensure the accurate segregation of chromosomes, anaphase progression must be coordinated with the onset of cytokinesis. In metazoans, the kinetochore passenger protein complex has been proposed to coordinate these events as they act both at centromeres (CENs) to establish a bipolar spindle and at the spindle midzone where they are required for cytokinesis (Adams et al., 2001; Vagnarelli and Earnshaw, 2004). Despite the long observed “transfer” of passenger proteins from chromosomes and kinetochores to the anaphase spindle and their requirement for successful cytokinesis, little is known concerning regulation of their distribution or their precise role in cytokinesis.
Homologues of metazoan passenger proteins in yeast (Aurora B and INCENP in metazoans; Ipl1p and Sli15p in yeast, respectively) were isolated in genetic screens for genes involved in mitotic fidelity (Chan and Botstein, 1993; Biggins et al., 1999; Kim et al., 1999; Yoon and Carbon, 1999). The analysis of Ipl1p and its binding partner Sli15p, isolated as a synthetic lethal mutation with an ipl1 mutant, focused on the role of this complex in regulating the kinetochore–microtubule attachments in metaphase (Biggins et al., 2001; Biggins and Murray, 2001; Tanaka et al., 2002). In ipl1 or sli15 mutants, unsegregated green fluorescent protein (GFP)-marked CENs are frequently observed, consistent with the failure of chromosomes to properly form bioriented attachments with the mitotic spindle. Biochemical and genetic evidence demonstrated that the DASH microtubule binding complex is phosphorylated by Ipl1p to allow its disassociation from microtubules, thus helping to correct sister kinetochores attached to microtubules from the same pole (Cheeseman et al., 2002; Li et al., 2002; Shang et al., 2003). The dissociation of kinetochores from microtubules creates unattached sites that activate the MAD2-dependent checkpoint and delay entry of cells into anaphase (Pinsky and Biggins, 2005). A similar role has been proposed for the human homologue Aurora B in resolving merotelic kinetochore attachments (Hauf et al., 2003; Lampson et al., 2004).
In contrast to its role at CENs, the function of passenger proteins in cytokinesis is less clear; inhibition of passenger proteins typically result in failure to complete cytokinetic furrow ingression. Furthermore, in several systems, Aurora B, INCENP, and Survivin have been shown to be in a complex (Adams et al., 2000; Wheatley et al., 2001; Bolton et al., 2002; Leverson et al., 2002; Honda et al., 2003); both INCENP and Survivin are substrates of Aurora B and thus link the enzymatic activity of the Aurora B kinase to the function of passenger complexes (Bishop and Schumacher, 2002; Bolton et al., 2002; Petersen and Hagan, 2003). Biochemical studies indicate that there are distinct INCENP–Aurora B and Survivin–INCENP–Aurora B complexes, raising the possibility that discrete passenger complexes have specific anaphase functions, although the precise function of each subcomplex is not known (Gassmann et al., 2004). Localization of passenger proteins to the equatorial cortex in anaphase before furrow initiation and the ability of Aurora B to modify the RhoA regulator MgcRacGAP suggests that passenger proteins function early, before furrow ingression (Earnshaw and Cooke, 1991; Eckley et al., 1997; Minoshima et al., 2003). Consistent with this early cytokinetic role, inhibition of the Aurora B homologue in Caenorhabditis elegans shows that it is required early in anaphase but dispensable later for cytokinesis completion (Severson et al., 2000).
In yeast, Ipl1p, Sli15p, and Bir1p localize to the interpolar microtubules of the anaphase spindle and their loading on the spindle requires the dephosphorylation of Sli15p early in anaphase (Buvelot et al., 2003; Pereira and Schiebel, 2003; Bouck and Bloom, 2005; Gillis et al., 2005). Many studies have shown that Bir1p interacts with the Ndc10p subunit of the core kinetochore complex, centromere binding factor 3 (CBF3) (Yoon and Carbon, 1999; Bouck and Bloom, 2005; Gillis et al., 2005). Recently, it has been suggested that a phosphorylated form of Bir1p is required to recruit Ndc10p to the anaphase spindle (Widlund et al., 2006). Interestingly, mutation of the Bir1p phosphorylation sites causes shorter anaphase spindles, whereas a loss of Ipl1p function results in longer anaphase spindles (Buvelot et al., 2003; Widlund et al., 2006). These findings raise the possibility that passenger proteins directly or indirectly modulate anaphase spindle behavior.
In yeast, many core kinetochore proteins have been observed on the anaphase spindle in addition to the passenger complexes already discussed. Remarkably, subunits of the CEN–DNA binding complex CBF3 have also been observed to associate with the anaphase spindle (Buvelot et al., 2003; Bouck and Bloom, 2005; Gillis et al., 2005; Widlund et al., 2006). CBF3 consists of three core subunits, Ndc10p, Cep3p, and Ctf13p; the stepwise assembly, as well as the turnover, of these subunits requires Skp1p, Sgt1p, and HSP90 (Stemmann et al., 2002; Rodrigo-Brenni et al., 2004). Assembly is required for CBF3 to bind stably to the specific CEN–DNA sequence CDEIII. Loss of CBF3 from the CEN prevents the recruitment of all known kinetochore subunits (McAinsh et al., 2003), highlighting the critical role in kinetochore nucleation played by this complex. Recently, we have shown that inhibiting CBF3 assembly or turnover leaves CBF3 intact on CEN–DNA but compromises cytokinesis by altering the organization of septins, arguing there are distinct roles for CBF3 in mitosis (Gillis et al., 2005). Septins are filamentous structures that form at the mother-daughter bud neck, and they are critical for polarized growth as well as cytokinesis (Faty et al., 2002; Longtine and Bi, 2003). Specifically, blocking CBF3 assembly was observed to inhibit the dynamic behavior of septins in anaphase, resulting in partially disassembled and improperly positioned septin rings (Gillis et al., 2005). Conditional alleles of BIR1 gave rise to a similar defect in septin organization, suggesting that CBF3 and Bir1p function together to regulate septins during anaphase. Together, these findings support the possibility that passenger complexes regulate multiple aspects of mitosis, including spindle behavior and septin dynamics.
To address how passenger complexes function to regulate anaphase events in yeast, we have performed a structure–function analysis of the Survivin homologue BIR1. Consistent with previously published results, we show that only the carboxy terminus of Bir1p is required for its essential function (Yoon and Carbon, 1999; Li et al., 2000; Widlund et al., 2006). This region of Bir1p mediates the formation of a passenger complex that includes Sli15p and Ipl1p. Biochemical analysis demonstrates that the majority of Bir1p associates with Sli15p but only with a minor fraction of total Ipl1p. Mutations that disrupt the Bir1p–Sli15p interaction prevent recruitment of Bir1p to kinetochores and the anaphase spindle, but they do not affect the localization of Sli15p or Ipl1p. We interpret this result to suggest that there is a hierarchy of complexes anchored by Sli15p through its interaction with metaphase and anaphase microtubules. Importantly, we find that loss of Bir1p from kinetochores and the anaphase spindle does not affect the resolution of mono-attached kinetochores but that it does compromise septin organization and cytokinesis. We propose that Bir1p and Sli15p form a passenger complex that functions independently of Ipl1p to regulate septin dynamics in anaphase.
MATERIALS AND METHODS
Yeast Growth and Strain Construction
BIR1 deletion constructs and site-directed mutants were generated by plasmid amplification using complimentary primer sets (sequences available upon request) containing the coding sequence change, and they were verified by DNA sequencing. A plasmid shuffle strategy was used to introduce BIR1 constructs on a LEU2 CEN/ARS plasmid into a haploid strain containing a chromosomal deletion of BIR1 and an episomal copy of BIR1 marked with URA3 (see strain list for more information; Supplemental Material). The wild-type copy of BIR1 was selected against on 5′-fluoroorotic acid (5′-FOA) minimal media plates lacking leucine and containing 2% dextrose. We did not observe suppressors when the BIR1 deletion was uncovered in our plasmid “shuffle” strain (i.e., loss of the wild-type BIR1 covering plasmid; Supplemental Figure S1C). Other strains were grown as indicated using standard yeast medium.
For yeast two-hybrid analyses, NDC10 and SLI15 were cloned into the GAL4 binding domain vector (pGBD-C1) (James et al., 1996) as described previously (pUD256 and pUD364, respectively) (Gillis et al., 2005). IPL1 and CEP3 were cloned into pGBD-C1 with BamHI and SalI (pUD404 and pUD406, respectively). The BIR1 deletion constructs and site-directed mutants were digested with BamHI and XhoI, and they were cloned into the GAL4 activation domain vector (pGAD-C1) (James et al., 1996) at BamHI and SalI (pUD347, pUD413, and pUD414). IPL1 and NDC10 truncation constructs were cloned into pGBD-C1 with BamHI and SalI (pUD455-459 and pUD445-449, respectively). The SLI15 truncation constructs were digested with BamHI and XhoI, and they were cloned into pGBD-C1 at BamHI and SalI (pUD450-454). Bait and prey plasmids were transformed into the two-hybrid strain AH109 (BD Biosciences Clontech, Palo Alto, CA) and isolated on minimal media plates lacking tryptophan and leucine. They were assayed for growth on plates also lacking histidine to detect two-hybrid interactions. We added 6 mM 3′-amino-1,2,4-triazole to plates for two-hybrid interactions involving BD–SLI15 constructs. IPL1 and SLI15 plasmids were generously provided by Clarence S.M. Chan (University of Texas). Lac O/I GFP fusion strains were generously provided by Sue W. Biggins (Fred Hutchinson Cancer Research Center, Seattle, WA).
For determining cell viability, the indicated strains were grown to log phase at 30°C. One thousand cells were plated to four plates, and colonies were counted after growth at 30°C. Cell viability was calculated as the average number of colonies produced per plate divided by the total number of cells plated, and the standard deviation was determined between the four plates.
Yeast Extracts and Immunopurifications
Cell extracts were prepared as described previously (Rodrigo-Brenni et al., 2004). Briefly, cells were washed in ice-cold water followed by extract buffer containing 50 mM Bis-Tris propane, 100 mM β-glycerol phosphate, 5 mM EDTA, 250 mM KCl, 10% glycerol, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 1 mM N-tosyl-l-phenylalanine chloromethyl ketone, and 10 μg/ml leupeptin, pepstatin, and chymostatin. Washed cells were resuspended 1:1 (vol/vol) with extract buffer and ground in the presence of liquid nitrogen by using a mortar and pestle (Kaplan and Sorger, 1997). Lysates were cleared by centrifugation at 21000 × g for 15 min. Protein concentrations were determined by Bradford assay per manufacturer's instructions (Bio-Rad, Hercules, CA). To purify the indicated epitope-tagged fusion protein, 5 mg of cell extract was diluted to 500 μl in extract buffer and incubated overnight at 4°C with 5 μl of the indicated antibody. Antibodies were captured using 15 μl of GammaBind Plus Sepharose beads (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom), incubating for 2 h at 4°C. Immunocomplexed beads were washed three times with extract buffer, and proteins were resolved on 7.5% SDS-polyacrylamide gel electrophoresis (PAGE) gels. For glycerol gradient sedimentation, 1.6 mg of the indicated yeast extract was layered to the top of a 2-ml 0–35% glycerol gradient made using extract buffer (see above) and centrifuged at 134,000 × g for 12 h at 4°C. Then, 50-μl fractions were collected from the top (0% glycerol) to the bottom (35% glycerol) of the gradient, and they were precipitated by bringing each fraction to 15% (wt/vol) trichloroacetic acid (TCA) and incubating at −20°C for 1 h. TCA precipitates were pelleted by centrifugation at 21,000 × g for 15 min, washed with 200 μl of acetone, and air-dried. Sample fraction pellets were resuspended in SDS-PAGE loading buffer and resolved on 7.5% SDS-PAGE gels. Immunoblotting was performed as described previously (Rodrigo-Brenni et al., 2004), using anti-Myc (9E10; Santa Cruz Biotechnology, Santa Cruz, CA) antibodies at 1/1000, anti-hemagglutinin (HA) (12CA5; Abcam, Cambridge, MA) antibodies at 1/4000, and polyclonal Bir1p and Sli15p sera, generously provided by Arshad Desai (University of California at San Diego, La Jolla, CA). Western blot quantification was conducted using an Odyssey infrared imaging system (LI-COR, Lincoln, NE) according to manufacturer's instructions.
Microscopy
Cells containing the indicated GFP gene fusions were placed on 1.7% agarose pads made with synthetic complete media containing the same carbon source as the culture media as described previously (Hoepfner et al., 2000). Z-sections (0.2 μm) were collected to capture the entire volume of the cell using a Nikon E600 epifluorescence microscope (Nikon, Tokyo, Japan) equipped with a Nikon 60× (numerical aperture 1.35) oil immersion lens, and recorded with a Hamamatsu Orca ER charge-coupled device camera (Hamamatsu, Bridgewater, NJ) controlled by Simple PCI software (www.cimaging.net). Images were formatted using Adobe Photoshop version 7.0 (Adobe Systems, Mountain View, CA). For time-lapse imaging, the indicated cells were placed on agarose pads as described above. A water-jacketed heated stage was used to maintain cells at 30°C. We collected 4-μm Z-stacks every 3 min using Z-sweep acquisition with a Deltavision microscope (Applied Precision, Issaquah, WA). Images were deconvolved using SoftWoRx software and rendered into avi movies at 4 frames per second by using Simple PCI software.
Chromosome Fragment Loss Assay
To evaluate chromosome fragment loss, the indicated BIR1 alleles were integrated into the BIR1 plasmid shuffle strain at the LEU2 locus containing a chromosomal tester fragment used to measure chromosome loss (Spencer and Hieter, 1992). The covering plasmid containing wild-type BIR1 was selected against by streaking cells onto 5′-FOA plates lacking histidine. Yeast strains were then grown to log phase in minimal media lacking histidine and containing 2% dextrose. Cells were plated on YEP-agar low adenine plates containing 2% dextrose. The number of sectored colonies (equal to or greater than ½ the colony) was determined, and the fragment loss -fold increase was calculated by subtracting the number of red colonies from the total number of colonies, and then dividing the number of sectored colonies by the total.
Flow Cytometry
Cells were harvested, washed once with 50 mM sodium citrate, pH 7.5, resuspended in 70% ethanol, and stored at 4°C. Fixed cells were washed into 1 ml of 50 mM sodium citrate, pH 7.5, containing 0.1 mg of RNase A and 0.5 mg of Proteinase K (Roche Diagnostics, Mannheim, Germany), and then they were incubated at 50°C for 1 h. We added 2.5 μl of 10% Triton X-100 to each sample and briefly sonicated. SYBR Green was added to samples according to manufacturer's instructions (Invitrogen, Carlsbad, CA). Samples were analyzed using a BD Biosciences FACSCalibur flow cytometer (BD Biosciences, Franklin Lakes, NJ).
RESULTS
The Carboxy Terminus of Bir1p Is Required for Proper Cell Growth
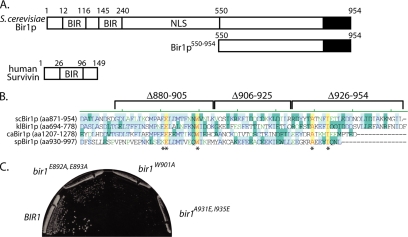
Functional analysis of Sli15p has shown that its interaction with the Ipl1p kinase is required for the proper biorientation of chromosomes during metaphase (Kang et al., 2001; Tanaka et al., 2002). Recent studies have shown that the passenger protein Bir1p interacts with Sli15p and with the CBF3 subunit Ndc10p (Yoon and Carbon, 1999; Bouck and Bloom, 2005; Gillis et al., 2005; Widlund et al., 2006). Although it has been suggested that the CBF3–Bir1p–Slip15 complex links kinetochores to microtubules in metaphase (Sandall et al., 2006), functional data have to date only implicated Bir1p in anaphase spindle and septin regulation (Gillis et al., 2005; Widlund et al., 2006). There are at least two explanations for these seemingly contradictory findings: 1) there are distinct Sli15p complexes that function in metaphase and in anaphase, or 2) there is a single Sli15p complex with multiple functions in both metaphase and anaphase. To begin to distinguish between these possibilities, we used deletion analysis to identify the regions of Bir1p required for its essential role in cell growth. Consistent with results that were published as this work was being carried out (Widlund et al., 2006), we observed that the BIR domains in the amino terminus are not required for the essential function of Bir1p and that a conserved region in the carboxy terminus (amino acids 880-954) is required (Figure 1A and Supplemental Figure S1). Therefore, we concentrated our analysis on the essential carboxy terminal region of Bir1p.
Figure 1.
The carboxy terminus of Bir1p is required for proper cell growth. (A) Bir1p domain diagram. Bir1p contains two BIR domains (amino acids [aa]12-116 and aa145-240), a proposed nuclear localization signal (aa288-295), and a carboxy-terminal region (black) well conserved in other yeast Bir1p proteins. In contrast, the human homologue Survivin is smaller and contains a single BIR domain (aa26-96). The region 550-954 represents the portion of Bir1p analyzed in subsequent experiments. (B) The carboxy termini of Bir1p homologues from S.cerevisiae, Kluyveromyces lactis, Candida albicans, and S.pombe were aligned; blue represents identity between at least two proteins, green represents a conservative change, and yellow represents identity between all proteins (the asterisk [*] indicates amino acids mutated). (C) Cells containing the indicated BIR1 alleles (pUD286 and 423-425) were grown at 30°C on 5-fluoroorotic acid-containing plates; strain growth with empty vector included in Supplemental Figure S1C (see Materials and Methods).
Although Saccharomyces cerevisiae BIR1 shares very little sequence homology with Survivin homologues in metazoans, the carboxy terminal 100 amino acids shares significant conservation with Bir1p from other budding yeast and more limited conservation with fission yeast Bir1p (Figure 1B). In particular, we noticed that four amino acids are very well conserved: E893, W901, A931, and I935 (S.cerevisiae numbering). To determine whether this conserved group of amino acids is essential for growth, we mutated them and we evaluated their ability to support growth in the absence of the wild-type BIR1 (see Materials and Methods). Changing the charged amino acids to alanines (bir1E892A, E893A) had a moderate effect on cell growth (Figure 1C); in contrast, changing the conserved tryptophan (bir1W901A) or substituting charged residues for alanine and isoleucine significantly inhibited cell growth (bir1A931E, I935E; Figure 1C). In data not shown, we observed that the bir1A931E, I935E allele is slow growing at lower temperatures (25–30°C) but that it is more compromised for growth at 37°C, consistent with this allele having a partial loss of the essential function of BIR1. We conclude that the highly conserved amino acids in the carboxy terminus are important for the essential function of Bir1p.
Because of the growth defect in the bir1A931E, I935E strains, we were concerned that the genetic suppression previously reported for BIR1 deletions may have given rise to suppressors in our analysis (Li et al., 2000). After backcrossing bir1A931E, I935E to a wild-type strain and analyzing the meiotic progeny, we observed reproducible segregation of bir1A931E, I935E defects, arguing that these strains do not have unlinked suppressing mutations. In addition, in multiple isolates of bir1A931E, I935E we observed very consistent growth behavior and normal ploidies, in contrast to suppressors isolated from BIR1 deletions that grew at variable rates and were always associated with whole genome increases in ploidy (data not shown; see Materials and Methods). Finally, as we demonstrate, the phenotypic characteristics of the BIR1 point mutants are similar to anaphase phenotypes observed in SLI15 mutants that are not prone to suppression, making it unlikely that the phenotypes we observed are modulated by suppression (Figure 7 and Supplemental Figure S3).
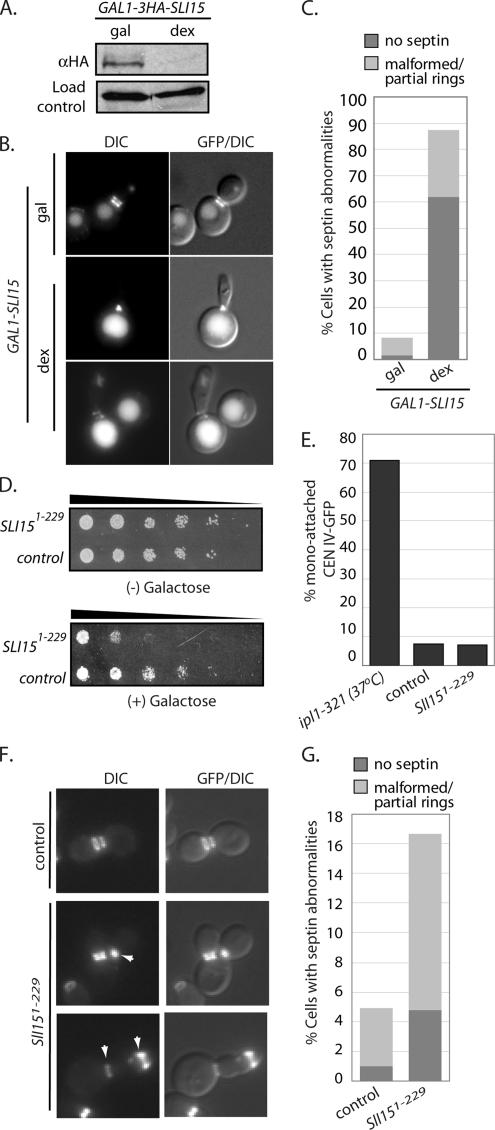
Figure 7.
Sli15p is required for proper septin organization. (A) Extracts were prepared after the GAL1–3HA-SLI15 strain was grown in medium containing galactose or after shifting to medium containing dextrose (inhibit transcription) for 5 h. The “load-control” represents a nonspecific HA-reactive band observed in all of our immunoblots. (B) Representative fluorescent (GFP) or DIC and fluorescent overlaid (GFP/DIC) images are shown from cells containing GAL-3HA-SLI15 and expressing Cdc11p-GFP that were grown in medium containing galactose (gal) and then shifted to dextrose (dex) for 8 h. (C) The percentages of cells with no septins, malformed, or partial septin rings were quantified after cells were grown under the indicated conditions. (D) A dilution series (indicated by the sloping line) of cells containing either pGAL-SLI151-229 (“Sli151-229”; pUD499) or pGAL (“control”; pUD108) were plated onto medium with (+) or without (−) galactose and grown at 30°C for 3 d. (E) After 18 h of growth in galactose, the percentage of large-budded cells with mono-attached CEN IV-GFP was determined in the indicated strains and compared with ipl1-321 cells at 37°C. A similar analysis was carried out after releasing cells from an α-factor induced arrest (see Supplemental Figure S3). (F) Representative fluorescent (GFP) or DIC and fluorescent overlaid (GFP/DIC) images from indicated strains expressing Cdc11p-GFP that were grown in media containing galactose for 18 h are shown. The arrowhead in the middle left panel indicates a partially formed septin ring. The arrowheads in the bottom left panel indicate malformed septin rings. (G) In cells from F, the percentage with septin abnormalities (no septins, dark gray; malformed/partial rings, light gray) was quantified.
Bir1p Interacts Independently with Ndc10p, Sli15p, and Ipl1p
We used yeast two-hybrid analysis to confirm that the BIR1 deletions we constructed behave as predicted from published work (Widlund et al., 2006; see Materials and Methods). As demonstrated previously, we found that amino acids 550-954 of Bir1p (AD-Bir1550-954) are sufficient to interact with the CBF3 subunit Ndc10p (BD-NDC10), and the region between amino acids 591-690 (AD-Bir1Δ591-690) is required for Bir1p to interact with Ndc10p (see summary in Table 1 and Supplemental Figure S1D and E). We also observed that Bir1p does not interact with the other CBF3 subunits (Cep3p and Ctf13p), demonstrating the specificity of the two-hybrid assay (data not shown). Bir1p550-954 was also able to interact with both Ipl1p (BD-IPL1) and Sli15p (BD-SLI15), a result that contrasts with biochemical purification data using a tandem affinity tag-Bir1p, which did not identify Ipl1p associated with Bir1p (Widlund et al., 2006). The single point mutant W901A (AD-BIR1W901A) prevented the interaction with Bir1p and Ipl1p, but it did not affect its ability to interact with Sli15p. In contrast, the double mutant, A931E and I935E (AD-BIR1A931E, I935E) abolished both the interaction with Sli15p and Ipl1p (Table 1 and Supplemental Figure S1). Together, these data suggest that separate domains of Bir1p can mediate its interaction with other passenger proteins, and they raise the possibility that Bir1p forms a larger passenger protein complex that includes both Sli15p and Ipl1p.
Table 1.
Yeast two-hybrid summary
| Strains tested | BD-NDC10 | BD-SLI15 | BD-IPL1 |
|---|---|---|---|
| AD-IPL1 | − | N.D.a | N.A. |
| AD-SLI15 | − | N.A. | N.D.a |
| AD-birΔ550-954 | + | + | + |
| AD-birΔ591-690 | − | + | + |
| AD-birW901A | + | + | − |
| AD-birA931E, I935E | + | − | − |
N.D., not determined; N.A., not applicable.
a From Kang et al. (2001).
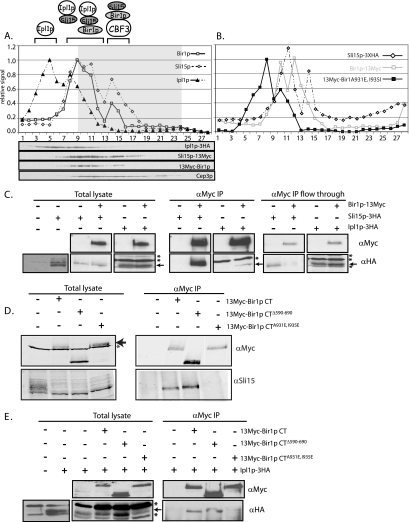
To characterize the types of passenger complexes that form in cells, we created a yeast strain containing epitope-tagged versions of Bir1p, Ipl1p, and Sli15p (Ipl1p–3HA, Sli15p–3HA, and episomal 13Myc–Bir1p; see Materials and Methods and Table 1), and we used glycerol gradient sedimentation to separate protein complexes based on their sedimentation coefficient. We were only able to isolate triple-tagged strains in the presence of wild-type Bir1p (i.e., 13Myc–Bir1p/Sli15p–3HA/Ipl1p–3HA; Bir1p). However, double-tagged strains with the wild-type BIR1 deleted gave similar sedimentation results and thereby validate our use of the triple-tagged strain (Supplemental Figure S2A and B). In extracts from cells grown in log phase, we observed three distinct populations of Ipl1p, Bir1p, and Sli15p (Figure 2A). Fractions containing only Ipl1p (2–5) likely represent a pool of uncomplexed protein, which is consistent with reports that there is a diffuse pool of Ipl1p in the nucleus (Kang et al., 2001). Fractions where Ipl1p comigrates with Sli15p and Bir1p (6–10) potentially represent complexes containing Ipl1p–Sli15p or Bir1p–Sli15p–Ipl1p. Finally, fractions where only Bir1p and Sli15p comigrate (12–15) may reflect a separate Bir1p–Sli15p complex. However, this faster migrating population raises the possibility that other proteins are part of this complex; the comigration of Cep3p (a CBF3 subunit) in these fractions is consistent with the possibility that interaction with CBF3 subunits gives rise to the faster migrating Bir1p–Sli15p complexes (Figure 2B; immunoblot and gray shaded region in graph). To show that the migration of these complexes reflects passenger protein interactions, we examined the behavior of complexes in a strain with Sli15p–3HA and 13myc–Bir1pA931E, I935E (an allele that fails to interact with Sli15p or Ipl1p by yeast two-hybrid; Table 1) in place of the wild-type BIR1. In this strain, we observed that the bulk of 13myc–Bir1pA931E, I935E migrates slower than wild-type Bir1p (Figure 2B). The migration of Sli15p–3HA is confined to a narrower peak, consistent with loss of only a subset of Sli15p complexes. Although it is possible that other interactions affect the migration of Bir1p and Sli15p complexes, these data are consistent with the presence of at least three distinct populations of passenger complexes (see diagram in Figure 2A).
Figure 2.
Bir1p interacts independently with Ndc10p, Sli15p, and Ipl1p. (A and B) Extracts from yeast were separated by glycerol gradient sedimentation and immunoblotted; fractions were collected from the top of the gradient (fraction 1), quantified after immunoblot, and expressed as relative signals. (A) Separation of extracts containing 13Myc–Bir1p (□), Sli15p–13Myc (◇), and Ipl1p–3HA (▴). Resolution of 13Myc–Bir1p and Sli15p–13Myc was possible due to their distinct molecular weights. The gray-boxed region indicates the migration of the CBF3 subunit, Cep3p (also shown as an immunoblot below). The cartoon of complexes indicates possible interactions that can be rationalized with two-hybrid and immunopurification data, and they are not meant to encompass all possibilities. (B) The migration of 13Myc–Bir1pA931E, I935E (■) and Sli15p–3HA (◇) on glycerol gradients is presented, and the position of wild-type Bir1p–13Myc (□) is overlaid for comparison. Immunopurifications were carried out as described in the text by using the monoclonal α-Myc antibody (9E10), and immunoblots were performed to identify Bir1p–13Myc (α-Myc), Sli15p–3HA (α-HA), untagged Sli15p (α-Sli15p), and Ipl1p–3HA (α-HA). An equivalent fraction of “flow through” (C) (extract after isolation of protein G-α-Myc–Sepharose beads; see Materials and Methods) and extract (C–E) was loaded. The indicated 13Myc–BIR1 constructs were expressed on an episomal plasmid (pUD359, 361, and 479; see strain list; Supplemental Material). Arrows indicate the Ipl1p–3HA band; the asterisk [*] indicates the position of nonspecific protein bands. The control panel in E compares the immunoblot signal from extracts in the absence or presence of the 3HA epitope tag to distinguish background bands from the specific signal.
To directly examine the interaction of Bir1p with other passenger protein homologues, we created the following strains expressing the indicated epitope fusions in place of the wild-type genes (see Materials and Methods): Sli15p–3HA alone, Ipl1p–3HA alone, Bir1p–13Myc and Sli15p–3HA, or Bir1p–13Myc and Ipl1p–3HA (Figure 2C, total lysate). As expected, neither Ipl1p–3HA nor Sli15p–3HA was detected after immunopurification by using the Myc monoclonal antibody in strains lacking Bir1p–13Myc. In contrast, immunopurification of Bir1p–13Myc depleted the majority of Bir1p–13Myc and codepleted a similar fraction of Sli15p–3HA (Figure 2C; α-Myc IP compare to α-Myc IP flow through). The immunopurifications are consistent with the behavior of these proteins in glycerol gradients; the bulk of Sli15p comigrates and copurifies with Bir1p, whereas a minority of Ipl1p comigrates and copurifies with Bir1p.
To test the effect of BIR1 mutants on complex formation, we first examined the association between Bir1p and Sli15p. We expressed the indicated BIR1 mutants in a strain with wild-type BIR1 deleted, and we used a polyclonal antibody to detect wild-type Sli15p (kindly provided by Sandall and Desai, Ludwig Institute for Cancer Research, UCSD, La Jolla, CA; Sandall et al., 2006). As expected, wild-type Sli15p did not copurfiy when a Myc-immunopurification was performed in a strain with untagged Bir1p; in the presence of 13Myc–Bir1p, 13Myc–Bir1pΔ591-690 (Figure 2D) or 13Myc–Bir1pW901A (data not shown), Sli15p was efficiently copurified. In contrast, Sli15p did not copurify with 13Myc–Bir1pA931E, I935E, supporting yeast two-hybrid and glycerol gradient analyses that together argue that these two amino acid changes disrupts the Bir1p–Sli15p complex (Figure 2D).
We next examined the effect of BIR1 mutants on the copurification of Ipl1p. To detect Ipl1p, we tagged it with HA in strains expressing the indicated BIR1 mutant. Copurification of Myc complexes showed that Ipl1p–3HA could interact with 13Myc–Bir1p and 13Myc–Bir1pΔ591-690 but not with 13Myc–Bir1pW901A (data not shown) or 13Myc–Bir1pA931E, I935E (Figure 2E). Despite attempts with multiple affinity tags, we failed to copurify Ndc10p with Bir1p; this contrasts with two-hybrid data and previously published in vitro binding assays by using rabbit reticulocyte lysates or purified Myc–Bir1p from yeast extracts (Gillis et al., 2005; Widlund et al., 2006). We speculate that the interaction between Ndc10p and Bir1p may be transient in vivo and labile in extracts and may be further compromised by the affinity tags used. When considered together, the yeast two-hybrid, the sedimentation analyses, and the immunopurifications are consistent with there being distinct domains in Bir1p that interact with multiple passenger proteins, resulting in the formation of at least three passenger protein complexes in yeast extract (see cartoon in Figure 2A).
Sli15p Can Mediate Multiple Passenger Complex Interactions
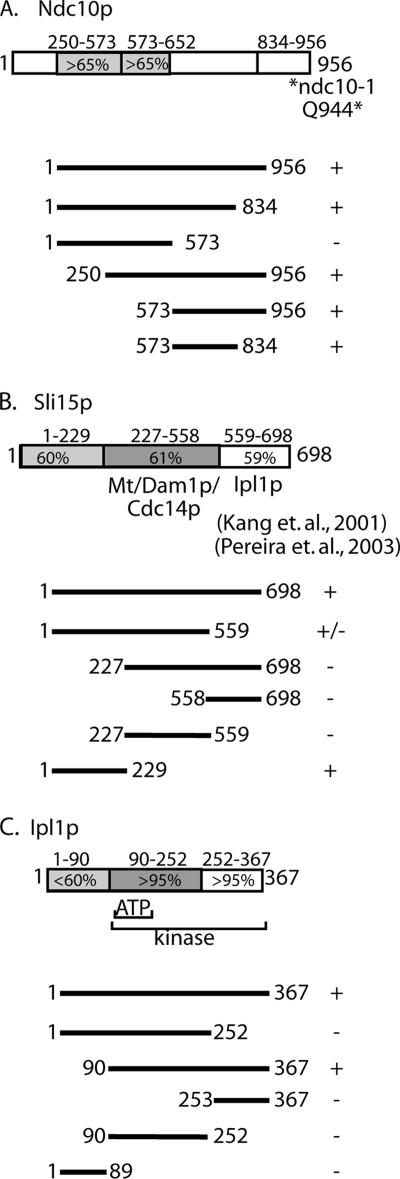
One implication of our interaction data are that Bir1p acts to link other passenger proteins to form one large complex (i.e., CBF3–Bir1p–Sli15p–Ipl1p). This predicts that each passenger protein is composed of distinct binding domains that interact with Bir1p. To test this prediction, we used yeast two-hybrid analysis to identify the Bir1p interaction domains on Ndc10p, Sli15p, and Ipl1p. Truncations of Ndc10p were constructed based on regions of homology with other budding yeast homologues (see diagram in Figure 3A). Yeast two-hybrid results indicate that sequences in a carboxy terminal block of conserved amino acids are critical for the interaction with Bir1p (amino acids 573-834; Figure 3B). Interestingly, the extreme carboxy terminus of Ndc10p (amino acids 834-956), which contains the amino acid changes associated with the ndc10-1 (Q944*) and ndc10-2 (A914T) conditional alleles (Pearson et al., 2003), is not required for its interaction with Bir1p (Figure 3A). This observation argues that the loss of function in the ndc10-1 allele results in chromosome segregation failures that are not linked to its interaction with Bir1p.
Figure 3.
Mapping the Bir1p binding domain in Ndc10p, Sli15p, and Ipl1p by directed two-hybrid experiments. (A) Schematic of the relevant Ndc10p domains as discussed in Results. Percentages indicate the level of similarity among different yeast homologues (Eremothecium gossypii and Candida glabrata). The relative position of the amino acid change associated with the ndc10-1 mutation is indicated by an asterisk (Pearson et al., 2003). The line diagrams below the schematic indicate the domains of Ndc10p included in the directed yeast two-hybrid experiments with Bir1p (pUD445 to pUD449); + indicates growth and − indicates no growth. (B) Schematic of Sli15p domains depicts domains determined from previously published mapping experiments (pUD450-454) (Kang et al., 2001; Pereira and Schiebel, 2003); the line diagrams and growth classifications are as described in A. (C) Schematic of domains found in Ipl1p based on homology with other yeast homologues (K. lactis, C.glabrata, and E.gossypii) and the human Aurora B; the central region (aa90-252) contains the predicted ATP binding site; the region from aa90 to aa367 comprises the serine/threonine kinase domain. The line diagrams of the IPL1 plasmids (pUD455-459) and growth classifications are as described in A. For the directed two-hybrid analysis, BIR1 was fused to the GAL4 activation domain and NDC10, SLI15 and IPL1 truncations were fused to the GAL4 binding domain. Cells were tested for two-hybrid interaction by plate growth.
Previous domain mapping of Sli15p is consistent with the ability of Sli15p to simultaneously mediate multiple protein–protein interactions (see diagram in Figure 3B); the middle region of Sli15p (227-559) was shown to interact with microtubules, Dam1p, and Cdc14p, whereas the carboxy terminus of Sli15p (558-698) was shown to interact with Ipl1p (Kang et al., 2001; Pereira and Schiebel, 2003). Interestingly, we found that the amino terminus (1-229) is sufficient for the interaction with Bir1p (Figure 3B). This result supports the possibility that Sli15p can simultaneously interact with Bir1p, Ipl1p, and microtubules.
Ipl1p is predicted to have a serine/threonine protein kinase domain beginning at amino acid 104 and spanning most of the carboxy terminus, and sequence alignment shows that this region is highly conserved among yeast versions of Ipl1p (see diagram in Figure 3C). In contrast, the first 90 amino acids is less conserved with other yeast Ipl1p or with human Aurora B (<12% identity). We found that the first 90 amino acids of Ipl1p is not necessary for Ip1p to interact with Bir1p. In contrast, truncating the kinase domain (construct amino acids 1-89) or splitting the kinase domain (constructs 1-252 and 253-367) eliminated the interaction with Bir1p (Figure 3C). This result could indicate that Bir1p interacts as a substrate with the kinase domain of Ipl1p. We conclude that Ndc10p, Sli15p, and Ipl1p have discrete Bir1p interacting domains, further supporting the ability of these proteins to form separate complexes composed of distinct groupings of subunits (e.g., CBF3–Bir1p–Sli15p–Ipl1p, Bir1p–Sli15p–Ipl1p, or Sli15p–Ipl1p).
Sli15p Separately Localizes Bir1p–CBF3 and Ipl1p to the Anaphase Spindle
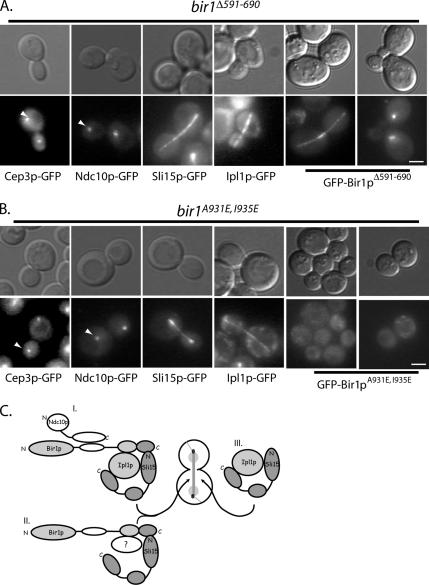
The ability of Sli15p to interact independently with Bir1p (amino terminus), microtubules (middle domain), and Ipl1p (carboxy terminus) and the ability of Bir1p to link Sli15p to the CBF3 complex (via Ndc10p) led us to ask whether the Bir1p–Sli15p core complex is required to localize all passenger protein components to the anaphase spindle. To address this question, we examined the hierarchy of spindle associated passenger proteins fused to GFP in strains that contain the BIR1 mutants. The bir1Δ591-690 mutation inhibits the interaction between Bir1p and Ndc10p based on yeast two-hybrid data (Table 1 and Supplemental Figure S1). Consistent with other localization studies (e.g., Widlund et al., 2006), we observed that neither of the CBF3 subunits, Cep3p-GFP or Ndc10p-GFP, were able to localize to the anaphase spindle in this strain (44.5 and 0% of large-budded cells [n > 50] exhibited Ndc10p-GFP on anaphase spindles in BIR1 and bir1Δ591-690, respectively; Figure 4A). However, we observed that Bir1pΔ591-690-GFP, Sli15p-GFP, and Ipl1p-GFP all localized to the anaphase spindle (Figure 4A). Similarly, Bir1pΔ591-690-GFP, Sli15p-GFP, and Ipl1p-GFP also localized to CEN-proximal clusters, suggesting that their association with kinetochores is independent of the Bir1p–Ndc10p interaction (Figure 4A; data not shown); this result is inconsistent with a CBF3–CEN linkage to microtubules being mediated by a Bir1p–Sli15p complex as proposed previously (Sandall et al., 2006; see Discussion). We conclude that amino acids 591-690 of Bir1p are required to recruit CBF3 subunits, but not Sli15p and Ipl1p, to the anaphase spindle.
Figure 4.
Sli15p separately localizes Bir1p–CBF3 and Ipl1p to the anaphase spindle. Fluorescent (GFP) or differential interference contrast (DIC) images of representative anaphase cells expressing GFP fusions to the indicated CBF3 passenger proteins in bir1Δ591-690 cells (pUD325) (A) and bir1A931E, I935E (pUD425) (B) cells are shown. Arrowheads indicate the position of the kinetochore-localized GFP signals. (C) Diagram of proposed passenger protein complexes as presented in Results and Discussion. Separate microtubule, Ipl1p, and Bir1p binding domains on Sli15p are depicted to independently link CBF3-Bir1p (complex I), Bir1p–Sli15p complexes (complex II) or Sli15p–Ipl1p complexes (complex III) to the anaphase spindle. All bars, 4 μm.
To determine whether the localization of the other passenger proteins depend on the Bir1p–Sli15p complex, we analyzed the effect of bir1A931E, I935E on the localization of Ipl1p and Sli15p. Bir1pA931E, I935E fails to interact with either Ipl1p or Sli15p and it fails to localize CBF3 subunits to the anaphase spindle (44.5 and 0% of large-budded cells [n > 50] exhibited Ndc10p-GFP on anaphase spindles in BIR1 and bir1A931E, I935E strains, respectively; Figure 4B). The failure of Bir1pA931E, I935E-GFP to localize to the spindle confirms the importance of the Bir1p–Sli15p interaction in vivo, and it argues that Sli15p brings Bir1p to spindle microtubules through its microtubule binding domain (Figure 4B). Interestingly, Bir1pA931E, I935E-GFP fails to localize to kinetochores as well as to spindle microtubules, indicating that Bir1p may localize to kinetochores and the anaphase spindle via a similar Sli15p linkage to microtubules. Finally, both Ipl1p and Sli15p properly localize to the anaphase spindle in bir1A931E, I935E arguing that a distinct Sli15p–Ipl1p complex can associate with the anaphase spindle independently of Bir1p (Figure 4B). These findings support our biochemical data that there are multiple, distinct Sli15p–passenger complexes in cells and is summarized in the model (Figure 4C).
The Bir1p–Sli15p Interaction Is Required for Proper Chromosome Segregation
As has been reported previously, Bir1p localizes near kinetochores as well as the anaphase spindle, and it has been implicated in chromosome segregation (Yoon and Carbon, 1999; Widlund et al., 2006). To relate the function of Bir1p in chromosome segregation with its ability to interact with other passenger proteins, we analyzed BIR1 deletions and point mutants in a chromosome loss assay (see Materials and Methods; Table 2). Deletions of essential carboxy terminal sequences gave the highest increase in chromosome loss (Table 2; 28.4- and 19.6-fold increase in chromosome loss for bir1Δ906-929 and bir1Δ930-954, respectively). These increases in chromosome loss were similar for the double point mutant bir1A931E, I935E that prevents the interaction of Bir1p with Sli15p. Surprisingly, cells expressing Bir1pΔ591-690, which fails to interact with Ndc10p, exhibit no increase in the rate of chromosome loss. We interpret this finding to mean that the interaction between Bir1p and Ndc10p is not required for the kinetochore function of CBF3.
Table 2.
Chromosome fragment loss
| Strain tested | Total colonies | Red colonies | Half-red sectored | Loss rate | Fold increase |
|---|---|---|---|---|---|
| BIR1 | 2782 | 1 | 2 | 7.2 × 10−4 | 1.0 |
| birΔ550-954 | 2872 | 0 | 2 | 7.0 × 10−4 | 1.0 |
| birΔ591-690 | 5077 | 0 | 0 | N.D. | N.D. |
| birΔ880-905 | 2113 | 32 | 14 | 67.3 × 10−4 | 9.4 |
| birΔ906-929 | 2912 | 131 | 57 | 204.0 × 10−4 | 28.4 |
| birΔ930-954 | 3190 | 62 | 44 | 140.7 × 10−4 | 19.6 |
| birA931E, I935E | 3360 | 113 | 42 | 129.4 × 10−4 | 18.0 |
N.D., not determined.
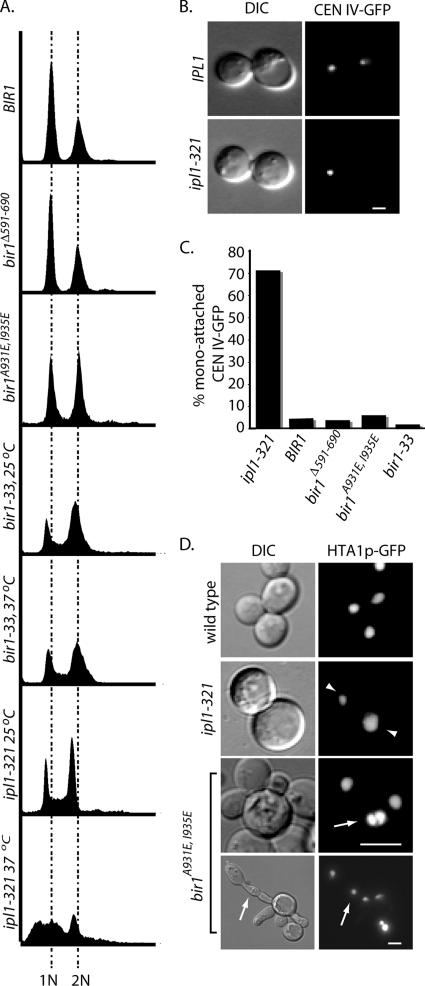
Although increases in chromosome loss can reflect kinetochore failure, the rates of loss that have been observed for BIR1 mutants are modest compared with core kinetochore subunits. For example, the increases in chromosome loss rates for ctf13-30 (50-fold; Doheny et al., 1993), skp1-4 (100-fold; Yoon and Carbon, 1999), and CTF19Δ (100-fold; Hyland et al., 1999) are more dramatic than BIR1 mutants, and in the case of CTF19Δ do not compromise cell growth. Furthermore, we observed no obvious defect in the ability of kinetochores in bir1A931E, I935E mutants to assemble and cluster CENs at the spindle pole, even though Bir1pA931E, I935E-GFP fails to localize to kinetochores (Figure 4B). One possible explanation for the elevated rates of chromosome loss is that BIR1 mutants alter the activity of Ipl1p, possibly by compromising the CBF3–Bir1p–Sli15p–Ipl1p complex. IPL1 mutants fail to resolve mono-attached kinetochores, resulting in dramatic changes in chromosome ploidy (Biggins et al., 2001; Tanaka et al., 2002). To test this possibility, we first compared the DNA content of the ipl1-321 allele with BIR1 mutants. As expected, at nonpermissive temperature ipl1-321 cells exhibited a large proportion of cells with less than 1N and greater than 2N DNA content (Figure 5A), consistent with severe aneuploidy and previously published results (Tanaka et al., 2002). bir1Δ591-690 is comparable with wild-type BIR1, and both strains exhibit distinct 1N and 2N peaks. In bir1A931E, I935E cells, we observe a slight enrichment of 2N cells with a very modest appearance of >2N cells (Figure 5A). Because bir1A931E, I935E cells grow poorly at all temperatures, we chose to also examine the more potent temperature sensitive bir1-33 allele. Consistent with decreased function of bir1-33, we observed a more dramatic enrichment of 2N cells and a modest enrichment of large-budded cells in this strain (data not shown). Importantly, the bir1 alleles exhibit profiles that are clearly distinct from ipl1-321, suggesting BIR1 and IPL1 functions in maintaining proper chromosome segregation are different.
Figure 5.
The carboxy terminus of Bir1p is required for proper cytokinesis. (A) Cells from bir1550-954, bir1Δ591-690, and bir1A931E, I935E were grown to log phase at 30°C. ipl1-321 and bir1-33 cells were grown at the indicated temperature for 3 h. Cells were analyzed for DNA content by flow cytometry (see Materials and Methods). 1N and 2N DNA content positions based on wild-type controls are indicated by the dashed lines. (B) Representative fluorescence and DIC images of wild-type (IPL1, 37°C; top) and mutant IPL1 (ip1l-321 37°C; bottom) cells containing CEN IV-GFP are shown. (C) The percentage of cells with mono-attached CEN IV-GFP was determined in large-budded cells of the indicated strains. ipl1-321 and bir1-33 cells were grown at 37°C, whereas BIR1, bir1Δ591-690, and bir1A931E, I935E cells were grown at 30°C. (D) Fluorescent or DIC images of the indicated strains containing Hta1p-GFP are shown. Arrowheads point to uneven segregation of Hta1p-GFP in ipl1-321 cells grown at 37°C. Arrows indicate cells with multiple nuclei. All bars, 4 μm.
To more directly examine the possibility that BIR1 mutants affect the ability of Ipl1p to resolve mono-attached kinetochores, we monitored the behavior of a GFP-tagged CEN on chromosome IV (see Materials and Methods). Consistent with previous studies, we observe >70% of large budded ipl1-321 cells grown at nonpermissive temperature have a mono-attached CEN-GFP phenotype; when the GFP signal is confined to a single large bud (Figure 5, B and C; Biggins et al., 1999). In contrast, <10% of BIR1, bir1Δ591-690, bir1A931E, I935E, and bir1-33 cells had mono-attached CEN-GFP (Figure 5C). Visualization of chromosomes in cells expressing Hta1p (histone H2A)-GFP in ipl1-321 cells highlight the high frequency of uneven chromosome content, presumably due to multiple misoriented chromosomes (Figure 5D, arrowheads). A similar phenotype was not observed when BIR1 alleles were examined; instead, we observed a modest number of bir1A931E, I935E cells that contained multiple nuclei, consistent with defective cytokinesis (Figure 5D, arrows). Although we cannot rule out that the BIR1 alleles we examined are selectively wild type for regulating kinetochore attachment, our results are more consistent with BIR1 having an IPL1-independent role in mitosis (see Discussion).
The Bir1p–Sli15p Complex Is Required for Proper Septin Organization
We have shown previously that inhibiting CBF3 assembly or turnover compromises septin organization in anaphase, preventing proper septin ring separation and disassembly (Gillis et al., 2005). Similarly, the conditional mutants bir1-30 and bir1-33 result in disorganized septins. In contrast, mutations in IPL1 or mutations in SLI15, isolated as synthetically lethal with ipl1-2, failed to affect septin organization (Kim et al., 1999; Gillis et al., 2005). We interpreted these results to suggest that CBF3 and Bir1p function together to regulate septin dynamics, independently of Sli15p and Ipl1p. One caveat noted at the time is that the SLI15 alleles we analyzed were likely to affect Ipl1p activity but not other Sli15p–passenger protein functions (see Discussion; Gillis et al., 2005). Using the alleles we generated in this study, we asked whether Sli15p–passenger protein interactions are necessary for the regulation of septins.
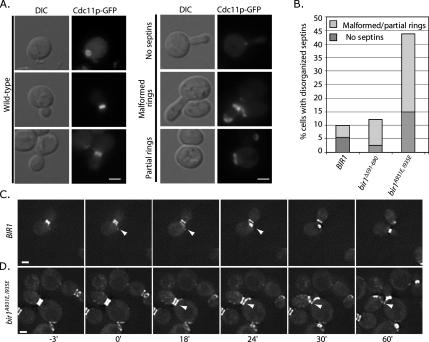
We used strains expressing the Cdc11p septin fused to GFP and carrying episomal copies of the indicated BIR1 mutants over a deletion of the chromosomal copy of BIR1 (see Materials and Methods). We quantified several classes of septin defects: 1) cells with either malformed septins (inappropriately positioned in the bud neck) or partially formed rings (see examples in Figure 6A) and 2) cells lacking a septin ring. Surprisingly, the loss of the Bir1p–Ndc10p interaction in bir1Δ591-690 cells resulted in only an ∼twofold increase in cells with partly assembled or malformed septin rings (Figure 6, A and B). This is a modest effect compared with that observed in mutants that block CBF3 assembly (>5-fold increase over control; Gillis et al., 2005), and it argues that blocking CBF3 assembly may indirectly affect the ability of Bir1p complexes to regulate septins. Consistent with this possibility, we noticed that inhibiting CBF3 assembly by turning off transcription of the core Ctf13p subunit reduces overall levels of Bir1p and Sli15p in extracts and immunopurifications, but it does not affect the apparent affinity of Bir1p for Sli15p (Supplemental Figure S2C and D). This result implies that Bir1p–Sli15p complexes are important and limiting for septin regulation (see Discussion).
Figure 6.
The Bir1p–Sli15p complex is required for proper septin organization. (A) Cells expressing Cdc11p-GFP were grown at 30°C; representative fluorescent or DIC images showing cells with wild type, disorganized (partial and malformed), and no septins are presented. (B) The percentages of cells with no septins (dark gray) or malformed/partial septins (light gray) were quantified in the indicated strains. Frames from movies of wild-type (C; BIR1) or mutant (D; bir1A931E, I935E) cells expressing Cdc11p-GFP are presented highlighting the behavior of septins during anaphase (also see Movies 1 and 2 in Supplemental Material). The zero time point was assigned as the first frame with visible septin ring separation. Arrowheads in C indicate the position of the axially assembled septin ring in the mother cell. Arrowheads in D indicate the stabilized septin ring in the mother cell. All bars, 4 μm.
To directly assess the importance of the Bir1p–Sli15p interaction for septin regulation, we analyzed septins in the bir1A931E, I935E strain. We observed a significant increase in the percentage of cells with defective septins (43%, >5-fold; Figure 6B), arguing that the interaction between Bir1p and Sli15p is required for septin regulation. One prediction based on our previous work is that this interaction will affect septins specifically during anaphase (Gillis et al., 2005). To test this prediction, we used live-cell imaging to characterize septin dynamics in BIR1 mutants during anaphase. Mitotic cells were identified by bud size and by septin morphology; early mitotic cells exhibit distinct but largely unseparated rings (e.g., Figure 6C, −3-min panels, our unpublished observations; for review, see Faty et al., 2002). We assigned the zero time point to the frame of the movie where septin ring separation was first observed. In wild-type cells, we observed the separation of rings, followed by the initiation of disassembly of the mother septin ring after 18 min (see arrow in wild type panel in Figure 6C and Movie 1 in Supplemental Material). Septin ring disassembly and the formation of a new septin ring in an axial position in the mother cell were completed by 24 min; a new bud was observed to form by 60 min. In contrast, bir1A931E, I935E cells exhibited a clear defect in septin ring disassembly. By 18 min when wild-type cells had begun disassembly, we observed no reduction in the mother septin ring signal in bir1A931E, I935E, even though a faint septin signal at an axial position is visible (see arrow in bir1A931E, I935E panel in Figure 6D and Movie 2 in Supplemental Material). The original mother cell septin ring remains even after a new bud has formed (Figure 6D, arrow, and bir1A931E, I935E panel, 60′ time point). Cells that retain hyperstabilized septin rings go on to attempt another round of cell division, and they exhibit dramatic polarity defects that results in elongated buds, mispositioned septin rings, and in many cases, cell lysis (see Movie 3 in Supplemental Material; our unpublished observations). These results are consistent with our previous observations of CBF3 mutants, and they argue that like CBF3 mutants, the Bir1p–Sli15p interaction is required for the proper timing of septin ring disassembly and reassembly in anaphase.
Sli15p Is Required for Septin Organization
Alleles of BIR1 that compromise its interaction with Sli15p prevent the normal anaphase regulation of septins. If this complex is really involved in septin regulation, we predict that perturbing Sli15p function should also give rise to defects in septin regulation. We noted above that alleles of SLI15 reported in the literature were likely to be specific to Ipl1p regulation based on how they were selected (Kim et al., 1999). To analyze septin phenotypes associated with loss of Sli15p, we placed the genomic copy of SLI15 under control of the regulated galactose promoter, and we fused it to three HA epitope tags (see Sandall et al., 2006 for a similar approach). The growth of cells in medium containing dextrose (promoter off) results in the loss of detectable 3HA–Sli15p (Figure 7A). We observed a very strong septin phenotype after loss of 3HA–Sli15p; a significant fraction of cells had partial or misformed septin rings (>25%) and a larger fraction of cells had no septins (>60%; Figure 7, B and C). The approximately twofold increase in septin defects relative to bir1A931E, I935E is likely to reflect the relative severity of the alleles, i.e., bir1A931E, I935E is a hypomorph, whereas transcriptional shutoff of Sli15p is close to null. As predicted, loss of 3HA–Sli15p also compromised chromosome segregation and mitotic spindle integrity (Sandall et al., 2006; data not shown). Together, our biochemical and genetic results argue that Sli15p plays multiple roles during mitosis and that a Sli15p–Bir1p complex is specifically required to regulate septins during anaphase.
Although our data argue for the importance of the Bir1p–Sli15p interaction, it remains possible that additional Bir1p or Sli15p interactions are required for septin regulation. A prediction from the mapping data is that overexpression of the amino terminus of Sli15p (amino acids 1-229) that interacts with Bir1p would disrupt the Sli15p–Bir1p complex and compromise cell growth (Figure 3). To test this prediction, we transformed Sli15p1-229 under control of the GAL promoter (pGAL-SLI151-229) or the empty GAL promoter vector (pGAL-empty) in wild-type cells, and we examined the effect on cells in the presence of galactose. Both the pGAL-SLI151-229 and pGAL-empty vectors had no effect on the growth of yeast when transcription was repressed (−galactose). In contrast, when grown on medium containing galactose (+galactose), cell growth was inhibited in the presence of pGAL-SLI151-229, whereas the pGAL-empty control had a negligible effect on cell growth (Figure 7D). Analysis of cell viability in the presence of this fragment suggests that it only minimally impacts the interaction between Sli15p and Bir1p (Supplemental Figure S3); perhaps not surprisingly, we could not detect a change in the levels of Sli15p–Bir1p complexes isolated from extracts in the presence of Sli15p1-229 (data now shown). Nonetheless, consistent with our prediction, overexpression of the Sli15p domain that interacts with Bir1p dominantly interferes with cell division.
One explanation for the inhibition of colony growth (Figure 7D) is that expression of Sli15p1-229 results in mitotic failures and cell death due to increases in mono-attached chromosomes. To test this possibility, we overexpressed Sli15p1-229 during a single cell cycle after release from an α-factor block; we observed no increase in mono-attached CENs under these conditions (Supplemental Figure S3C). To address whether long-term expression of the Sli15p1-229 fragment increased mono-attached CENs, we also monitored cells grown in galactose over the course of multiple doublings (∼18 h) and similarly observed no increase in mono-attached CENs (Figure 7E). As expected, ipl1-321 exhibited a high rate of mono-attached CENs after 3 h at nonpermissive temperature (>70%; Figure 7E). These results suggest that the lethality caused by overexpression of the amino-terminal fragment of Sli15p is not due to increases in mono-attached chromosomes.
As our results with CBF3 depletion hinted, we reasoned that even subtle effects on the levels of Sli15p–Bir1p complex could result in septin defects. To test this possibility, we grew strains expressing Cdc11p–GFP and containing either pGAL-empty or pGAL-SLI151-229 in medium supplemented with galactose. A majority of cells with the pGAL-empty vector had normal septin organization, with <5% exhibiting malformed or partial septin rings (Figure 7, F and G). In contrast, when Sli15p1-229 was overexpressed, we observed a modest increase in cells with multi- or elongated buds (>16%; Figure 7, E and F, pGAL-SLI151-229) and malformed septins (Figure 7E, arrowhead in bottom panel) or partially formed septins (Figure 7E, arrowhead in top panel). A similar increase in disorganized septins was observed when cells were released from an α-factor arrest and monitored through mitosis in the presence or absence of Sli15p1-229 (Supplemental Figure S3A and B), arguing that inhibiting the Sli15p–Bir1p interaction has a rapid impact on septin regulation. Although the septin defect is modest compared with bir1A931E, I935E cells, we suspect this is because the dominant fragment has a limited ability to compete for Bir1p binding. Together, these results suggest that septins are extremely sensitive to the total levels of Sli15p–Bir1p in the cell. In total, the similarities between Sli15p depletion and overexpression of the Bir1p-interaction domain to BIR1 mutants are consistent with a role for the Bir1p–Sli15p complex in anaphase septin regulation.
DISCUSSION
We have used budding yeast to address the function of kinetochore–passenger proteins in mitosis. A long-held precept in the study of kinetochore–passenger proteins is that their multiple subcellular locations allows them to coordinate early and late mitotic events, thus ensuring their proper temporal sequence. Our findings are consistent with this notion. We show that there are multiple, distinct kinetochore–passenger complexes. We propose that Sli15p and Bir1p can mediate multiple sets of kinetochore–passenger interactions. We envision that there are minimally Sli15p–Ipl1p, CBF3–Bir1p–Slip15p–Ipl1p and Bir1–Sli15p complexes (see model in Figure 4C). Genetic analyses show that the Bir1p–Sli15p interaction is critical for septin regulation in anaphase. However, neither conditional alleles of BIR1 nor alleles that fail to localize to the kinetochore affect the resolution of mono-attached kinetochores, arguing for a separation of kinetochore–passenger protein function during mitosis. We speculate that the formation of distinct kinetochore passenger complexes allows for a separation of mitotic activities to ensure that the physical events of mitosis occur in their proper order.
Multiple Mitotic Functions of Kinetochore–Passenger Complexes
Findings from studies in animal cells and the work presented here support the idea that distinct kinetochore–passenger complexes have specific functions in mitosis. In studies from animal cells, a proportion of Aurora B (Ipl1p in yeast) is found associated with INCENP (Sli15p in yeast) and Survivin (Bir1p in yeast), whereas a second population of Aurora B is associated only with INCENP (Gassmann et al., 2004). This result is consistent with our findings that show a small proportion of Ipl1p is found associated with Sli15p and Bir1p, whereas a separate pool is found associated only with Sli15p. We propose that the Bir1p–Sli15p complex functions in anaphase to regulate cytokinesis through septins, whereas the Sli15p–Ipl1p complex carries out the metaphase function of resolving mono-attached kinetochores.
The evidence for multiple kinetochore–passenger complexes comes from a combination of yeast two-hybrid, biochemical studies, targeted mutation analyses, and localization studies that serve as in vivo confirmation of the biochemistry. The two-hybrid and localization data argue that Bir1p links CBF3 subunits to a larger passenger complex containing Slip15p and Ipl1p (complex I in model, Figure 4C). These findings are consistent with previous biochemical studies demonstrating the interaction between Ndc10p and Bir1p (Yoon and Carbon, 1999; Gillis et al., 2005; Montpetit et al., 2006; Widlund et al., 2006). Consistent with their physical interaction, we show that Bir1p is required for CBF3 subunits to localize to the anaphase spindle via Sli15p; the ability of multiple CBF3 subunits to associate with the anaphase spindle but only Ndc10p to interact with Bir1p argues that assembled CBF3 complexes are on interpolar microtubules in anaphase. This complex can be functionally separated from other passenger complexes by bir1Δ591-690, which prevents Ndc10p and Cep3p from associating with the anaphase spindle but does not alter septin organization (see below for discussion of CBF3 function and septin regulation). We speculate that the CBF3–Bir1p complex may regulate other anaphase events; indeed, we have observed a decrease in the rate of anaphase spindle elongation when CBF3 is compromised or in bir1Δ591-690 cells (Hansen, Rozelle, and Kaplan, unpublished data). It is therefore possible that CBF3–Bir1p–Sli15p complexes distinctly affect spindle microtubules.
The second distinct complex we observe is a complex that minimally includes Bir1p and Sli15p (complex II in model, Figure 4C). This complex is biochemically distinct in that it does not require CBF3 (bir1Δ591-690) or Ipl1p (bir1W901A; our unpublished observations) for assembly or for anaphase spindle localization. Like the CBF3–Bir1p–Sli15p complex, the Bir1p–Sli15p complex seems to be functionally distinct as well; elimination of the Bir1p–Sli15p interaction (bir1A931E, I935E) prevents proper regulation of septin dynamics in anaphase. Our analysis does not rule out the possibility that there are other unidentified proteins that also contribute to the function of this complex in anaphase. Interestingly, the behavior of Sli15p on glycerol gradients in the absence of Bir1p interaction suggests there may be other Sli15p complexes (Figure 2B). As shown in the model (Figure 4C), we propose that Bir1p–Sli15p forms the core of multiple distinct passenger complexes that regulate multiple events in anaphase.
Finally, we suggest that the Bir1p–Sli15p–Ipl1p module is functionally distinct from the Slip15p–Ipl1p complex that has a well-characterized role in resolving mono-attached chromosomes (complex III in model, Figure 4D). This argument is based on two lines of evidence: 1) our biochemical analyses of complexes and 2) our functional analyses of BIR1 alleles. Analysis of complexes by immunopurifications or glycerol gradient sedimentation is consistent with there being a major fraction of Bir1p-associated with Sli15p, a minor fraction that also includes Ipl1p and a separate Sli15p–Ipl1p complex (Figure 2). Importantly, we did not observe an increase in mono-attached kinetochores in the BIR1 alleles we analyzed, arguing that Bir1p is not an important participant with Ipl1p in resolving mono-attached kinetochores. Although we cannot rule out that different alleles of BIR1 might compromise resolution of mono-attached kinetochores, current data are not consistent with such a role.
Recently, Sandall et al. (2006) proposed a model whereby CBF3–Bir1p–Sli15p links metaphase chromosomes to microtubules before stable attachment, acting as a tension sensor. However, our finding that Bir1pΔ591-690 still localizes to kinetochores and does not affect the resolution of mono-attached kinetochores argues against this model. Interestingly, the ability of Sli15p and Ipl1p to remain associated with the anaphase spindle in the absence of Bir1p suggests Sli15p–Ipl1p may function independently in anaphase cells. This possibility is supported by our finding that loss of Ipl1p from the Bir1p–Sli15p complex (bir1W901A) does not affect resolution of mono-attached chromosomes (Thomas and Kaplan, unpublished observations), but it is nevertheless compromised for cell growth and chromosome segregation. This additional role of Ipl1p may reflect the recently described requirement for Ipl1p in the “no-cut” pathway (Norden et al., 2006) or it may represent the reported role for Ipl1p in regulating anaphase spindle elongation (Buvelot et al., 2003). In summary, the work presented here represents an important step in linking passenger protein complexes to the regulation of discrete mitotic events.
CBF3–Passenger Complexes and Septin Regulation
How do CBF3–Bir1p passenger protein complexes contribute to the regulation of anaphase progression? Our previous results demonstrated that inhibiting CBF3 assembly caused defects in the regulation of septin dynamics in anaphase (Gillis et al., 2005). Here, we show that mutants that block the Ndc10p–Bir1p interaction paradoxically have a minor effect on septin regulation. Instead, our data argue that the interaction between Bir1p and Sli15p is critical for regulating anaphase septin dynamics. To explain this paradox, we propose that multiple kinetochore–passenger complexes exist in a dynamic relationship with each other. Thus, compromising one complex may impact a separate but related complex because individual subunits are exchanged or shared. Consistent with this type of dependent relationship, we find that inhibiting the assembly of CBF3 reduces the levels of Bir1p and Sli15p but not their affinity for each other (Supplemental Figure S2). Although there may be multiple reasons why protein levels would change when the balance of complexes is altered, it is intriguing to note the connection between these complexes and the heat-shock protein of 90 kDa (HSP90)–Sgt1p chaperone complex. Mutations that compromise HSP90–Sgt1p result in severe defects in septin organization (Gillis et al., 2005), and we have reported previously that CBF3 assembly and turnover is dependent on this chaperone complex (Lingelbach and Kaplan, 2004; Rodrigo-Brenni et al., 2004). An important question for the future is to understand how HSP90 chaperones affect kinetochore–passenger complexes and mitotic progression.
The Role of Bir1p in Chromosome Segregation
The increases in chromosome loss frequency that have been observed raise the possibility that BIR1 has a separate role in maintaining genomic stability (Table 1; Yoon and Carbon, 1999; Widlund et al., 2006). Clearly, the localization of Bir1p to kinetochores makes a compelling case for its involvement earlier in mitosis. One possibility is that Bir1p, together with Sli15p, stabilizes or clusters metaphase spindle microtubules, helping to enrich for plus-ends near unattached kinetochores. Consistent with Sli15p being a microtubule linker protein, deletion of the microtubule binding domain of Sli15p prevents it from localizing to either anaphase spindles or kinetochores in metaphase (Kang et al., 2001). One intriguing implication of a Bir1p–Sli15p microtubule organizer is that its role may not be limited to mitosis. Indeed, except for anaphase, Bir1p and Sli15p localize to kinetochores throughout the cell cycle. Clustering microtubules at kinetochores may be more important in G1 when unreplicated chromosomes are attached to microtubules at the spindle pole through the interaction of a single microtubule/chromatid. Little is known about the role of kinetochore–passenger proteins outside of mitosis, but it is worth keeping in mind nonmitotic roles of these microtubule-associated proteins, especially in budding yeast.
What accounts for the increased chromosome loss frequency in BIR1 mutants? One possibility is that defective anaphase segregation or cytokinetic failures result in loss of the chromosome fragment. As suggested above, BIR1 may play nonmitotic roles that are important for chromosome maintenance. In this regard, it is suggestive that psf2+, a component of the GINS complex involved in DNA replication, suppresses the bir1-46 mutant allele in fission yeast (Huang et al., 2005). Finally, evidence from a number of systems suggests that at least Aurora B and INCENP play an important role in recruiting condensin to chromosomes, and this function seems to require Bir1 in Schizosaccharomyces pombe (Giet and Glover, 2001; Morishita et al., 2001; Murnion et al., 2001; Crosio et al., 2002). It will be important to conduct a more detailed analysis of chromosome behavior in BIR1 mutants to address a possible role for Bir1p in chromosome maintenance independently of segregation.
Conserved Passenger Complex Module
In metazoans, evidence supports the view that multiple passenger complexes can form. In human cells, Gassmann et al. (2004) identified an INCENP–Aurora B complex that does not contain Survivin or other passenger proteins. This finding is consistent with our observation that the majority of Ipl1p (Aurora B) is not associated with Bir1p (Survivin) and fractionates in smaller molecular weight fractions after glycerol gradient sedimentation (Figure 2). This potentially uncomplexed pool of Ipl1p could represent a rapidly exchanging pool or a population that has low affinity for its binding partners. In human cells, it seems that the passenger protein Borealin interacts with Survivin and INCENP via distinct regions, but it does not directly associate with Aurora B. Although yeast do not have a Borealin homologue, yeast Bir1p is a much larger protein than Survivin and contains multiple unique domains, including the essential carboxy terminus that mediates its interaction with Sli15p (Figure 1A); this distinction raises the possibility that Bir1p acts analogously to Borealin, forming a complex containing INCENP and Aurora B.
In summary, we have used budding yeast as a model system to understand kinetochore–passenger proteins. We have characterized a unique passenger complex that contains the core CEN-DNA binding complex, CBF3, as well as conserved homologues of metazoan passenger proteins. Findings that CBF3–passenger complexes also affect anaphase spindle length (Buvelot et al., 2003; Bouck and Bloom, 2005; Widlund et al., 2006) suggest that passenger complexes may in general provide an important link between the physical separation of chromosomes in anaphase and the timing of specific cytokinetic events (i.e., septin ring dynamics). Further studies in yeast are necessary to shed light on the full range of mitotic events regulated by passenger complexes and to ultimately identify the downstream targets of passenger complexes.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Kaplan laboratory, the University of California at Davis Cytoskeletal club, and Dr. Frank McNally for important feedback on the manuscript. This work was supported by funding from American Cancer Society grants RSG-02-035-01 and RSG-02-035-05-CCG (to K.B.K.).
Abbreviations used:
- AD
activating domain
- BD
binding domain
- CBF3
centromere binding factor 3
- CEN
centromere
- DIC
differential interference contrast
- 5′-FOA
5′-fluoroorotic acid
- GFP
green fluorescent protein
- HA
hemagglutinin.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-03-0201) on July 25, 2007.

 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Adams R. R., Carmena M., Earnshaw W. C. Chromosomal passengers and the (aurora) ABCs of mitosis. Trends Cell Biol. 2001;11:49–54. doi: 10.1016/s0962-8924(00)01880-8. [DOI] [PubMed] [Google Scholar]
- Adams R. R., Wheatley S. P., Gouldsworthy A. M., Kandels-Lewis S. E., Carmena M., Smythe C., Gerloff D. L., Earnshaw W. C. INCENP binds the Aurora-related kinase AIRK2 and is required to target it to chromosomes, the central spindle and cleavage furrow. Curr. Biol. 2000;10:1075–1078. doi: 10.1016/s0960-9822(00)00673-4. [DOI] [PubMed] [Google Scholar]
- Biggins S., Bhalla N., Chang A., Smith D. L., Murray A. W. Genes involved in sister chromatid separation and segregation in the budding yeast Saccharomyces cerevisiae. Genetics. 2001;159:453–470. doi: 10.1093/genetics/159.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S., Murray A. W. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev. 2001;15:3118–3129. doi: 10.1101/gad.934801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S., Severin F. F., Bhalla N., Sassoon I., Hyman A. A., Murray A. W. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 1999;13:532–544. doi: 10.1101/gad.13.5.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop J. D., Schumacher J. M. Phosphorylation of the carboxyl terminus of inner centromere protein (INCENP) by the Aurora B Kinase stimulates Aurora B kinase activity. J. Biol. Chem. 2002;277:27577–27580. doi: 10.1074/jbc.C200307200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton M. A., Lan W., Powers S. E., McCleland M. L., Kuang J., Stukenberg P. T. Aurora B kinase exists in a complex with survivin and INCENP and its kinase activity is stimulated by survivin binding and phosphorylation. Mol. Biol. Cell. 2002;13:3064–3077. doi: 10.1091/mbc.E02-02-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouck D. C., Bloom K. S. The kinetochore protein Ndc10p is required for spindle stability and cytokinesis in yeast. Proc. Natl. Acad. Sci. USA. 2005;102:5408–5413. doi: 10.1073/pnas.0405925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buvelot S., Tatsutani S. Y., Vermaak D., Biggins S. The budding yeast Ipl1/Aurora protein kinase regulates mitotic spindle disassembly. J. Cell. Biol. 2003;160:329–339. doi: 10.1083/jcb.200209018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C. S., Botstein D. Isolation and characterization of chromosome-gain and increase-in-ploidy mutants in yeast. Genetics. 1993;135:677–691. doi: 10.1093/genetics/135.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I. M., Anderson S., Jwa M., Green E. M., Kang J., Yates J. R., 3rd, Chan C. S., Drubin D. G., Barnes G. Phospho-regulation of kinetochoremicrotubule attachments by the Aurora kinase Ipl1p. Cell. 2002;111:163–172. doi: 10.1016/s0092-8674(02)00973-x. [DOI] [PubMed] [Google Scholar]
- Crosio C., Fimia G. M., Loury R., Kimura M., Okano Y., Zhou H., Sen S., Allis C. D., Sassone-Corsi P. Mitotic phosphorylation of histone H3: spatiotemporal regulation by mammalian Aurora kinases. Mol. Cell. Biol. 2002;22:874–885. doi: 10.1128/MCB.22.3.874-885.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doheny K. F., Sorger P. K., Hyman A. A., Tugendreich S., Spencer F., Hieter P. Identification of essential components of the S. cerevisiae kinetochoreS. Cell. 1993;73:761–774. doi: 10.1016/0092-8674(93)90255-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw W. C., Cooke C. A. Analysis of the distribution of the INCENPs throughout mitosis reveals the existence of a pathway of structural changes in the chromosomes during metaphase and early events in cleavage furrow formation. J. Cell Sci. 1991;98(Pt 4):443–461. doi: 10.1242/jcs.98.4.443. [DOI] [PubMed] [Google Scholar]
- Eckley D. M., Ainsztein A. M., Mackay A. M., Goldberg I. G., Earnshaw W. C. Chromosomal proteins and cytokinesis: patterns of cleavage furrow formation and inner centromere protein positioning in mitotic heterokaryons and midanaphase cells. J. Cell Biol. 1997;136:1169–1183. doi: 10.1083/jcb.136.6.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faty M., Fink M., Barral Y. Septins: a ring to part mother and daughter. Curr. Genet. 2002;41:123–131. doi: 10.1007/s00294-002-0304-0. [DOI] [PubMed] [Google Scholar]
- Gassmann R., Carvalho A., Henzing A. J., Ruchaud S., Hudson D. F., Honda R., Nigg E. A., Gerloff D. L., Earnshaw W. C. Borealin: a novel chromosomal passenger required for stability of the bipolar mitotic spindle. J. Cell Biol. 2004;166:179–191. doi: 10.1083/jcb.200404001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giet R., Glover D. M. Drosophila aurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J. Cell Biol. 2001;152:669–682. doi: 10.1083/jcb.152.4.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis A. N., Thomas S., Hansen S. D., Kaplan K. B. A novel role for the CBF3 kinetochore-scaffold complex in regulating septin dynamics and cytokinesis. J. Cell Biol. 2005;171:773–784. doi: 10.1083/jcb.200507017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf S., Cole R. W., LaTerra S., Zimmer C., Schnapp G., Walter R., Heckel A., van Meel J., Rieder C. L., Peters J. M. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol. 2003;161:281–294. doi: 10.1083/jcb.200208092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoepfner D., Brachat A., Philippsen P. Time-lapse video microscopy analysis reveals astral microtubule detachment in the yeast spindle pole mutant cnm67. Mol. Biol. Cell. 2000;11:1197–1211. doi: 10.1091/mbc.11.4.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda R., Korner R., Nigg E. A. Exploring the functional interactions between Aurora B, INCENP, and survivin in mitosis. Mol. Biol. Cell. 2003;14:3325–3341. doi: 10.1091/mbc.E02-11-0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H. K., Bailis J. M., Leverson J. D., Gomez E. B., Forsburg S. L., Hunter T. Suppressors of Bir1p (Survivin) identify roles for the chromosomal passenger protein Pic1p (INCENP) and the replication initiation factor Psf2p in chromosome segregation. Mol. Cell. Biol. 2005;25:9000–9015. doi: 10.1128/MCB.25.20.9000-9015.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyland K. M., Kingsbury J., Koshland D., Hieter P. Ctf19p: a novel kinetochore protein in Saccharomyces cerevisiae and a potential link between the kinetochore and mitotic spindle. J. Cell Biol. 1999;145:15–28. doi: 10.1083/jcb.145.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P., Halladay J., Craig E. A. Genomic libraries and a host strain designed for highly efficient two- hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J., Cheeseman I. M., Kallstrom G., Velmurugan S., Barnes G., Chan C. S. Functional cooperation of Dam1, Ipl1, and the inner centromere protein (INCENP)-related protein Sli15 during chromosome segregation. J. Cell Biol. 2001;155:763–774. doi: 10.1083/jcb.200105029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan K. B., Sorger P. K. Purification of sequence-specific DNAbinding proteins. In: Creighton T.E., editor. Protein Function: A Practical Approach. Oxford: Oxford University Press; 1997. pp. 245–278. [Google Scholar]
- Kim J. H., Kang J. S., Chan C. S. Sli15 associates with the ipl1 protein kinase to promote proper chromosome segregation in Saccharomyces cerevisiae. J. Cell Biol. 1999;145:1381–1394. doi: 10.1083/jcb.145.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson M. A., Renduchitala K., Khodjakov A., Kapoor T. M. Correcting improper chromosome-spindle attachments during cell division. Nat. Cell Biol. 2004;6:232–237. doi: 10.1038/ncb1102. [DOI] [PubMed] [Google Scholar]
- Leverson J. D., Huang H. K., Forsburg S. L., Hunter T. The Schizosaccharomyces pombe aurora-related kinase Ark1 interacts with the inner centromere protein Pic1 and mediates chromosome segregation and cytokinesis. Mol. Biol. Cell. 2002;13:1132–1143. doi: 10.1091/mbc.01-07-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Flanary P. L., Altieri D. C., Dohlman H. G. Cell division regulation by BIR1, a member of the inhibitor of apoptosis family in yeast. J. Biol. Chem. 2000;275:6707–6711. doi: 10.1074/jbc.275.10.6707. [DOI] [PubMed] [Google Scholar]
- Li Y., Bachant J., Alcasabas A. A., Wang Y., Qin J., Elledge S. J. The mitotic spindle is required for loading of the DASH complex onto the kinetochore. Genes Dev. 2002;16:183–197. doi: 10.1101/gad.959402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingelbach L. B., Kaplan K. B. The Interaction between Sgt1p and Skp1p is regulated by HSP90 chaperones and is required for proper CBF3 assembly. Mol. Cell. Biol. 2004;24:8938–8950. doi: 10.1128/MCB.24.20.8938-8950.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., Bi E. Regulation of septin organization and function in yeast. Trends Cell Biol. 2003;13:403–409. doi: 10.1016/s0962-8924(03)00151-x. [DOI] [PubMed] [Google Scholar]
- McAinsh A. D., Tytell J. D., Sorger P. K. Structure, function, and regulation of budding yeast kinetochores. Annu. Rev. Cell Dev. Biol. 2003;19:519–539. doi: 10.1146/annurev.cellbio.19.111301.155607. [DOI] [PubMed] [Google Scholar]
- Minoshima Y., et al. Phosphorylation by aurora B converts MgcRacGAP to a RhoGAP during cytokinesis. Dev. Cell. 2003;4:549–560. doi: 10.1016/s1534-5807(03)00089-3. [DOI] [PubMed] [Google Scholar]
- Montpetit B., Hazbun T. R., Fields S., Hieter P. Sumoylation of the budding yeast kinetochore protein Ndc10 is required for Ndc10 spindle localization and regulation of anaphase spindle elongation. J. Cell Biol. 2006;174:653–663. doi: 10.1083/jcb.200605019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita J., Matsusaka T., Goshima G., Nakamura T., Tatebe H., Yanagida M. Bir1/Cut17 moving from chromosome to spindle upon the loss of cohesion is required for condensation, spindle elongation and repair. Genes Cells. 2001;6:743–763. doi: 10.1046/j.1365-2443.2001.00459.x. [DOI] [PubMed] [Google Scholar]
- Murnion M. E., Adams R. R., Callister D. M., Allis C. D., Earnshaw W. C., Swedlow J. R. Chromatin-associated protein phosphatase 1 regulates aurora-B and histone H3 phosphorylation. J. Biol. Chem. 2001;276:26656–26665. doi: 10.1074/jbc.M102288200. [DOI] [PubMed] [Google Scholar]
- Norden C., Mendoza M., Dobbelaere J., Kotwaliwale C. V., Biggins S., Barral Y. The NoCut pathway links completion of cytokinesis to spindle midzone function to prevent chromosome breakage. Cell. 2006;125:85–98. doi: 10.1016/j.cell.2006.01.045. [DOI] [PubMed] [Google Scholar]
- Pearson C. G., Maddox P. S., Zarzar T. R., Salmon E. D., Bloom K. Yeast kinetochores do not stabilize Stu2p-dependent spindle microtubule dynamics. Mol. Biol. Cell. 2003;14:4181–4195. doi: 10.1091/mbc.E03-03-0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G., Schiebel E. Separase regulates INCENP-Aurora B anaphase spindle function through Cdc14. Science. 2003;302:2120–2124. doi: 10.1126/science.1091936. [DOI] [PubMed] [Google Scholar]
- Petersen J., Hagan I. M. S. pombe aurora kinase/survivin is required for chromosome condensation and the spindle checkpoint attachment response. Curr. Biol. 2003;13:590–597. doi: 10.1016/s0960-9822(03)00205-7. [DOI] [PubMed] [Google Scholar]
- Pinsky B. A., Biggins S. The spindle checkpoint: tension versus attachment. Trends Cell Biol. 2005;15:486–493. doi: 10.1016/j.tcb.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Rodrigo-Brenni M. C., Thomas S., Bouck D. C., Kaplan K. B. Sgt1p and Skp1p modulate the assembly and turnover of CBF3 complexes required for proper kinetochore function. Mol. Biol. Cell. 2004;15:3366–3378. doi: 10.1091/mbc.E03-12-0887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandall S., Severin F., McLeod I. X., Yates J. R., 3rd, Oegema K., Hyman A., Desai A. A Bir1-Sli15 complex connects centromeres to microtubules and is required to sense kinetochore tension. Cell. 2006;127:1179–1191. doi: 10.1016/j.cell.2006.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson A. F., Hamill D. R., Carter J. C., Schumacher J., Bowerman B. The aurora-related kinase AIR-2 recruits ZEN-4/CeMKLP1 to the mitotic spindle at metaphase and is required for cytokinesis. Curr. Biol. 2000;10:1162–1171. doi: 10.1016/s0960-9822(00)00715-6. [DOI] [PubMed] [Google Scholar]
- Shang C., Hazbun T. R., Cheeseman I. M., Aranda J., Fields S., Drubin D. G., Barnes G. Kinetochore protein interactions and their regulation by the Aurora kinase Ipl1p. Mol. Biol. Cell. 2003;14:3342–3355. doi: 10.1091/mbc.E02-11-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer F., Hieter P. Centromere DNA mutations induce a mitotic delay in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 1992;89:8908–8912. doi: 10.1073/pnas.89.19.8908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmann O., Neidig A., Kocher T., Wilm M., Lechner J. Hsp90 enables Ctf13p/Skp1p to nucleate the budding yeast kinetochore. Proc. Natl. Acad. Sci. USA. 2002;99:8585–8590. doi: 10.1073/pnas.082223899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T. U., Rachidi N., Janke C., Pereira G., Galova M., Schiebel E., Stark M. J., Nasmyth K. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 2002;108:317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- Vagnarelli P., Earnshaw W. C. Chromosomal passengers: the fourdimensional regulation of mitotic events. Chromosoma. 2004;113:211–222. doi: 10.1007/s00412-004-0307-3. [DOI] [PubMed] [Google Scholar]
- Wheatley S. P., Carvalho A., Vagnarelli P., Earnshaw W. C. INCENP is required for proper targeting of Survivin to the centromeres and the anaphase spindle during mitosis. Curr. Biol. 2001;11:886–890. doi: 10.1016/s0960-9822(01)00238-x. [DOI] [PubMed] [Google Scholar]
- Widlund P. O., Lyssand J. S., Anderson S., Niessen S., Yates J. R., 3rd, Davis T. N. Phosphorylation of the chromosomal passenger protein Bir1 is required for localization of Ndc10 to the spindle during anaphase and full spindle elongation. Mol. Biol. Cell. 2006;17:1065–1074. doi: 10.1091/mbc.E05-07-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon H. J., Carbon J. Participation of Bir1p, a member of the inhibitor of apoptosis family, in yeast chromosome segregation events. Proc. Natl. Acad. Sci. USA. 1999;96:13208–13213. doi: 10.1073/pnas.96.23.13208. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.