Abstract
In Saccharomyces cerevisiae, minichromosome maintenance protein (Mcm) 10 interacts with DNA polymerase (pol)-α and functions as a nuclear chaperone for the catalytic subunit, which is rapidly degraded in the absence of Mcm10. We report here that the interaction between Mcm10 and pol-α is conserved in human cells. We used a small interfering RNA-based approach to deplete Mcm10 in HeLa cells, and we observed that the catalytic subunit of pol-α, p180, was degraded with similar kinetics as Mcm10, whereas the regulatory pol-α subunit, p68, remained unaffected. Simultaneous loss of Mcm10 and p180 inhibited S phase entry and led to an accumulation of already replicating cells in late S/G2 as a result of DNA damage, which triggered apoptosis in a subpopulation of cells. These phenotypes differed considerably from analogous studies in Drosophila embryo cells that did not exhibit a similar arrest. To further dissect the roles of Mcm10 and p180 in human cells, we depleted p180 alone and observed a significant delay in S phase entry and fork progression but little effect on cell viability. These results argue that cells can tolerate low levels of p180 as long as Mcm10 is present to “recycle” it. Thus, human Mcm10 regulates both replication initiation and elongation and maintains genome integrity.
INTRODUCTION
DNA replication is a highly regulated process in which multiprotein complexes generate an exact copy of a cell's DNA. These multiprotein complexes are assembled into replication forks. Fork assembly takes place at replication origins that are spread throughout the genome in all eukaryotic cells. The first step is the formation of a prereplicative complex (pre-RC) (Diffley et al., 1994), which is subsequently transformed into a preinitiation complex, and finally into a pair of functional replication forks that have the ability to unwind parental DNA and synthesize new daughter strands (Mendez and Stillman, 2003). DNA unwinding is likely catalyzed by the minichromosome maintenance (Mcm) 2-7 complex (Aparicio et al., 1997; Labib et al., 2000; Shechter et al., 2004) in conjunction with two coactivators, Cdc45 and GINS (Pacek and Walter, 2004; Gambus et al., 2006; Moyer et al., 2006; Pacek et al., 2006). The association of Cdc45 and GINS with DNA is interdependent (Kubota et al., 2003; Takayama et al., 2003), and it requires yet another factor, Mcm10 (Wohlschlegel et al., 2002; Gregan et al., 2003; Sawyer et al., 2004). Despite its name, Mcm10 is not a member of the MCM protein family, although it was isolated in the same genetic screen in budding yeast that identified the MCM2-7 genes (Solomon et al., 1992; Merchant et al., 1997).
Mcm10 is a constitutively nuclear DNA binding protein (Merchant et al., 1997; Burich and Lei, 2003) that is essential in yeast (Burich and Lei, 2003). Besides its role in DNA replication, Mcm10 has also been implicated in transcriptional silencing (Liachko and Tye, 2005). It is important to note that the general protein domain structure of Mcm10 is not unique in Saccharomyces cerevisiae, but it is highly conserved among eukaryotic species (Izumi et al., 2000; Ricke and Bielinsky, 2006). Mcm10 has a central oligonucleotide/oligosaccharide binding (OB)-fold (Ricke and Bielinsky, 2006), a signature motif of RNA- and single-stranded (ss) DNA binding proteins. Thus, it is not surprising that, in vitro, Mcm10 prefers ss over double-stranded (ds) DNA (Fien et al., 2004). Within the central OB-fold resides a proliferating cell nuclear antigen (PCNA) interacting protein motif (Fien et al., 2004; Das-Bradoo et al., 2006), or PIP-box, as well as a hydrophobic stretch, similar to the one found in the mobile loop of heat-shock factor (Hsp) 10 (Ricke and Bielinsky, 2006). Although the PIP box mediates the interaction with PCNA (Das-Bradoo et al., 2006), the Hsp10-like domain forms part of the interaction site with the catalytic subunit of DNA polymerase (pol)-α (Ricke and Bielinsky, 2004, 2006). Because purified S. pombe Mcm10 coimmunoprecipitates purified p180, the catalytic subunit of pol-α in humans, it is reasonable to conclude that the binding between Mcm10 and pol-α is direct and that it does not require any other mediators. Because Mcm10 is necessary for chromatin association of the entire pol-α/primase complex in budding yeast (Ricke and Bielinsky, 2004), it probably has a crucial role in lagging strand synthesis. The initiation of individual Okazaki fragments requires the repeated action of pol-α/primase (Nasheuer and Grosse, 1987) to generate a primed template onto which PCNA can be loaded, and it is conceivable that Mcm10 facilitates the RNA-DNA primer synthesis by pol-α/primase and may also be involved in the subsequent recruitment of PCNA (Das-Bradoo et al., 2006). Alternatively, Mcm10 itself may function as a primase. Schizosaccharomyces pombe Mcm10 purified from Escherichia coli has been reported to possess primase activity by the virtue of a C-terminal nucleotide transfer domain, which seems to be conserved among species (Fien and Hurwitz, 2006). The picture that is emerging for the role of Mcm10 in DNA replication is that it is involved in the initiation as well as the elongation steps. During initiation, Mcm10 promotes chromatin association of Cdc45, which, in turn, is likely required to activate the Mcm2-7 helicase (Wohlschlegel et al., 2002; Gregan et al., 2003; Sawyer et al., 2004). On unwinding, replication protein A stabilizes the ssDNA, which allows for the Mcm10-dependent loading of pol-α/primase (Walter and Newport, 2000). Mcm10 is further necessary to maintain DNA binding of pol-α/primase but not that of Cdc45 (Ricke and Bielinsky, 2004; Yang et al., 2005). This is most easily explained by the finding that Mcm10 controls the stability of the catalytic subunit of pol-α, regardless of whether pol-α is bound to chromatin or not (Ricke and Bielinsky, 2004). Consistent with this notion, we have observed that Mcm10 is in a complex with pol-α/primase throughout the cell cycle in budding yeast (Ricke and Bielinsky, 2006). Furthermore, overexpression of the catalytic subunit of pol-α, Cdc17/Pol1 in the absence of stoichiometric amounts of Mcm10 leads to rapid protein degradation in S. cerevisiae. In contrast, Cdc17/Pol1 is stable when Mcm10 is co-overexpressed (Ricke and Bielinsky, 2006). Whether this function of Mcm10 is conserved in other species has not been explored. Similarly, it has remained unclear how Mcm10 contributes to DNA replication in human cells. In HeLa cells, Mcm10 associates with chromatin at the G1/S transition and dissociates after S phase completion (Izumi et al., 2000, 2001). Nuclear localization studies using green fluorescent protein (GFP)-tagged Mcm10 further suggested that Mcm10 is involved in the activation of pre-RCs but that it may not have an active role at the replication fork (Izumi et al., 2004).
To better understand the role of Mcm10 during DNA replication in human cells, we introduced Mcm10-specific small interfering RNA (siRNA) into HeLa cells to deplete cells of Mcm10. After treatment, cells accumulated DNA damage and arrested in late S/G2 phase. Interestingly, depletion of Mcm10 caused a decrease in the catalytic subunit of pol-α, p180, but it had no effect on steady-state levels of the regulatory pol-α subunit, p68, or Cdc45. Together, our results argue that Mcm10 is not only required for replication initiation but also for DNA elongation and that the combined loss of Mcm10 and p180 impedes lagging strand synthesis, resulting in replication fork arrest, DNA damage, and cell death.
MATERIALS AND METHODS
Cell Culture and RNA Interference
HeLa cells were maintained in DMEM (Invitrogen, Carlsbad, CA), supplemented with 1% penicillin and streptomycin and 10% fetal bovine serum (FBS). Apoptosis was induced by adding staurosporine (STS) to 5 × 106 cells at a final concentration of 5 μM for up to 8 h (Han et al., 1999). Cells were irradiated using a 137Cs γ-ray source at a dose rate of 2.5 Gy/min.
Mcm10- and p180-specific siRNAs were synthesized by Dharmacon RNA Technologies (Lafayette, CO). The control siRNA (with at least 4 mismatches to all known human or mouse mRNA sequences) sense strand was 5′-UAGCGACUAAACACAUCAAUU-3′. Lamin A/C siRNA sense strand was 5′-GGUGGUGACGAUCUGGGCUUU-3′. Mcm10 siRNA sense strands were Mcm10si-1, 5′-GAACGGGACGGAAUGCUAAUU-3′, Mcm10si-2, 5′-GCACAAACUUGAUCAUCCAUU-3′, Mcm10si-3, 5′-GGAGGUGUGUUUAUCUAUCUU-3′ and Mcm10si-4, 5′-GGACGAAUUCCAAAGAAGUUU-3′. Mcm10 SMARTpool was a combination of siRNA1-4. For p180 depletion, a combination of four siRNAs (SMARTpool) was used. The sequences of the sense strands were as follows: p180-1, 5′-GCAGUAACAUCGAUUGUAAUU-3′, p180-2, 5′-GACAUUAGACGUUUCAUUAUU-3′, p180-3, 5′-UAACAUCGCUGGGAACAUUUU-3′, and p180-4, 5′-GCUCAAAGGAUUAGAUAUAUU-3′. SMARTpool siRNAs targeting Mcm10 or p180 were used with the experiments shown in Figures 3, 6, 7, and 8.
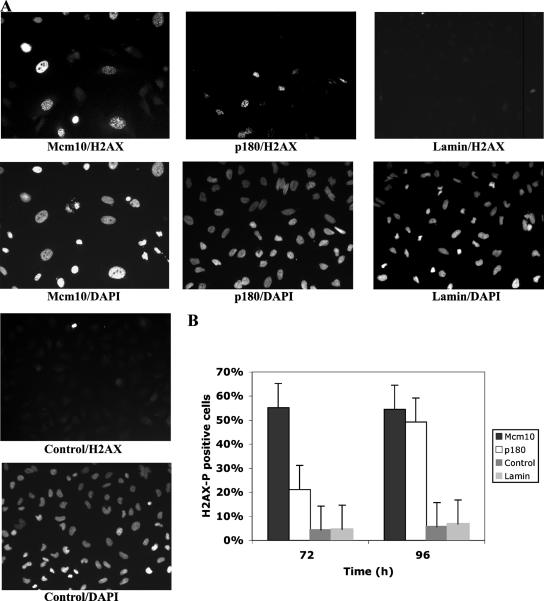
Figure 3.
Mcm10 and p180 knockdown cause an increase in γ-H2AX foci. (A) Fluorescence microscopy was applied to detect γ-H2AX foci in HeLa cells treated with Mcm10 siRNA1, p180-, lamin-, or control siRNA 72 h after transfection. (B) The percentage of γ-H2AX-positive cells in Mcm10-, p180-, lamin-, and control siRNA-treated cells was plotted at 72 and 96 h. Ten fields with 50 cells each were counted.
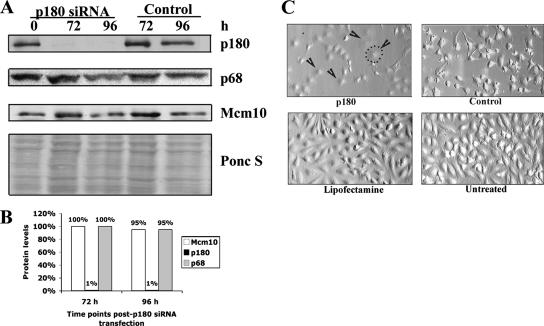
Figure 6.
The catalytic subunit of DNA polymerase-α (p180) is not required to stabilize Mcm10. (A) HeLa cells were transfected three times at 24-h intervals with siRNA against p180 or with control siRNA. Cells were harvested at the indicated time points, and immunoblotting was performed with anti-p180, anti-p68, and anti-Mcm10 antibodies. Ponceau S staining served as a loading control. (B) The percentage of each protein remaining at indicated time points was plotted when HeLa cells were treated with siRNA against p180. The amount of protein was normalized with respect to the loading control, and then it was expressed as a fraction of the amount in control siRNA-treated samples. (C) Images of p180-depleted and control HeLa cells at 96 h post-siRNA transfection are shown. Arrowheads point to some of the cells that exhibited a phenotype similar to Mcm10/p180-codepleted HeLa cells. The dotted line circles the cell membrane of one representative cell.
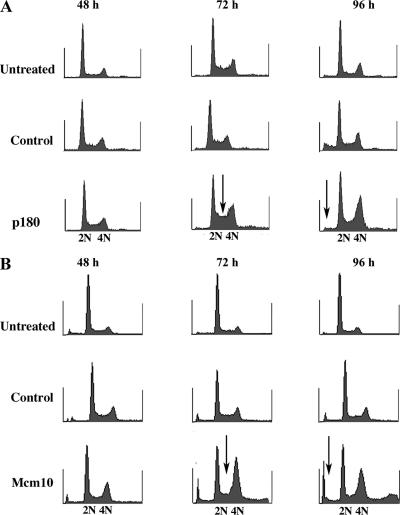
Figure 7.
Cells lacking the catalytic subunit of DNA polymerase-α accumulate in S phase, but they do not undergo apoptosis. HeLa cells were treated with siRNA as described previously, and then they were harvested at indicated time points and stained with PI. (A) DNA content profiles are shown for cells that were left untreated or treated with control siRNA or p180 siRNA. Arrows point to cells accumulated in early S phase (72 h) and the missing apoptotic fraction (96 h), respectively. (B) DNA content profiles are shown for cells that were left untreated or treated with control siRNA or Mcm10 SMARTpool siRNA. Arrows point to cells in early S phase (lack of accumulation at 72 h) and the increased apoptotic fraction (96 h), respectively.
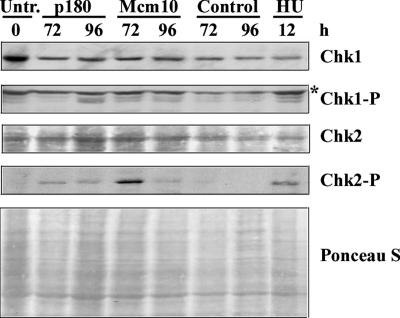
Figure 8.
Mcm10-depleted cells activate Chk2. HeLa cells were treated with siRNA as described previously or with 3 mM hydroxyurea for 12 h, and then they were harvested at indicated time points. Samples were then immunoblotted for Chk1, phospho-Chk1 (Ser 345), Chk2, and phospho-Chk2 (Thr 68). Ponceau S staining served as a loading control. The Chk1-P immunoblot has a nonspecific band marked by an asterisk (*).
Transfections were performed at a starting confluence of 30% for three consecutive times, 24 h apart (Prasanth et al., 2004), with 100 nM siRNA and Lipofectamine 2000 reagent according to the manufacturer's instructions (Invitrogen).
Antibodies
Anti-Mcm10 antibody was purchased from Bethyl Laboratories (Montgomery, TX), and it was used at a 1:1000 for Western blotting. This antibody only recognizes the unmodified form of Mcm10. Anti-DNA polymerase-α (against the 180-kDa subunit) was a gift from Dr. Irene Dornreiter (Heinrich-Pette-Institut für Experimentelle Virologie and Immunologie, Universität Hamburg, Hamburg, Germany), and it was used at a 1:10 dilution for Western blotting (Dehde et al., 2001). Anti-p68 (against the B-subunit of DNA polymerase-α) was a gift from Dr. Heinz-Peter Nasheuer (Heinz-Peter Nasheuer, Department of Biochemistry, National University of Ireland, Galway, Ireland), and it was used at a 1:3000 dilution for Western blotting. Anti-lamin A and C antibody (Covance, Princeton, NJ) was used at a 1:1000 dilution for Western blotting. Anti-α-tubulin antibody (Covance) was used at a 1:6000 dilution for Western blotting. Anti-Cdc45 antibody (Santa Cruz Biotechnology, Santa Cruz, CA) was used at a 1:200 dilution for Western blotting. Anti-poly(ADP-ribose) polymerase (PARP) (BD Biosciences PharMingen, San Diego, CA) was used at a 1:2000 for Western blotting. Anti-Chk1 antibody (Santa Cruz Biotechnology) was used at a 1:250 dilution for Western blotting. Anti-Chk2 antibody (Santa Cruz Biotechnology) was used at a 1:250 dilution for Western blotting. Anti-phosphorylated Chk1 (Cell Signaling Technology, Danvers, MA) was used at a 1:1000 dilution for Western blotting. Anti-phosphorylated Chk2 (Cell Signaling Technology) was used at a 1:1000 dilution for Western blotting. Anti-phospho-histone-H2AX (Upstate Biotechnology, Lake Placid, NY) was used at a concentration of 2 μg/ml for immunofluorescence studies. Alexa Fluor 647-coupled anti-histone H3 antibody (against phosphorylated serine 28) was purchased from BD Biosciences PharMingen, and anti-5-bromo-2′-deoxyuridine (BrdU) antibody was from BD Biosciences (San Jose, CA).
Cell Cycle Analysis and Flow Cytometry
For detection of BrdU incorporation and fluorescence-activated cell sorting (FACS) analysis, 106 HeLa cells were labeled with BrdU at a final concentration of 10 μM after treatment with siRNA and incubated for 22 h. For immunostaining of BrdU-incorporated DNA, recovered cells were fixed, permeabilized, and stained with anti-BrdU antibody according to the manufacturer's protocol (BD Biosciences). This was followed by incubation with 7-amino-actinomycin D (7-AAD) for 30 min and subsequent resuspension in staining buffer (3% FBS and 0.09% sodium azide in PBS). BrdU-associated emission (fluorescein isothiocyanate [FITC]; 530 ± 20-nm band-pass filter) and DNA-associated fluorescence (7-AAD; 660 ± 20-nm band-pass filter) were measured by FACSCalibur (BD Biosciences).
Total DNA content was also measured by incubation with propidium iodide (20 μg/106 cell sample) for 30 min at room temperature. Briefly, cells were treated with 100 μg of RNase A (Sigma-Aldrich, St. Louis, MO) for 30 min at 37°C and incubated with 20 μg of propidium iodide (PI) for 30 min at room temperature. DNA-associated fluorescence for PI (620-nm long-pass filter) was measured by FACSCalibur (BD Biosciences). Histograms for DNA content were quantified by ModFit LT (Verity Software House, Topsham, ME).
For staining of phosphorylated histone H3, cells were fixed in ice-cold 70% ethanol for 1 h. After fixation, cells were washed once with ice-cold phosphate-buffered saline (PBS) and then permeablized in PBS with 0.1% Tween 20. Cells were then washed twice with cold PBS containing 1% FBS and resuspended in 100 μl of this buffer with 20 μl of the anti-histone H3 antibody (BD Biosciences PharMingen) per 106 cells for 30 min on ice. Cells were washed once in 1% FBS in PBS buffer, treated with 100 μg of RNase A for 30 min at 37°C, and incubated with 20 μg of propidium iodide for 30 min at room temperature. Histone phosphorylation-associated fluorescence for AlexaFluor (647 nm) and DNA-associated fluorescence for PI (620-nm long-pass filter) were measured by FACSCalibur (BD Biosciences).
Immunofluorescence Staining of γ-H2AX
HeLa cells were washed three times in PBS, fixed in 2% paraformaldehyde in PBS for 45 min at room temperature, permeabilized in 0.2% Triton-X-100 in PBS for 5 min at room temperature, blocked in 3% bovine serum albumin/PBS for 30 min at 37°C in a humidified box. Cells were incubated with 2 μg/ml γ-H2AX antibody for 1 h in the dark. Then, cells were washed in PBS and stained with 500 ng/ml DAPI for 20 min at room temperature. This was followed by a PBS wash and subsequently, cells were mounted with 10% glycerol in PBS and analyzed with an inverted fluorescence microscope (Eclipse TE 200; Nikon, Tokyo, Japan).
Coimmunoprecipitation
HeLa cells were lysed in buffer A (120 mM NaCl, 0.5% NP-40, 0.1 mM EDTA, 0.5 mM Tris-HCl, pH 8.0, 0.1 mM NaF, 0.1 mM Na3VO4, and 0.1 mM dithiothreitol) and incubated with or without 12.5 μg/ml ethidium bromide for 30 min on ice (Gomez et al., 2006) or with 100 μg/ml DNase I in the presence of 10 mM MgCl2 for 30 min on ice (Ricke and Bielinsky, 2004). The lysate was centrifuged at 14,000 × g in a precooled centrifuge for 10 min. The supernatant was incubated with anti-p180 (2 μg/100–500 μg of total protein; Santa Cruz Biotechnology), anti-Mcm10 (4 μg/mg lysate; Bethyl Laboratories), or immunoglobulin (Ig)G (4 μg) for 2 h at 4°C on an orbital shaker. Protein G-Sepharose beads (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) were then added to the lysate and incubated overnight at 4°C. Beads were pelleted, washed three times with PBS and resuspended in 1X Laemmli buffer.
Analysis of Apoptosis by Flow Cytometry Analysis
Double staining of HeLa cells with Annexin V-FITC and PI were used to discriminate viable cells from early and late apoptotic or dead cells. FITC-conjugated Annexin V and PI were obtained as a kit from BD Biosciences PharMingen. Cultured cells treated with siRNA were collected at indicated time points by centrifugation and washed twice with cold PBS, resuspended in binding buffer, and incubated together with Annexin V-FITC and PI as per manufacturer's protocol. Cells were gently vortexed, incubated at room temperature for 15 min, and then analyzed by flow cytometry within 1 h. Annexin V-associated emission (FITC; 530 ± 20-nm band-pass filter) and DNA associated fluorescence for PI (620-nm long-pass filter) were measured by FACSCalibur (BD Biosciences) (Vermes et al., 1995).
RESULTS
Mcm10 Regulates the Stability of p180 in Human Cells
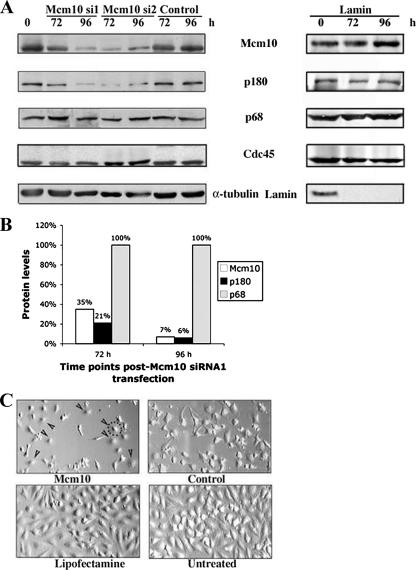
Mcm10 is essential for DNA replication in yeast (Solomon et al., 1992; Merchant et al., 1997), and it is likely that this extends to higher eukaryotes as well, although this has not been experimentally confirmed. To define the role of human Mcm10 in DNA replication, we used a siRNA knockdown strategy to deplete Mcm10 in HeLa cells. We observed a reproducible decrease of Mcm10 only when we performed three consecutive transfections, following a previously described protocol (Prasanth et al., 2004). A significant reduction in the amount of Mcm10 was detected 72- and 96 h after the first transfection for two different Mcm10-specific siRNAs (Figure 1A). Interestingly, however, the kinetics of Mcm10 depletion differed slightly depending upon which siRNA was used, peaking at 96 h for siRNA 1 and at 72 h for siRNA 2 (Figure 1A). This is not surprising, because different siRNAs exhibit different efficiencies toward their target sequences. In contrast, Mcm10 levels remained unchanged in cells that were treated with control siRNA (Figure 1A). We obtained similar results when we transfected U2OS cells (data not shown).
Figure 1.
Mcm10 depletion results in destabilization of the catalytic subunit of DNA polymerase-α. (A) HeLa cells were transfected with Mcm10 siRNA (si1 and si2) or control siRNA, as shown and harvested at different time points. At a starting confluence of 30%, cells were transfected three times at 24-h intervals, as described previously (Prasanth et al., 2004). Efficacy of RNAi was measured by immunoblotting with anti-Mcm10 antibodies (BL531). Immunoblots were also developed with anti-p180, anti-p68, and anti-Cdc45 antibodies. α-Tubulin served as a loading control. The same blot was stripped and reprobed. Right, immunoblots developed with anti-Mcm10, anti-p180, anti-p68, anti-Cdc45, and anti-lamin antibodies for HeLa cells treated with lamin A/C siRNA in the same manner as described above. (B) The percentage of each protein remaining at indicated time points was plotted when HeLa cells were treated with siRNA 1 (si1) against Mcm10. The amount of protein was normalized with respect to the loading control, and then it was expressed as a fraction of the amount in control siRNA-treated samples. (C) HeLa cells were transfected with Mcm10 siRNA1 or control siRNA, as indicated and photographed 96 h after transfection. Arrowheads point to some of the cells that exhibited an abnormal phenotype. The dotted line circles the cell membrane of one representative cell.
Because our earlier studies in budding yeast had established that Mcm10 regulates the stability of the catalytic subunit of pol-α (p180 in mammals), we were particularly interested in understanding the relationship between Mcm10 and p180 in human cells (Ricke and Bielinsky, 2004, 2006). As shown in Figure 1B, depletion of Mcm10 caused the codepletion of p180. Moreover, p180 levels very closely mimicked those of Mcm10 and mirrored the different depletion kinetics that we described above for siRNA 1 and siRNA 2. With siRNA 1, Mcm10 was depleted to 35% at 72 h, and p180 was reduced to 21%. At 96 h, Mcm10 had decreased to 7% and p180 to 6% (Figure 1). For siRNA 2, Mcm10/p180 levels were reduced to ∼7% at 72 h, but amounts for both proteins increased to ∼23% at 96 h (Figure 1A; data not shown). Because transfection with siRNA 1 seemed to result in a more robust down-regulation of Mcm10 than transfection with siRNA 2, we continued to use the former for subsequent experiments. Importantly, expression of p68, the regulatory subunit of pol-α, remained unchanged for both siRNA 1 and 2. The same was true for expression levels of Cdc45, which is recruited to replication origins in a Mcm10-dependent manner (Wohlschlegel et al., 2002; Gregan et al., 2003; Sawyer et al., 2004). In contrast, the control siRNA did not affect the expression of any of the proteins that we analyzed. Moreover, cells treated with lamin-specific siRNA retained normal steady-state levels of all four replication factors, including p180 (Figure 1A, right). This indicated to us that loss of p180 was not just a consequence of triggering the RNA interference (RNAi) pathway. Thus, we conclude that down-regulation of Mcm10 causes degradation of p180, either in a direct or indirect manner, but that it has no effect on p68 or Cdc45 protein stability.
Mcm10 Depletion Inhibits Proliferation and Causes Cell Cycle Arrest
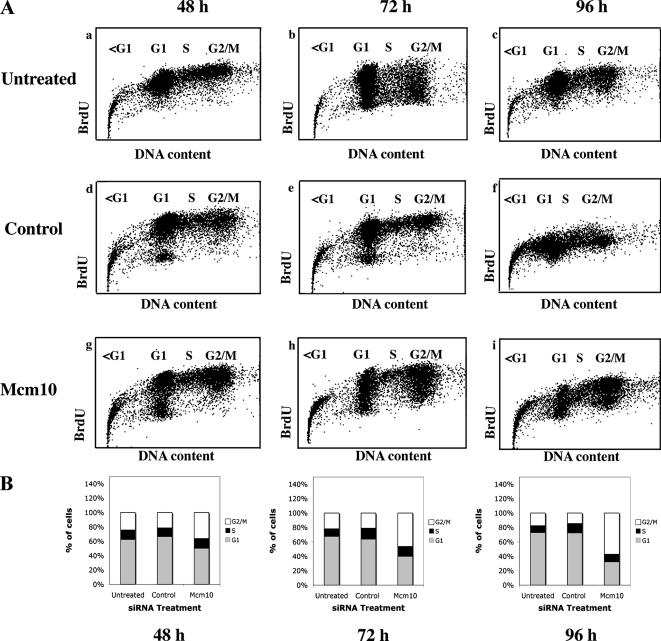
The most obvious consequence of Mcm10 down-regulation was a drastic reduction in the cell number, consistent with a proliferation defect. We also noticed morphological changes (Figure 1C), because the majority of Mcm10-depleted cells displayed an unusual phenotype. Approximately 60% of cells treated with Mcm10-specific siRNA 1 displayed this abnormal phenotype compared with 9% in control siRNA-treated cells 96 h posttransfection. This differs from the phenotype of Orc2-depleted HeLa cells (Prasanth et al., 2004) and suggests that it is not just a by-product of inducing the RNA silencing pathway, but rather specific to the codepletion of Mcm10 and/or p180 (Figure 1C). To further substantiate that down-regulation of Mcm10 or p180-inhibited cell cycle progression, we labeled cells with BrdU for 22 h before harvest at 48, 72, and 96 h after siRNA treatment. Under the assumption that most BrdU was incorporated during DNA replication, we first determined the number of BrdU-positive, actively cycling cells (Table 1). We observed a dramatic decline in proliferating cells as levels of Mcm10 and p180 decreased (Table 1). This is consistent with roles for Mcm10 and p180 in proliferation. To provide a more accurate measure of the cell cycle distributions in the differently treated cultures over time, we quantified both BrdU-positive and -negative cells in G1, S, and G2/M phase. The results of one representative experiment are shown in Figure 2. Although untreated and control cells showed no or little change in their cell cycle profiles (we noticed a slight delay in S phase in control siRNA-treated cells), the codepletion of Mcm10 and p180 caused an increasing accumulation of cells in G2/M phase (Figure 2B). The G2/M peak showed a broad S phase shoulder in the DNA content profiles (Supplemental Figure S1). In addition, few cells entered S phase between 72 and 96 h (39 vs. 33%), which corresponds to the time window with the highest level of depletion (Figure 1). To ensure that these cell cycle aberrations were not triggered unspecifically, we analyzed lamin-depleted cells and found them to be almost indistinguishable from untreated or control siRNA-treated cells (Supplemental Figure S2). These results argue that codepletion of Mcm10 and p180 inhibits S phase entry in G1 cells and completion of DNA synthesis in S phase cells. The latter notion predicted that Mcm10 siRNA-treated cells might not enter mitosis because of incomplete DNA replication or DNA damage. To directly test this hypothesis, we determined the percentage of cells in which serine 28 of histone H3 was phosphorylated. Unlike the phosphorylation of serine 10, which occurs before cells enter mitosis, phosphorylation at serine 28 is a metaphase-specific marker (Goto et al., 1999). FACS analysis revealed that the percentage of mitotic cells in untreated, Mcm10 siRNA-treated and control cultures was almost identical and ranged between 2.1 and 2.9%, 72 and 96 h post-siRNA transfection (data not shown). Thus, the majority of Mcm10-depleted cells with apparent 4N DNA content had not entered metaphase. Finally, we also noticed a significant increase in apoptotic cells upon loss of Mcm10 and p180, as indicated by the growing sub-G1 peak in the normal cell cycle profiles (Supplemental Figure S1). The results of our quantification of BrdU-negative cells with less than 2N DNA content are listed in Table 1 and indicate that 96 h after transfection, Mcm10 siRNA-treated cells had a significant increase in cell death above background. These results were confirmed in an independent assay by measuring the amount of Annexin V-positive cells (Supplemental Figure S3). Both experiments showed a higher than threefold increase in apoptosis above background.
Table 1.
Mcm10 siRNA-treated HeLa cells show a decrease in the actively cycling cell population and an increase in apoptosis
| A. Actively cycling cell population | |
|---|---|
| siRNA treatment | % Actively cycling cells |
| Mcm10 siRNA 48 h | 81.2 |
| Mcm10 siRNA 72 h | 58.1 |
| Mcm10 siRNA 96h | 59.7 |
| Control siRNA 48 h | 88.0 |
| Control siRNA 72 h | 83.4 |
| Control siRNA 96 h | 84.5 |
| Untreated cells 48 h | 90.6 |
| Untreated cells 72 h | 93.9 |
| Untreated cells 96 h | 88.3 |
| B. BrdU-negative cells with less than 2N DNA content | |
| siRNA treatment | % Apoptotic |
| Mcm10 siRNA 48 h | 7.5 |
| Mcm10 siRNA 72 h | 23.1 |
| Mcm10 siRNA 96h | 18.3 |
| Control siRNA 48 h | 4.9 |
| Control siRNA 72 h | 9.3 |
| Control siRNA 96 h | 12.4 |
| Untreated cells 48 h | 4.8 |
| Untreated cells 72 h | 2.9 |
| Untreated cells 96 h | 4.1 |
A, HeLa cells were labeled with BrdU for 22 h before harvest at the indicated time points to monitor actively proliferating cells. B, HeLa cells were analyzed by flow cytometry to measure the fraction of apoptotic cells (cells with <2N DNA content). Lamin siRNA-treated HeLa cells showed actively proliferating and apoptotic cells comparable to untreated cultures (Supplemental Figure S2).
Figure 2.
Mcm10 depletion results in cell cycle arrest and increased apoptosis. (A) Profiles of HeLa cells incorporating BrdU at various times are shown. a–i show profiles of cells that were untreated (a–c) or treated with either control (d–f) or Mcm10 (g–i) siRNA and then BrdU labeled for 22 h. Cells were harvested at the indicated time points for cell cycle analysis. (B) The fraction of cells (BrdU positive and BrdU negative) in various phases of the cell cycle was quantified based on DNA content and plotted. The relative distribution was similar in BrdU-positive cells, and the data are presented in Table 1, A and B.
Because of the high incidence of apoptosis, we next explored whether codepletion of Mcm10 and p180 caused DNA damage. Both stalled replication forks and double-strand breaks (DSBs) are known to trigger phosphorylation of histone H2AX (Burma et al., 2001; Ward and Chen, 2001). Indirect immunofluorescence studies revealed that 55% of Mcm10 siRNA-treated cells displayed phosphorylated (γ) H2AX foci 72 and 96 h after transfection. For p180-depleted cultures, we detected 21 and 49% γ-H2AX-positive cells, whereas in lamin and control siRNA treated cultures only between 5 and 7% of cells exhibited γ-H2AX foci (Figure 3). Thus, codepletion of Mcm10 and p180 as well as loss of p180 by itself causes activation of a DNA damage/replication stress response. A more detailed analysis of p180-depleted cells is described further below.
Loss of p180 in Mcm10 siRNA-treated Cells Is Not Due to the Induction of Apoptosis
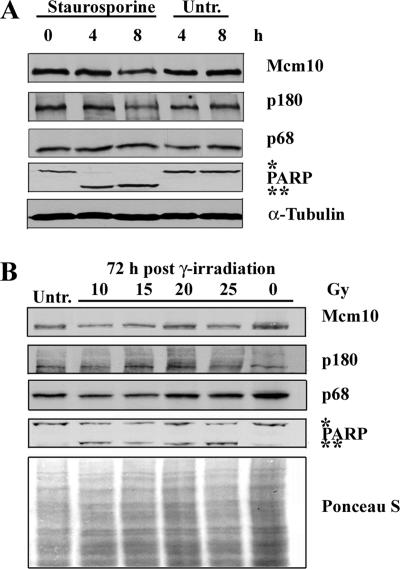
So far, we have established that loss of Mcm10 triggers loss of p180 and inhibits proper DNA replication, because cells accumulate DNA damage and arrest in late S/G2 phase or undergo apoptosis. This scenario prompted us to consider the possibility that loss of p180 was not a direct consequence of Mcm10 depletion, but rather a secondary event triggered by the induction of apoptosis. To determine whether this was the case, we exposed cells to STS, a very efficient inducer of apoptosis. Cells were treated for 4 or 8 h before they were harvested to analyze the status of PARP, Mcm10, p180, and p68. PARP is a well-established caspase target, and it is cleaved within a few hours after apoptotic induction (Patel et al., 1996). In parallel, we analyzed control cells that remained untreated. As expected, we observed PARP cleavage as soon as 4 h after the addition of STS, but not in untreated cells (Figure 4). Mcm10 and p180 levels were readily detected and remained fairly constant at 4 and 8 h after STS addition, although the majority of cells had clearly triggered apoptosis, as indicated by PARP cleavage (Figure 4A). Because the data in Figure 3 suggested that apoptosis in Mcm10 siRNA-treated cells was triggered by DNA damage, we tried to more accurately mimic this condition and asked whether DNA damage-induced apoptosis would affect the stability of p180. We exposed cells to different doses of gamma-irradiation or left them untreated and analyzed whole cell extracts 72 h later. Although PARP cleavage was not quite as pronounced as in staurosporine-treated cells, apoptosis was clearly induced (Figure 4B). Protein levels of Mcm10, p180, and p68, however, remained unchanged. Based on these two experiments, we excluded the possibility that the overall loss of p180 in Mcm10-depleted cells was a nonspecific, secondary effect triggered by the induction of apoptosis.
Figure 4.
Down-regulation of DNA polymerase-α (p180) is triggered by loss of Mcm10 and not by induction of apoptosis. (A) HeLa cells were treated with 5 μM staurosporine for 4 or 8 h. Whole cell lysates were prepared and immunoblotted for Mcm10, p180, p68, and PARP. The single asterisk indicates full-length PARP-1 (116 kDa), and the double asterisk indicates the cleavage product of PARP-1 (85 kDa). α-Tubulin served as a loading control. (B) HeLa cells were irradiated using a 137Cs gamma-ray source at a dose rate of 2.5 Gy/min. Cells were irradiated with 10, 15, 20, and 25 Gy or left untreated (lane 6), and they were harvested at 72 h after irradiation. Untreated HeLa cells were harvested at 0 h (lane 1) and immunoblotted for Mcm10, p180, p68, and PARP-1. Ponceau S staining served as a loading control. The single asterisk indicates full-length PARP-1 (116 kDa), and the double asterisk indicates the cleavage product of PARP-1 (85 kDa).
Mcm10 and pol-α Interact in Human Cell Extracts
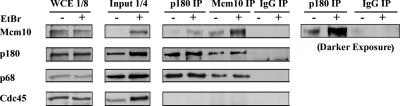
Because steady-state levels of Mcm10 and p180 followed the same kinetics after Mcm10 siRNA treatment (Figure 1), we reasoned that they might be associated with each other. To test this hypothesis, we performed coimmunoprecipitation experiments with whole cell extracts from asynchronous HeLa cells. Because most of these cells are in G1, a portion of Mcm10 is likely chromatin bound (Izumi et al., 2004). To ensure that an interaction between Mcm10 and p180 was independent of DNA, we treated one half of the extract with ethidium bromide (EtBr) to release proteins from DNA (Lai and Herr, 1992), and the other half was left untreated. Moreover, the EtBr treatment proved essential to release sufficiently high amounts of Mcm10 into the soluble fraction (Figure 5, input). Mcm10- or p180-specific antibodies were added, and immunoprecipitates were analyzed by Western blot. We found that Mcm10 was associated with p180 and p68, but neither Mcm10 nor p180 was in a complex with Cdc45 (Figure 5). Similar results were obtained when we performed coimmunoprecipitation experiments in the presence and absence of DNase I (data not shown). These results suggest that human Mcm10 forms a complex with pol-α.
Figure 5.
Mcm10 and DNA polymerase-α interact in human cells. Whole cell lysate from HeLa cells were incubated in the presence (+) or absence (−) of EtBr and immunoprecipitated with anti-p180, anti-Mcm10, or IgG as indicated. Antibodies against Mcm10, p180, p68, and Cdc45 were used for Western blotting. A darker exposure of the Mcm10 blot is shown on the right.
Loss of p180 in the Presence of Mcm10 Slows Down S Phase Progression, but It Does Not Compromise Cell Viability
Based on the observations that Mcm10 and p180 protein levels were coregulated and that the two proteins interacted with each other, we asked whether depletion of p180 would alter protein levels of Mcm10. To this end, we used the same knock down strategy as described above and introduced p180-specific siRNA into HeLa cells. Steady-state levels of p180 were markedly reduced 72- and 96 h post-siRNA transfection, whereas neither Mcm10 nor p68 was affected by the loss of p180, because their protein levels were similar to those detected in HeLa cells that were treated with control siRNA (Figure 6, A and B). We also attempted to deplete p68 to investigate any possible effects on Mcm10 and p180; however, these experiments were unsuccessful (data not shown). Therefore, although it remains unclear whether p68 has any role in regulating Mcm10, our results clearly demonstrate that p180 does not control Mcm10 levels. Further observation of the cell morphology revealed that p180-depleted cells showed a similar abnormal phenotype that we had previously observed in Mcm10 siRNA-treated cultures (compare Figures 1C and 6C). This is consistent with the result that Mcm10-depleted cells have severely diminished levels of p180.
To evaluate whether all of the phenotypes exhibited by Mcm10-depleted cells were in fact due to the loss of p180, we analyzed p180-depleted cultures in more detail. Interestingly, the cell cycle profile of p180 siRNA-treated cultures revealed a pronounced S phase delay 72 h after transfection (Figure 7A). For a better comparison between p180 and Mcm10 siRNA-treated cells, we have included a cell cycle profile of the latter that was obtained using the identical DNA staining protocol (without BrdU labeling; Figure 7B). The accumulation of early S phase cells in p180-depleted cultures was in stark contrast to cultures that were depleted for both Mcm10 and p180 (Figure 7B). Moreover, it became obvious that cells continued to progress through S phase when p180 levels were low (Figure 7A, 96 h after transfection). Another striking difference was the lack of apoptosis in p180-depleted cells, because we were unable to detect cell death rates above background, neither by DNA (Figure 7A) nor by Annexin V staining (Supplemental Figure S3). These results indicate that cell death was triggered by loss of Mcm10 and not by loss of p180. Cell cycle progression and cell viability are affected differently in the presence or “absence” of Mcm10. In the presence of Mcm10, loss of p180 causes an overall S phase delay without compromising cell viability. In the absence of Mcm10, p180 depletion inhibits S phase entry (visualized by the complete absence of early S phase cells in Figure 7B) as well as timely replication fork progression causing DNA damage and apoptosis.
Chk2 Is Activated in Mcm10-depleted Cells
To gain better insight into the question of how loss of Mcm10 triggered apoptosis, we examined the activation status of the checkpoint kinases Chk1 and Chk2. It is commonly thought that, during S phase, Chk1 is phosphorylated by the ataxia telangiectasia mutated and Rad3 related (ATR) kinase primarily in response to stalled replication forks, whereas Chk2 is rapidly activated by ataxia telangiectasia mutated (ATM) after DSB induction, and to a lower extend by ATR after replication fork arrest (Osborn et al., 2002). If apoptosis of Mcm10-depleted cells was truly triggered by DSBs, we predicted to observe pronounced Chk2 phosphorylation. This was indeed the case, at least 72 h post-siRNA transfection (Figure 8). However, at the later time point Chk2 was no longer activated, likely because the majority of cells (∼70% according to the Annexin V staining in Supplemental Figure S3) had already triggered apoptosis. Moreover, ATM but not ATR is a caspase target (Smith et al., 1999). In contrast, p180-depleted cells exhibited very little Chk2 activation even 96 h post-siRNA transfection, at a time when ∼50% of cells exhibited γ-H2AX foci (Figure 3B and Supplemental Figure S4), and Chk1 was markedly phosphorylated (Figure 8). We take these results as evidence that despite the similarities in γ-H2AX foci formation, the actual damage inflicted onto the DNA differs depending on whether p180 is down-regulated in combination with Mcm10 or not.
DISCUSSION
In this study, we provide evidence that specific aspects of Mcm10 function are conserved from yeast to human cells, and we provide further insight into the regulation of pol-α by Mcm10. Most notably, we demonstrate that human Mcm10 has a role in DNA elongation by regulating p180, the catalytic subunit of DNA pol-α. Previous localization studies on Mcm10 in HeLa cells found that Mcm10 foci formation only partly overlapped with nuclear regions in which DNA synthesis occurred and that localization of Mcm10 preceded that of PCNA by 30–60 min (Izumi et al., 2004). Their observations suggested a role for Mcm10 in replication initiation rather than elongation, and they concluded that the primary role of Mcm10 in DNA replication is in pre-RC activation. Although this claim is consistent with the finding that human Mcm10 colocalizes with ORC and other initiation factors (Izumi et al., 2000), our data extend these findings and suggest that Mcm10 is indeed required to complete DNA replication in human cells. A role for human Mcm10 in elongation is also consistent with previous reports that argue that Mcm10 stays associated with chromatin throughout S phase in HeLa cells (Izumi et al., 2000, 2001). Moreover, the lack of complete colocalization with BrdU-labeled DNA or PCNA has previously been documented for components of the Mcm2-7 complex (Madine et al., 1995), which constitutes the core of the replicative helicase in eukaryotic cells (Schwacha and Bell, 2001; Moyer et al., 2006). Therefore, the incomplete overlap between Mcm10 and PCNA foci does not necessarily mean that Mcm10 is not involved in elongation. Like subcomponents of the Mcm2-7 complex, Mcm10 is highly abundant (Izumi et al., 2004), and probably in large excess of the number of replication origins, which might explain why not all of the Mcm10 foci overlapped with PCNA foci in the previous study (Izumi et al., 2004), although both are involved in DNA elongation.
The findings that Mcm10 and p180 are coregulated (Figure 1) and that they are physically associated with each other (Figure 5) provide evidence that human Mcm10 might be involved in lagging strand synthesis. Consistent with this hypothesis, we have recently reported that the core of Mcm10 in S. cerevisiae contains an OB-fold domain (Ricke and Bielinsky, 2006), which mediates binding to ssDNA (Fien et al., 2004). This OB-fold domain is evolutionarily conserved, and it could allow human Mcm10 to bind to the lagging stand template, facilitating Okazaki fragment initiation by pol-α/primase. Furthermore, Mcm10 could have a role in recycling pol-α in between the initiation of consecutive Okazaki fragments (especially when pol-α levels are low). This model would be consistent with our finding that low levels of p180 are sufficient to support S phase progression (albeit slowly) in the presence but not in the “absence” of Mcm10. Codepletion of p180 and Mcm10 causes DNA damage and apoptosis. DNA damage likely arises from stalled replication forks after uncoupling of leading and lagging strand synthesis (Figure 9). We envision the following scenario to explain these results: in the presence of Mcm10, only p180 is limiting at replication forks. Recycling of p180 by Mcm10 could supply each fork with sufficient p180 to eventually progress through S phase. However, the combined loss of p180 and Mcm10 makes both limiting at replication forks, inhibiting lagging strand synthesis at a large number of forks. These forks can essentially not be restarted as long as Mcm10 and p180 levels remain low, thus causing irreversibly arrested replication forks, which could be processed into DSBs, consistent with the γ-H2AX foci and Chk2 activation we observed in this study and the occurrence of apoptosis. However, we have also considered the fact that Mcm10 seems to act further upstream in replication activation than p180 (Ricke and Bielinsky, 2004), and this may contribute to the differences between Mcm10- and p180 siRNA-treated cultures. Because human Mcm10 likely has a role in pre-RC activation (Wohlschlegel et al., 2002; Izumi et al., 2004), loss of Mcm10 could result in a decreased number of activated replication origins. In combination with the elongation defect caused by the loss of p180, Mcm10-depleted cells might simply not have a sufficiently high number of replication forks to duplicate their genome in a timely manner, explaining the accumulation in late S/G2. The only caveat is that this model does not provide an explanation for the DNA damage and apoptosis we observed, because a delay in replication does not necessarily cause these phenotypes (Bielinsky 2003; Christensen and Tye 2003; this study).
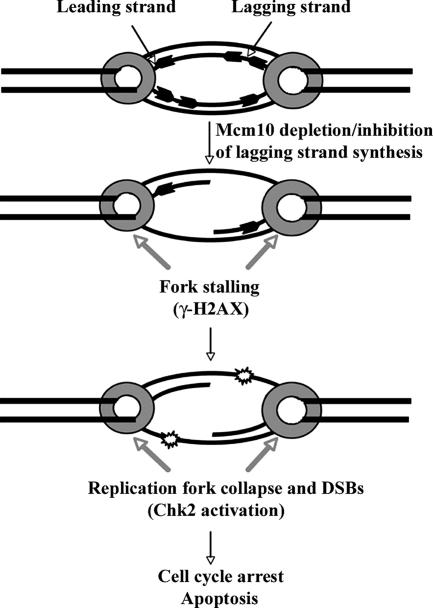
Figure 9.
Loss of Mcm10 and p180 causes lagging strand inhibition. Leading and lagging strands are shown (bold arrows) as well as the replicative helicase (gray circle). Depletion of Mcm10 and p180 blocks lagging strand synthesis, which causes irreversible fork stalling and subsequent replication fork collapse, giving rise to DSBs (stars). This triggers cell cycle arrest and apoptosis.
In a previous study, Christensen and Tye (2003) depleted Mcm10 in embryonic Drosophila melanogaster KC cells. These cells exhibited no cell cycle defect, although they have a functional S phase checkpoint system (MacAlpine et al., 2004), and they entered mitosis, unlike Mcm10/p180-codepleted HeLa cells. Interestingly however, a subset of Mcm10-depleted KC cells had defects in chromosome condensation (Christensen and Tye, 2003), likely due to incomplete replication as cells were entering mitosis (Bielinsky, 2003; Christensen and Tye, 2003). The differences between the two experimental systems can be explained by two factors. First, initiation of DNA replication might require less Mcm10 in embryonic cells than in HeLa cells, because initiation in the latter is largely inhibited in G1 cells upon Mcm10 depletion (Figure 2). Second, the small replicon size (∼25 kilobases [kb] in KC cells vs. multiple 100 kb in mammals) typically found in embryonic systems (Huberman and Riggs, 1968; Hyrien et al., 1995; Walter and Newport, 1997; Ermakova et al., 1999; MacAlpine et al., 2004) makes cells less sensitive toward elongation defects, and although initiation events might be reduced, most cells fully replicate their genome and undergo normal mitoses. In contrast, genomes with large replicons are much more prone to fork arrest and DNA damage, explaining the differences in phenotype.
One important question that arises from our study is how Mcm10 regulates p180. Our data support the idea that pol-α is in a complex with Mcm10 (Figure 5). Therefor, it is conceivable that Mcm10 interacts directly with the catalytic subunit, as shown for S. pombe Mcm10 (Fien et al., 2004). In budding yeast, a hydrophobic stretch similar to a domain in the mobile loop of Hsp10 is required to stabilize the catalytic subunit of pol-α, Cdc17/Pol1 (Ricke and Bielinsky, 2006). Importantly, the Hsp10-like domain in Mcm10 is evolutionarily conserved, and a single amino acid change within this domain drastically reduces the stability of Cdc17/Pol1 (Ricke and Bielinsky, 2006). We propose that the same domain in human Mcm10 is important for the interaction with p180. Although we do not yet understand the molecular mechanism underlying the association between Mcm10 and p180, our studies firmly establish that Mcm10 is required to stabilize p180 and that this occurs independently of p68 (Figure 1). This is further supported by the finding that p68 has no effect on the half-life of overexpressed p180 in mammalian cells although p68 facilitates the nuclear localization of p180 (Mizuno et al., 1998, 1999), and the same is true for the homologues in budding yeast (Ricke and Bielinsky, 2006). Clearly, future studies are needed to explore how exactly Mcm10 contributes to replication fork progression and maintains genome integrity.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. Irene Dornreiter and Heinz P. Nasheuer for antibodies. We acknowledge the assistance of the Flow Cytometry Core Facility at the University of Minnesota. We also thank Drs. Yongbao Wang and Goutam Ghosh for help with cell culture protocols. We thank all Bielinsky laboratory members and Dr. Eric A. Hendrickson for critical reading of the manuscript. This work was supported by MMF 3534-9201 and a Leukemia Research Fund LRF 20091506 from the University of Minnesota and National Institutes of Health grant GM-074917 (to A.K.B.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-12-1148) on August 15, 2007.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Aparicio O. M., Weinstein D. M., Bell S. P. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- Bielinsky A. K. Replication origins: why do we need so many? Cell Cycle. 2003;2:307–309. [PubMed] [Google Scholar]
- Burich R., Lei M. Two bipartite NLSs mediate constitutive nuclear localization of Mcm10. Curr. Genet. 2003;44:195–201. doi: 10.1007/s00294-003-0443-y. [DOI] [PubMed] [Google Scholar]
- Burma S., Chen B. P., Murphy M., Kurimasa A., Chen D. J. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 2001;276:42462–42467. doi: 10.1074/jbc.C100466200. [DOI] [PubMed] [Google Scholar]
- Christensen T. W., Tye B. K. Drosophila MCM10 interacts with members of the prereplication complex and is required for proper chromosome condensation. Mol. Biol. Cell. 2003;14:2206–2215. doi: 10.1091/mbc.E02-11-0706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das-Bradoo S., Ricke R. M., Bielinsky A. K. Interaction between PCNA and diubiquitinated Mcm10 is essential for cell growth in budding yeast. Mol. Cell. Biol. 2006;26:4806–4817. doi: 10.1128/MCB.02062-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehde S., Rohaly G., Schub O., Nasheuer H. P., Bohn W., Chemnitz J., Deppert W., Dornreiter I. Two immunologically distinct human DNA polymerase alpha-primase subpopulations are involved in cellular DNA replication. Mol. Cell. Biol. 2001;21:2581–2593. doi: 10.1128/MCB.21.7.2581-2593.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley J. F., Cocker J. H., Dowell S. J., Rowley A. Two steps in the assembly of complexes at yeast replication origins in vivo. Cell. 1994;78:303–316. doi: 10.1016/0092-8674(94)90299-2. [DOI] [PubMed] [Google Scholar]
- Ermakova O. V., Nguyen L. H., Little R. D., Chevillard C., Riblet R., Ashouian N., Birshtein B. K., Schildkraut C. L. Evidence that a single replication fork proceeds from early to late replicating domains in the IgH locus in a non-B cell line. Mol. Cell. 1999;3:321–330. doi: 10.1016/s1097-2765(00)80459-1. [DOI] [PubMed] [Google Scholar]
- Fien K., Cho Y. S., Lee J. K., Raychaudhuri S., Tappin I., Hurwitz J. Primer utilization by DNA polymerase alpha-primase is influenced by its interaction with Mcm10p. J. Biol. Chem. 2004;279:16144–16153. doi: 10.1074/jbc.M400142200. [DOI] [PubMed] [Google Scholar]
- Fien K., Hurwitz J. Fission yeast Mcm10p contains primase activity. J. Biol. Chem. 2006;281:22248–222460. doi: 10.1074/jbc.M512997200. [DOI] [PubMed] [Google Scholar]
- Gambus A., Jones R. C., Sanchez-Diaz A., Kanemaki M., van Deursen F., Edmondson R. D., Labib K. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat. Cell. Biol. 2006;8:358–366. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- Gomez M., Wu J., Schreiber V., Dunlap J., Dantzer F., Wang Y., Liu Y. PARP1 Is a TRF2-associated poly(ADP-ribose)polymerase and protects eroded telomeres. Mol. Biol. Cell. 2006;17:1686–1696. doi: 10.1091/mbc.E05-07-0672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto H., et al. Identification of a novel phosphorylation site on histone H3 coupled with mitotic chromosome condensation. J. Biol. Chem. 1999;274:25543–25549. doi: 10.1074/jbc.274.36.25543. [DOI] [PubMed] [Google Scholar]
- Gregan J., Lindner K., Brimage L., Franklin R., Namdar M., Hart E. A., Aves S. J., Kearsey S. E. Fission yeast Cdc23/Mcm10 functions after pre-replicative complex formation to promote Cdc45 chromatin binding. Mol. Biol. Cell. 2003;14:3876–3887. doi: 10.1091/mbc.E03-02-0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z., Bhalla K., Pantazis P., Hendrickson E. A., Wyche J. H. Cif (cytochrome c efflux-inducing factor) activity is regulated by Bcl-2 and caspases and correlates with the activation of Bid. Mol. Cell. Biol. 1999;19:1381–1389. doi: 10.1128/mcb.19.2.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huberman J. A., Riggs A. D. On the mechanism of DNA replication in mammalian chromosomes. J. Mol. Biol. 1968;32:327–341. doi: 10.1016/0022-2836(68)90013-2. [DOI] [PubMed] [Google Scholar]
- Hyrien O., Maric C., Mechali M. Transition in specification of embryonic metazoan DNA replication origins. Science. 1995;270:994–997. doi: 10.1126/science.270.5238.994. [DOI] [PubMed] [Google Scholar]
- Izumi M., Yanagi K., Mizuno T., Yokoi M., Kawasaki Y., Moon K. Y., Hurwitz J., Yatagai F., Hanaoka F. The human homolog of Saccharomyces cerevisiae Mcm10 interacts with replication factors and dissociates from nuclease-resistant nuclear structures in G(2) phase. Nucleic Acids Res. 2000;28:4769–4777. doi: 10.1093/nar/28.23.4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi M., Yatagai F., Hanaoka F. Cell cycle-dependent proteolysis and phosphorylation of human Mcm10. J. Biol. Chem. 2001;276:48526–48531. doi: 10.1074/jbc.M107190200. [DOI] [PubMed] [Google Scholar]
- Izumi M., Yatagai F., Hanaoka F. Localization of human Mcm10 is spatially and temporally regulated during the S phase. J. Biol. Chem. 2004;279:32569–32577. doi: 10.1074/jbc.M314017200. [DOI] [PubMed] [Google Scholar]
- Kubota Y., Takase Y., Komori Y., Hashimoto Y., Arata T., Kamimura Y., Araki H., Takisawa H. A novel ring-like complex of Xenopus proteins essential for the initiation of DNA replication. Genes Dev. 2003;17:1141–1152. doi: 10.1101/gad.1070003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib K., Tercero J. A., Diffley J. F. Uninterrupted MCM2–7 function required for DNA replication fork progression. Science. 2000;288:1643–1647. doi: 10.1126/science.288.5471.1643. [DOI] [PubMed] [Google Scholar]
- Lai J. S., Herr W. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc. Natl. Acad. Sci. USA. 1992;89:6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liachko I., Tye B. K. Mcm10 is required for the maintenance of transcriptional silencing in Saccharomyces cerevisiae. Genetics. 2005;171:503–515. doi: 10.1534/genetics.105.042333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine D. M., Rodriguez H. K., Bell S. P. Coordination of replication and transcription along a Drosophila chromosome. Genes Dev. 2004;18:3094–3105. doi: 10.1101/gad.1246404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madine M. A., Khoo C. Y., Mills A. D., Musahl C., Laskey R. A. The nuclear envelope prevents reinitiation of replication by regulating the binding of MCM3 to chromatin in Xenopus egg extracts. Curr. Biol. 1995;5:1270–1279. doi: 10.1016/s0960-9822(95)00253-3. [DOI] [PubMed] [Google Scholar]
- Mendez J., Stillman B. Perpetuating the double helix: molecular machines at eukaryotic DNA replication origins. Bioessays. 2003;25:1158–1167. doi: 10.1002/bies.10370. [DOI] [PubMed] [Google Scholar]
- Merchant A. M., Kawasaki Y., Chen Y., Lei M., Tye B. K. A lesion in the DNA replication initiation factor Mcm10 induces pausing of elongation forks through chromosomal replication origins in Saccharomyces cerevisiae. Mol. Cell. Biol. 1997;17:3261–3271. doi: 10.1128/mcb.17.6.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T., Ito N., Yokoi M., Kobayashi A., Tamai K., Miyazawa H., Hanaoka F. The second-largest subunit of the mouse DNA polymerase alpha-primase complex facilitates both production and nuclear translocation of the catalytic subunit of DNA polymerase alpha. Mol. Cell. Biol. 1998;18:3552–3562. doi: 10.1128/mcb.18.6.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T., Yamagishi K., Miyazawa H., Hanaoka F. Molecular architecture of the mouse DNA polymerase alpha-primase complex. Mol. Cell. Biol. 1999;19:7886–7896. doi: 10.1128/mcb.19.11.7886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer S. E., Lewis P. W., Botchan M. R. Isolation of the Cdc45/Mcm2–7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc. Natl. Acad. Sci. USA. 2006;103:10236–10241. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasheuer H. P., Grosse F. Immunoaffinity-purified DNA polymerase alpha displays novel properties. Biochemistry. 1987;26:8458–8466. doi: 10.1021/bi00399a064. [DOI] [PubMed] [Google Scholar]
- Osborn A., Elledge S., Zou L. Checking on the fork: the DNA-replication stress-response pathway. Trends Cell Biol. 2002;12:509–516. doi: 10.1016/s0962-8924(02)02380-2. [DOI] [PubMed] [Google Scholar]
- Pacek M., Tutter A. V., Kubota Y., Takisawa H., Walter J. C. Localization of MCM2–7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol. Cell. 2006;21:581–587. doi: 10.1016/j.molcel.2006.01.030. [DOI] [PubMed] [Google Scholar]
- Pacek M., Walter J. C. A requirement for MCM7 and Cdc45 in chromosome unwinding during eukaryotic DNA replication. EMBO J. 2004;23:3667–3676. doi: 10.1038/sj.emboj.7600369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel T., Gores G. J., Kaufmann S. H. The role of proteases during apoptosis. FASEB J. 1996;10:587–597. doi: 10.1096/fasebj.10.5.8621058. [DOI] [PubMed] [Google Scholar]
- Prasanth S. G., Prasanth K. V., Siddiqui K., Spector D. L., Stillman B. Human Orc2 localizes to centrosomes, centromeres and heterochromatin during chromosome inheritance. EMBO J. 2004;23:2651–2663. doi: 10.1038/sj.emboj.7600255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricke R. M., Bielinsky A. K. Mcm10 regulates the stability and chromatin association of DNA polymerase-alpha. Mol. Cell. 2004;16:173–185. doi: 10.1016/j.molcel.2004.09.017. [DOI] [PubMed] [Google Scholar]
- Ricke R. M., Bielinsky A. K. A conserved Hsp10-like domain in Mcm10 is required to stabilize the catalytic subunit of DNA polymerase-alpha in budding yeast. J. Biol. Chem. 2006;281:18414–18425. doi: 10.1074/jbc.M513551200. [DOI] [PubMed] [Google Scholar]
- Sawyer S. L., Cheng I. H., Chai W., Tye B. K. Mcm10 and Cdc45 cooperate in origin activation in Saccharomyces cerevisiae. J. Mol. Biol. 2004;340:195–202. doi: 10.1016/j.jmb.2004.04.066. [DOI] [PubMed] [Google Scholar]
- Schwacha A., Bell S. P. Interactions between two catalytically distinct MCM subgroups are essential for coordinated ATP hydrolysis and DNA replication. Mol. Cell. 2001;8:1093–1104. doi: 10.1016/s1097-2765(01)00389-6. [DOI] [PubMed] [Google Scholar]
- Shechter D., Ying C. Y., Gautier J. DNA unwinding is an Mcm complex-dependent and ATP hydrolysis-dependent process. J. Biol. Chem. 2004;279:45586–45593. doi: 10.1074/jbc.M407772200. [DOI] [PubMed] [Google Scholar]
- Smith G., D'Adda di Fagagna F., Lakin N., Jackson S. Cleavage and inactivation of ATM during apoptosis. Mol. Cell. Biol. 1999;19:6076–6084. doi: 10.1128/mcb.19.9.6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon N. A., Wright M. B., Chang S., Buckley A. M., Dumas L. B., Gaber R. F. Genetic and molecular analysis of DNA43 and DNA 52, two new cell-cycle genes in Saccharomyces cerevisiae. Yeast. 1992;8:273–289. doi: 10.1002/yea.320080405. [DOI] [PubMed] [Google Scholar]
- Takayama Y., Kamimura Y., Okawa M., Muramatsu S., Sugino A., Araki H. GINS, a novel multiprotein complex required for chromosomal DNA replication in budding yeast. Genes Dev. 2003;17:1153–1165. doi: 10.1101/gad.1065903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermes I., Haanen C., Steffens-Nakken H., Reutelingsperger C. A novel assay for apoptosis. Flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled Annexin V. J. Immunol. Methods. 1995;184:39–51. doi: 10.1016/0022-1759(95)00072-i. [DOI] [PubMed] [Google Scholar]
- Walter J., Newport J. Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase alpha. Mol. Cell. 2000;5:617–627. doi: 10.1016/s1097-2765(00)80241-5. [DOI] [PubMed] [Google Scholar]
- Walter J., Newport J. W. Regulation of replicon size in Xenopus egg extracts. Science. 1997;275:993–995. doi: 10.1126/science.275.5302.993. [DOI] [PubMed] [Google Scholar]
- Ward I. M., Chen J. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J. Biol. Chem. 2001;276:47759–47762. doi: 10.1074/jbc.C100569200. [DOI] [PubMed] [Google Scholar]
- Wohlschlegel J. A., Dhar S. K., Prokhorova T. A., Dutta A., Walter J. C. Xenopus Mcm10 binds to origins of DNA replication after Mcm2–7 and stimulates origin binding of Cdc45. Mol. Cell. 2002;9:233–240. doi: 10.1016/s1097-2765(02)00456-2. [DOI] [PubMed] [Google Scholar]
- Yang X., Gregan J., Lindner K., Young H., Kearsey S. E. Nuclear distribution and chromatin association of DNA polymerase alpha-primase is affected by TEV protease cleavage of Cdc23 (Mcm10) in fission yeast. BMC Mol. Biol. 2005;6:13. doi: 10.1186/1471-2199-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.