Abstract
Expression of smooth muscle myosin heavy chain (SM-MHC) is tightly controlled depending on the differentiated state of smooth muscle cells (SMCs). To better understand the mechanisms that regulate transcription of the SM-MHC gene in vivo, we tested the function of several conserved CArG elements contained within the –4200 to +11600 region of this gene that we had previously shown to drive SMC-specific expression in transgenic mice. CArG1 in the 5′-flanking sequence was required for all SMCs, while CArG2 and a novel intronic CArG element were differentially required in SMC subtypes. Of particular note, mutation of the intronic CArG selectively abolished expression in large arteries. A promoter construct containing three repeats of a conserved 227-bp intronic CArG-containing region was sufficient to direct transcription in vascular SMCs in transgenic mice, although this construct was also expressed in skeletal and cardiac muscle. These results support a model in which transcriptional regulation of SM-MHC is controlled by multiple positive and negative modular control regions that differ between SMCs and non-SMCs and among SMC subtypes. We also demonstrated that the CArG elements of the endogenous SM-MHC gene were bound by SRF in chromatin.
Introduction
Transcription of smooth muscle myosin heavy chain (SM-MHC) is strictly restricted within smooth muscle cells (SMCs) throughout development, which makes the gene one of the best markers for studying control of SMC lineage (1–3). Expression of SM-MHC isoforms is differentially regulated, depending on the differentiated state of SMCs during development (4) and also in the course of vascular lesion formation, maturation, and healing processes (5). It is likely that SM-MHC transcription is controlled by transcription factors that play central roles in differentiation and phenotypic modulation of SMCs. Therefore, elucidation of transcriptional controls of the SM-MHC gene are likely to contribute to a better overall understanding of the molecular mechanisms of SMC differentiation, phenotypic modulation, and proliferation.
Cell specification and differentiation can be considered as sequential and coordinate expression of an array of cell-specific/selective genes required for specialized cellular function. However, little is known regarding the mechanisms that control transcription during SMC differentiation. Several transcription factors have been suggested to be involved in SMC differentiation, including SRF, MEF2, and dHAND (6, 7). These factors are also required for skeletal and/or cardiac muscle differentiation, and relatively little is known regarding their unique roles in controlling expression of SMC-differentiation marker genes. A number of other transcription factors have also been shown to be involved in transcription of SMC-differentiation marker genes (8, 9). However, most of them are expressed in non-SMCs as well as SMCs. As such, a major unresolved issue in the field is the circuitry of transcription factors that control SMC-specific transcription.
Transcription of a number of SMC-specific/selective genes in cultured SMCs has been shown to be dependent on a cis element, CC(A/T)6GG, or CArG element (reviewed in ref. 10). However, there are major limitations in using cultured SMCs for studies of transcriptional regulation of SMC-specific genes. It is well established that mature SMCs can change their phenotype and differentiated state in vitro and in vivo responding to changing environmental cues (1, 11). Due to large differences in the environmental cues present in vivo versus in culture dishes, the phenotype of cultured SMCs is highly modulated with virtually all differentiated SMC-selective genes, including SM-MHC being downregulated (1, 11). Indeed, we have shown previously that SM-MHC promoter sequences that had strong SMC-specific activity in cultured SMCs (e.g., pCAT-4220 in ref. 12) had no activity in SMCs in vivo (12). As such, elucidation of the transcriptional regulatory mechanisms that control expression of SMC-differentiation genes must rely very heavily on use of in vivo experimental approaches, including studies in transgenic mice and employing in vivo footprinting methods in intact SM tissues (8, 12). In this regard, recent observations showing that expression of two SMC-differentiation marker genes SM22α and SM α-actin are dependent on multiple CArG elements in transgenic mice are of considerable significance (13–16).
We have identified recently SM-MHC genomic sequence sufficient to drive a reporter gene in transgenic mice in vivo (12, 17). A transgene harboring 4.2 kb of the 5′-flanking region, the entire first exon, and 11.5 kb of the first intronic sequence was capable of driving the reporter β-galactosidase gene in virtually all SM tissues. In contrast, 4.2 kb of the 5′-flanking region was not able to express β-galactosidase in any SMC subtype in multiple transgenic lines, clearly demonstrating the requirement of the first intronic sequence for expression of the SM-MHC gene in vivo. Although we and others have extensively characterized cis-regulatory elements within the 5′-flanking region of the SM-MHC gene, no transcriptional regulatory regions have been identified in the first intron. The proximal promoter region contains a TATA box and two CCTCCC boxes bound by Sp1-like factors (18). There is also a highly conserved 227-bp domain between –1321 and –1095 that we showed was required for maximal transcriptional activity in cultured SMCs (8, 19). This domain was shown to contain multiple cis elements including two positive-acting CArG elements (CArG1 and CArG2) that were important for transcriptional activity in cultured SMCs (8, 19). However, the functionality and roles of these CArG elements in vivo have not been investigated. In the present studies, we characterized multiple CArG elements of the SM-MHC gene, including a novel intronic CArG element in transcriptional regulation in vivo in transgenic mice. Results of the studies indicate that the SM-MHC gene is differentially controlled by multiple transcriptional regulatory modules in SMC subtypes in vivo.
Methods
Plasmid construction and transfection.
Mutant transgenic constructs of SM-MHC CArG elements were made in the context of –4200 to +11600 promoter/intron LacZ transgene (SM-MHC 4.2 + intron-LacZ plasmid; ref. 12). For simplicity, this construct is referred to as SM-MHC –4200/+11600 LacZ in this article. Site-directed mutagenesis was performed on small fragments subcloned in pBluescript II using GeneEditor (Stratagene, La Jolla, California, USA). The integrity of mutated fragments was confirmed by sequencing, and the fragments were subcloned back into the parental plasmid. The resultant mutant transgenic plasmids were tested for integrity by sequencing and restriction-enzyme mapping. To minimize the possibility of errors in DNA amplification, at least two independently constructed clones were tested for activity in cultured SMCs. Mutant sequences were as follows: CArG1, ttCCTTTTATGG to ggATCCTATGG; CArG2, CCTTTTTGGG to ATCCTTTGGG (19); intronic CArG, CCTTGTATGG to AGGCCTATGG.
The minimal thymidine kinase (TK) promoter taken from pBLCAT5 (20) was subcloned into pAUG LacZ (12). Subsequently a BstXI/BglI (+1447 to +1673) fragment of the SM-MHC first intron was subcloned into the TK LacZ vector so that the fragment was repeated three times upstream of the TK promoter (3×ICR-TK LacZ).
Transfection of the plasmids was performed using DOTAP (Roche Molecular Biochemicals, Indianapolis, Indiana, USA) as described previously (19). At least two independent clones were used for transfection, and the transfection of each plasmid was done at least in duplicate. Reporter activity was assayed by using ONPG as a substrate (21). The activity was normalized to the protein concentration of each cell lysate as measured by a DC protein assay kit (Bio-Rad, Hercules, California, USA). The endogenous β-galactosidase activity was determined by transfecting a nonfunctional DNA (pBluescript II) and was subtracted from the measured activity of each construct. Subsequently the activity was normalized to that of a promoterless construct, pAUG LacZ. ANOVA followed by the Bonferroni method was used for data analysis. Values of P less than 0.05 were considered statistically significant.
Generation and analysis of transgenic mice.
Transgenic mice were generated using standard methods as described previously (12) within the Transgenic Core Facility at the University of Virginia. Transgenic mice were used to establish breeding founder lines. Analysis of transgenic mice and histological analysis were performed as described previously (12). To determine possible position effects of transgene insertional sites on transgene expression, multiple independent founder lines were analyzed for each transgene construct. All animal procedures used in these studies were reviewed and approved by the University of Virginia Animal Use and Care Committee.
Preparation of nuclear extracts and electrophoretic mobility shift assays.
Preparation of nuclear extracts from cultured SMCs was described previously (19). Nuclear extracts from rat tissues were prepared as described previously (22) with the following modifications. In brief, tissues were taken from male Sprague-Dawley rats. Non-SMC layers were removed from the aorta, stomach, and bladder, and tissues were immediately frozen in liquid nitrogen. The frozen tissues were powdered and washed once with modified buffer A (10 mM HEPES, pH 7.9, 13 mM KCl, 0.1 mM EDTA, 0.5 mM DTT, 0.05% NP-40) with complete EDTA-free protease inhibitor (Roche Molecular Biochemicals). The samples were resuspended in 10 ml of buffer A and incubated on ice for 5 minutes. The samples were centrifuged and resuspended in a 10-packed-cell volume of buffer A. NP-40 was added to the final concentration of 0.3%. The samples were then homogenized using a Dounce homogenizer. Disruption of cell membranes was confirmed by microscopic observation. The centrifuged samples were resuspended in modified buffer C (20 mM HEPES, pH 7.9, 420 mM NaCl, 0.2 mM EDTA, 25% glycerol, 0.5 mM DTT, 0.01% NP-40, complete EDTA-free) and incubated on ice with gentle agitation for 30 minutes. Cell debris was removed by centrifugation. The samples were changed for buffer and were enriched using Ultrafree-4 concentrator (Millipore Corp., Bedford, Massachusetts, USA). Recombinant SRF was produced using in vitro transcription/translation system (Promega Corp., Madison, Wisconsin, USA).
The sequences of sense strands of electrophoretic mobility shift assay (EMSA) probes were as follows: CArG1, 5′-gacttccttttatggcctga-3′; CArG2, 5′-cctggcctttttgggttgtt-3′; and intronic CArG, 5′-catgcccttgtatggtagtg-3′. EMSAs were performed as described previously (21). In brief, 20,000 cpm of 32P-labeled probe was incubated with nuclear extracts in 20 μl of binding buffer (10 mM Tris-HCl, pH7.5, 50 mM NaCl, 0.5 mM DTT, 10% glycerol, and 0.05% NP-40) with 0.25 μg of poly(dI-dC)·(dI-dC). Reactions were incubated on ice for 20 minutes. For supershift assays, 1 μl of anti-SRF Ab was added after the 20-minute incubation period, and the reactions were incubated for an additional 10 minutes. The reactions were run on 5% polyacrylamide gels.
Chromatin immunoprecipitation.
L6 rat skeletal myoblasts were cultured in α-MEM (Life Technologies Inc., Rockville, Maryland, USA) supplemented with 2% FBS for 7 days to induce myotube formation. L6 myotubes, L6 myoblasts, Rat1 fibroblasts, and rat aortic SMCs in 100-mm dishes were fixed directly by adding 280 μl of 37% formaldehyde to 10 ml of culture media and incubating at 37°C for 10 minutes. The fixed cells were harvested and prepared for immunoprecipitation using the protocol of chromatin immunoprecipitation (ChIP) assay kit (Upstate Biotechnology Inc., Lake Placid, New York, USA) with minor modifications. A quarter of the sample was precleared with salmon sperm DNA/protein A agarose (Upstate Biotechnology Inc.) and subsequently incubated with either 2 μl of anti-SRF Ab (Santa Cruz Biotechnology Inc., Santa Cruz, California, USA) or no Ab at 4°C overnight. Chromatin samples were immunoprecipitated using salmon sperm DNA/protein A (Upstate Biotechnology Inc.). Samples were washed two times with 1 ml of wash buffer A (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.1, 150 mM NaCl), once with wash buffer B (0.25 M LiCl, 0.5% NP-40, 0.5% sodium deoxycholate, 1 mM EDTA, 10 mM Tris-HCl, pH 8.1), and two times with TE (10 mM Tris-HCl, pH8.0, 1 mM EDTA). Immune complexes were eluted and subsequently reverse cross-linked and purified by phenol/chloroform extraction. Ethanol-precipitated DNA pellets were redissolved in 40 μl of TE buffer. The supernatant of an immunoprecipitation reaction done in the absence of SRF Ab was purified and used as a control to show total input DNA. The supernatant DNA was diluted 1:100 before PCR analysis. One microliter of each sample was subjected to PCR amplification. PCR analysis was carried out using primers from different regions of the SM-MHC gene and promoter regions of a number of control genes that are silent in SMCs (skeletal α-actin, insulin, and β-globin). The sequences of PCR primers are shown in Table 1. After 32 (all primer sets except insulin) or 35 cycles (insulin) of amplification, PCR products were run on 2% agarose gels and analyzed by GelStar (BioWhittaker Molecular Applications, Rockland, Maine, USA) staining. As additional controls, the promoter regions of the genes either silent or lacking CArG elements were also amplified by PCR. The PCR samples of these promoters showed a low level of background chromatin immunoprecipitation. The sequences of the PCR primers were as follows: insulin, 5′-GCCAAAACTCTAGGGACTTTAGGAAGGATG-3′, 5′-GCCGGGCAACCTCCAGTGCCAAGGTCTGAAGATC-3′; β-globin, 5′-CAGCGTTTTCTTCAGAGGGAGTACCCAGAG-3′, 5′-TCAGAAGCAAATGTGAGGAGCGACTGATCC-3′; skeletal α-actin, 5′-CAGGCTGAGAAGCAGCCGAAGGGACTCTAG-3′, 5′-ACCTCCACCCTACCTGCTGCTCTGACTCTG-3′; SM-MHC –4000, 5′-ATGTCAGATGTCCTCTCACTGCTTTATTCC-3′,5′-AGCAAACAGCTTTAAATACGTATTGGCTTC-3′; 5′-CArG, 5′-CTGGAGCTCTTATTAGTACTGGGGTCCC-3′, 5′-ACTCAGGCCATAAAAGGAAGTCGAGGCAGAGTTGG-3′; intronic CArG, 5′-GGCCAAGCCACCCTGGAGAAACCTGGAC-3′, 5′-CCCAGAACTCAAGCCAGTCAGGCTGCATCG-3′. Due to the relatively low resolution of the ChIP method, we designed the PCR primers for the 5′-flanking CArG region for amplification of the region containing both CArG1 and CArG2.
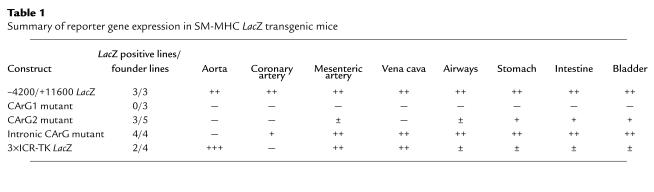
Table 1.
Summary of reporter gene expression in SM-MHC LacZ transgenic mice
Results
The SM-MHC first intron contained a conserved CArG element required for maximal promoter activity in cultured SMCs.
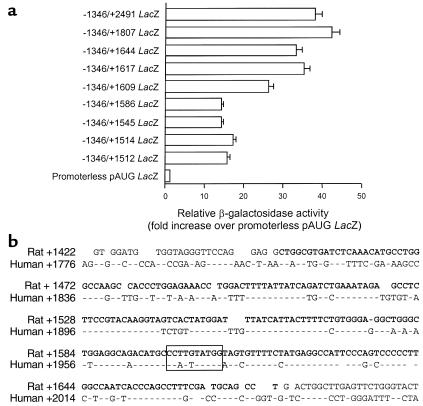
Our previous studies using transgenic mice clearly demonstrated the requirement of the first intronic sequence for transcription of the SM-MHC gene in vivo (12). Addition of the first intronic sequence to the 4,200-bp of the 5′-flanking sequence increased reporter activity in cultured SMCs approximately fourfold (57-fold vs. 14-fold activity over promoterless pAUG LacZ). Thus, we first examined transcriptional activity of the first intron in cultured rat aortic SMCs by using a series of 3′-deletion constructs as a means to identify putative cis-regulatory elements that would subsequently be tested in vivo in transgenic mice. Deletion of the region from +2491 to +417 decreased activity significantly (56-fold vs. 17-fold activity over pAUG LacZ). To identify possible cis elements in the region +2491 to +417, we further analyzed this region using finer deletion mutants. A series of finer deletion mutants was constructed in the context of 1,346 bp of the 5′-flanking region and transfected into cultured SMCs (Figure 1a). Significant reductions in reporter activity were observed when the sequence from +1617 to +1586 was deleted (Figure 1a). Within this region there is a CArG-like element at +1599 that is also present at an equivalent region in the human SM-MHC intron (Figure 1b).
Figure 1.
The intronic CArG element. (a) A series of 3′-end deletion mutants of the SM-MHC LacZ sequence was generated and assayed for reporter activity in cultured rat SMCs. The β-galactosidase activity of each construct is expressed relative to the activity of the promoterless pAUG LacZ. Error bars show SE. (b) The nucleotide sequence (+1422 to +1696 from the transcription start site) of a portion of the rat SM-MHC first intron was compared with the corresponding human genomic sequence (GenBank accession no. U91323). The intronic CArG element is boxed. Note that the human intronic CArG lacks a G substitution within the central A/T-rich sequence and perfectly matches the CArG consensus. Bold letters indicate the region used in 3×ICR-TK LacZ construct. Nucleotides conserved with the rat sequence are indicated by dashes.
We have previously identified two CArG elements in the 5′-flanking sequence of the SM-MHC gene that are functional in the context of –1346 to +88 region in cultured SMCs (8, 19). However, since the 5′-flanking sequence alone is completely inactive in SMC in vivo in transgenic mice, it is critical to retest the functionality of these cis elements within the context of the –4200/+11600 LacZ construct shown previously to be active in SMCs in vivo (12). To ensure the efficacy of mutations in abrogating transcription factor binding, we first performed a series of EMSA experiments with each CArG element. Consistent with our previous results (8, 19), both CArG1 and CArG2 probes bound SRF in nuclear extracts prepared from cultured SMCs (see Figure 5, lanes 21 and 22). In addition, as expected based on sequence analysis, the SM-MHC intronic CArG also exhibited SRF binding (see Figure 5, lanes 23; complexes A and B). Mutations of each CArG element completely abolished SRF-binding activity in EMSAs (ref. 19 and data not shown). We then tested the effects of these same CArG mutations on transcriptional activity of the –4200/+11600 LacZ SM-MHC construct in cultured SMCs. The mutations of CArG1, CArG2, and intronic CArG reduced reporter activity by 46%, 49%, and 74%, respectively.
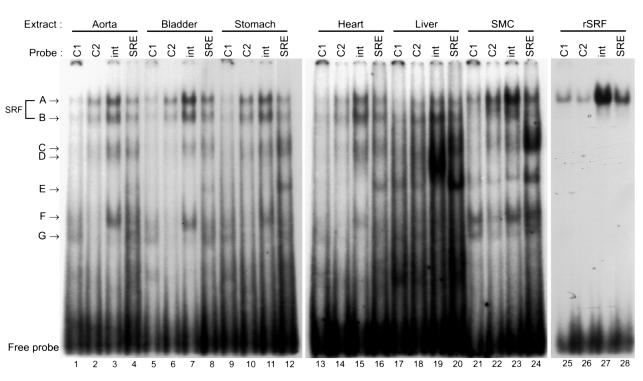
Figure 5.
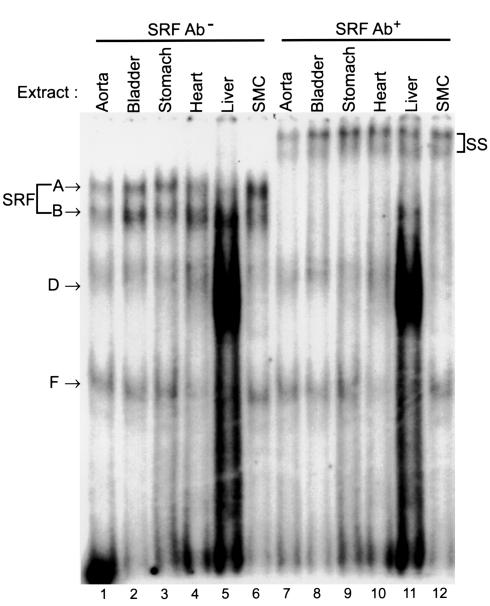
EMSA analysis of the CArG elements using tissue nuclear extracts. Radiolabeled 20-bp of double-stranded oligonucleotides encompassing CArG1, CArG2, intronic CArG, and c-fos SRE were incubated with either nuclear extracts prepared from tissues or SMCs or recombinant SRF. The amount of nuclear extracts was determined to produce SRF shift bands of similar intensity: 4 μg aortic, 3 μg bladder, 3 μg stomach, 7 μg heart, 3 μg liver, and 5 μg rat SMC nuclear extracts. One microliter of programmed lysate was used for recombinant SRF.
Mutations of CArG elements demonstrated differential requirements of the CArG elements in SMC-subtypes in vivo.
To examine the functional roles of the CArG elements in vivo, the –4200/+11600 LacZ SM-MHC CArG-mutant constructs were used to generate transgenic mice. The expression patterns observed are summarized in Table 1. Mutation of CArG1 resulted in abrogation of LacZ expression in all SM tissues in all three independent transgenic founder lines analyzed. In contrast, all three transgenic founder lines of the wild-type SM-MHC –4200/+11600 LacZ construct showed reporter expression in virtually all SMC tissues (Figure 2). These data clearly demonstrate that CArG1 was required for expression of the SM-MHC gene in vivo in all SM tissues.
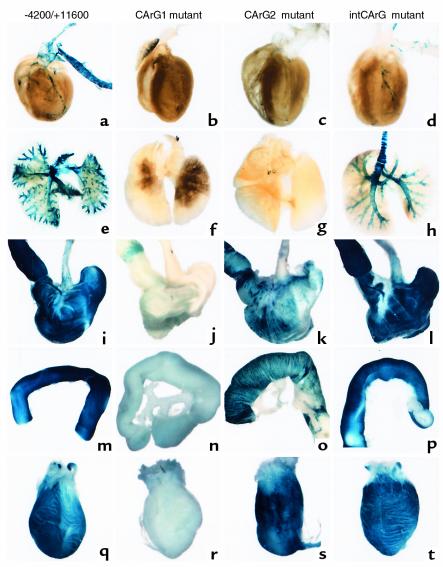
Figure 2.
Macroscopic examination of reporter-gene expression in wild-type and mutant SM-MHC LacZ transgenic mice. Transgenic mice (4 to 6 weeks old) were perfusion-fixed with a 2% formaldehyde/0.2% glutaraldehyde solution. Pictures show LacZ reporter expression in various tissues from wild-type –4200/+11600 LacZ mice (a, e, i, m, q), CArG1-mutant mice (b, f, j, n, r), CArG2-mutant mice (c, g, k, o, s), and intronic CArG-mutant mice (d, h, l, p, t). (a–d) Anterior view of the heart and aorta. (e–h) The lung. (i–l) The esophagus, stomach, and duodenum. (m–p) A portion of small intestine. (q–t) The bladder. Tissues were cleared by benzyl benzoate/benzyl alcohol in a–h.
Mutation of CArG2 resulted in differential reductions in reporter activity in SM tissues. LacZ expression in the gastrointestinal (GI) tract was decreased but was easily detectable in adult mice (Figure 2, i and m vs. k and o). Expression in the bladder was similar to that observed in wild-type mice (Figure 2, q vs. s). No expression was observed in large blood vessels, including the aorta, pulmonary, coronary, carotid, celiac, and femoral arteries, and the vena cava (Figure 2, a and e vs. c and g). However, very weak reporter expression was observed in smaller arteries, including small mesenteric arteries (data not shown). Mutation of CArG2 also virtually abolished expression in the trachea and bronchi (Figure 2, e vs. g).
Mutation of the intronic CArG resulted in a vascular SMC-specific phenotype. Reporter expression in the GI tract, urinary tract, and airways was equivalent to that of wild-type transgenic mice in adults and embryos of four independent intronic CArG-mutant lines (Figure 2, e, i, m, q vs. h, l, p, t). Expression in veins was also equivalent to that of the wild-type mice. However, expression in large arteries, including the aorta, common carotid arteries, and the main trunks of subclavian arteries was completely silenced in all lines (Figure 2, a and e vs. d and h). Interestingly, the small branching arteries from the thoracic aorta, including the intercostal arteries, showed transgene expression equivalent to that of the wild-type mice (Figure 3, c and d). In the carotid arteries no expression was observed in the proximal portion (Figure 3e), whereas in the distal common carotid arteries a few cells were stained positive and the internal and external carotid arteries were strongly stained. Strong expression of the intronic CArG mutant was also observed in arteries in the head including the basilar artery, arteries of the circle of Willis, and cerebral arteries (data not shown). Reporter expression was not detected in the abdominal aorta, whereas the branching arteries from the abdominal aorta, including the celiac, renal, and adrenal arteries were stained strongly positive (Figure 3, a vs. b). Indeed, the abrupt transition in expression from nondetectable to a high level between the conduit arteries and branch arteries was quite remarkable (see insets, Figure 3, b and f). Histological sectioning of blood vessels in the abdomen clearly showed selective abrogation of reporter expression in the aorta in intronic CArG-mutant transgenic mice (Figure 3, g–j). In the common iliac arteries expression was barely detectable, whereas expression was strong in their branches, including the femoral arteries (Figure 3f).
Figure 3.
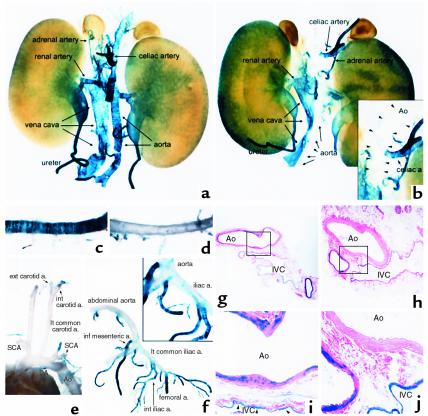
Large artery-specific silencing of the reporter gene in intronic CArG-mutant mice. Abdominal organs removed en block showed reporter expression in the blood vessels and urinary tract in the wild-type (a) and intronic CArG-mutant (b) transgenic mice. To better illustrate transgene expression in large arteries, several smaller arteries and connective tissues were removed, and the tissues were cleared. The supramesenteric artery, which was stained positive, was removed from the intronic CArG-mutant mouse tissues. A portion of the tissues is expanded in the inset in b. Arrowheads indicate the position of aorta that is not visible because of the lack of staining. Note that the blood vessels within the kidneys were not stained in either wild-type or intronic CArG mutants. (c and d) The thoracic aorta and branching arteries of the wild-type (c) and the intronic CArG-mutant (d) transgenic mice. (e) View of the large arteries in the cervicothoracic region of the intronic CArG-mutant transgenic mouse. (f) The large arteries and their branches in the abdomen of the intronic CArG mutant. A portion of the arteries is expanded in the inset in (f). (g–j) Histological examination of the abdominal aorta and inferior vena cava of the wild-type (g, i) and the intronic CArG mutant (h, j). The boxed areas (g, h) are shown by a higher magnification (i, j). Ao, aorta; DA, ductus arteriosus; IVC, inferior vena cava; SCA, subclavian artery.
Transgene expression in the coronary arteries was somewhat varied among the intronic CArG-mutant lines presumably due to position effects of transgene insertion sites. In two lines some expression was detectable in the coronary arteries, while little or no expression was observed in the other lines. However, even in the former two lines, overall transgene expression was clearly much weaker than that of the wild-type transgenic lines. Positive staining was restricted within the main trunks and a few major branches in the intronic CArG mutants, while in the wild-type mice expression was detectable in smaller branches. However, due to the qualitative nature of β-galactosidase staining and variability in expression level among the lines we could not conclude the extent of necessity of the intronic CArG in the coronary arteries.
Similarly, expression in the pulmonary arteries and veins was varied in mice containing the mutant intronic CArG transgene. Two lines, which showed transgene expression in the coronary arteries, had detectable transgene expression in the pulmonary blood vessels, whereas the other lines showed no expression. Even in two expressing lines, transgene expression was very weak, which made the staining of the lung look sparse as compared with that of the lung of wild-type mice as depicted in Figure 2. However, microscopically, some SMCs in the pulmonary blood vessels were stained positive (data not shown). Expression of the wild-type transgene in the pulmonary vessels was also somewhat varied among transgenic lines, and the expression level, especially that in the pulmonary veins, was generally weak as compared with other vascular beds. Given the variability and weakness of transgene expression in the pulmonary circulation, no definite conclusions can be made regarding the role of the intronic CArG in pulmonary blood vessels, although results showing no expression in two founder lines suggest that it may have some function. Reporter expression in intronic CArG-mutant transgenic mice showed that the intronic CArG was indispensable for transgene expression in large arteries, while it was dispensable in smaller arteries, veins, and visceral SMCs. The large arteries that absolutely required the intronic CArG largely fit the classification of elastic artery.
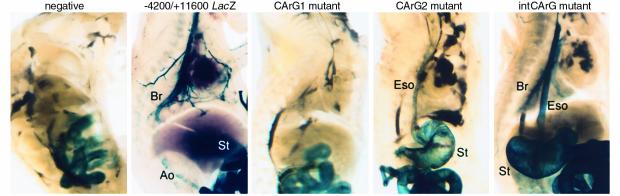
The SM-MHC gene is a marker of later-stage SM differentiation, and we showed previously that expression of the wild-type –4200/+11600 LacZ transgene was relatively weak in many SMC tissues until embryonic day (ED) 17.5–19.5 (12). Thus, to facilitate analysis of effects of CArG mutations during development, we restricted analyses to ED 19.5. Results showed the transgene expression pattern in each CArG-mutant transgenic mouse was largely consistent with that in adult mice (Figure 4). No expression was observed in embryos of CArG1-mutant transgenic mice. In the CArG2-mutant transgenic mice, reporter expression was observed only in the GI tract. In intronic CArG-mutant transgenic mice, the expression in the GI tract and airways was equivalent to that in the wild-type transgenic lines. While reporter expression in smaller arteries was easily detectable, no expression was detected in the large arteries in the intronic CArG-mutant transgenic mouse embryos.
Figure 4.
Transgene expression in embryos. Embryos were harvested at ED 19.5 and skinned and sectioned sagittally along the midline to permit dye penetration. The embryos were stained and cleared. The staining seen on the intestines in the negative and CArG2 mutant transgenic mice is due to endogenous β-galactosidase activity and limited within the epithelial layer. Eso, esophagus; H, heart; St, stomach; Br, bronchus.
In summary, the transgenic mouse data demonstrate that each CArG element is differentially required in SMC subtypes in vivo in transgenic mice. CArG1 is crucial for transcription in all SMCs, CArG2 is indispensable in large blood vessels but had a relatively minor role in the GI and urinary tracts, and the intronic CArG is absolutely required only in large elastic arteries.
SMCs in intact tissues expressed SRF and other proteins binding to the CArG elements.
As an initial step to determine mechanisms that control SMC subtype–specific transcriptional regulation through multiple CArG elements, we examined protein-binding properties of each CArG element using EMSAs. Since there were no SMC culture cell lines that had been shown to emulate faithfully differentiated phenotypes of SMC subtypes in terms of SM-MHC transcriptional control, we prepared nuclear extracts from intact rat tissues. As shown in Figure 5, each CArG probe formed several DNA-protein complexes with tissue nuclear extracts. The mobility of major shift bands (complexes A and B) formed with tissue nuclear extracts was the same as that with cultured SMC nuclear extracts. The mobility of complex A seen in tissues and culture cells was identical to that formed with recombinant SRF. Supershift assays using anti-SRF Ab showed that both complexes A and B contained SRF (Figure 6). Several non–SRF shift bands that were specifically competed by cold self-competitors, but not by unrelated sequences (data not shown), were also formed in the EMSA experiment (complexes C–G). Each probe formed largely similar shift-band patterns with the SM tissue nuclear extracts. However, the shift bands formed with liver or cultured SMC extracts were somewhat different from those formed with the SM tissue extracts. For example, CArG1, CArG2, and intronic CArG probes formed complex E with liver and cultured SMC extracts, while this complex was not formed with the SM tissue extracts. Conversely, complex F formed the intronic CArG probe, and SM tissue samples were not present in the liver samples. Although a further analysis is necessary to determine the significance of these non–SRF DNA-binding proteins in transcriptional regulation in cells, the data suggest that non–SRF DNA-binding proteins capable of binding to the CArG probes may be differentially expressed in SM and non-SM tissues.
Figure 6.
Supershift analysis of the intronic CArG-binding proteins. One microliter of anti-SRF Ab was added to the binding reaction of an intronic CArG probe and nuclear extracts after 20 minutes of incubation on ice, and the reactions were further incubated for 10 minutes on ice. Addition of the Ab resulted in supershift of SRF-containing complexes (A, B). Complexes A and B formed with other CArG probes were also supershifted (data not shown). SS indicates supershifted complexes.
SRF bound the CArG elements of the SM-MHC gene within intact chromatin under physiological conditions.
Although there is extensive evidence that SRF can bind to CArG elements in in vitro assays and that SRF is involved in transcriptional regulation through CArG elements in reporter assays in cultured SMCs (10, 19), there is a lack of direct evidence for involvement of SRF in transcription of the endogenous SMC marker genes within the context of intact chromatin. A number of studies have shown significant differences in transcription factor–binding to cis elements between episomal reporter genes and chromosomal genes (23). It has also been demonstrated that protein binding to cis elements in chromatin can be quite different from that observed with oligonucleotide probes in EMSAs (24). Previously, we used in vivo methylation footprinting methods to show that the 5′-flanking CArG elements of the endogenous SM-MHC gene in chromatin were occupied in vivo in the intact aorta (12). In vivo footprinting experiments likewise showed evidence that the intronic CArG was also occupied in the intact aorta (Manabe and Owens, unpublished observations). However, these sorts of experiments do not permit identification of the CArG-binding protein.
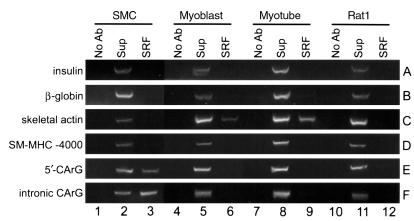
To directly address whether SRF bound the endogenous SM-MHC CArG elements, we employed ChIP assays. Intact cultured rat aortic SMCs, L6 rat myoblasts, L6 myotubes, and Rat1 fibroblasts were fixed directly with formaldehyde. Cross-linked chromatin was immunoprecipitated with anti-SRF Ab. The precipitated chromatin DNA was then purified and subjected to PCR analysis for enrichment of the target sequences. The gene promoters of insulin, β-globin, and skeletal α-actin genes (Figure 7, rows A–C), which are silent in SMCs, and a region (–4133 to –3832) of the SM-MHC 5′-flanking sequence (Figure 7, row D), which lacks CArG elements, were used in control reactions. Amplification of these sequences showed a background level of ChIP and PCR amplification (Figure 7, rows A–D). However, anti-SRF Ab specifically enriched the 5′-flanking CArG region (CArG1 and CArG2) and the intronic CArG regions of the SM-MHC gene (Figure 7, rows E and F, lane 3) in SMC chromatin as compared with the background amplifications of the promoters of negative control genes (Figure 7, rows A–D; lane 3). Importantly, the same SM-MHC regions were not enriched in immunoprecipitation samples of L6 or Rat1 cells that do not express the SM-MHC gene (Figure 7, rows E and F; lanes 6, 9, and 12). Since the promoter region of the skeletal α-actin has been shown to contain CArG elements active in skeletal myocytes, this promoter was used as a positive control for L6 cells. As expected, the skeletal actin promoter was enriched in SRF immunoprecipitation samples from L6 myoblasts and myotubes (Figure 7, row C, lanes 6 and 9), but not in SMC or Rat1 cells, further demonstrating the specificity of the SRF Ab in these experiments. It is important to note that the PCR detection methods used in these ChIP experiments are not quantitative. As such, it is impossible to determine the stoichiometry of SRF binding to the SM-MHC CArG elements. Nevertheless, the ChIP experiments indicated that at least some SM-MHC CArG regions were bound by SRF in chromatin in intact cultured SMCs. In addition, observations that SRF bound the CArG regions of endogenous SM-MHC gene in chromatin in intact SMCs, but not in L6 skeletal myocytes or Rat1 fibroblasts, provide evidence that mechanisms exist in vivo to control SRF binding to the SM-MHC CArGs in a cell-specific manner.
Figure 7.
ChIP analysis of SRF binding to the endogenous CArG regions. PCR was carried out to detect the endogenous CArG regions in immunoprecipitated chromatin fragments. Lanes 1, 4, 7, and 10 show PCR amplification of control precipitation samples with no Ab. Lanes 2, 5, 8, and 11 show amplification of 1:100 dilution samples of total input DNA for immunoprecipitation. Lanes 3, 6, 9, and 12 show amplification of target sequences in immunoprecipitated chromatin fragments with anti-SRF Ab.
A conserved region containing the intronic CArG conferred muscle type-selective transcription in vivo.
The data of transgenic mouse experiments clearly demonstrated that the intronic CArG element was necessary for transcription of the SM-MHC gene in the large arteries in vivo. As shown in Figure 1b, we found that the region containing the intronic CArG is highly conserved between the rat and human genes. To test if the intronic region could work as a distinct transcriptional regulatory module in vivo, three copies of the 227-bp sequence containing the highly homologous region (+1447 to +1673) were cloned in tandem 5′ to a minimal thymidine kinase (TK) promoter LacZ construct (3×ICR-TK LacZ). The construct showed very high activity (13.7-fold activity over the minimal TK LacZ construct) in cultured SMCs. This construct was used to produce transgenic mice. In one founder line (line 7240) among four founder lines, very strong reporter expression was observed in vascular SMCs (Figure 8). Another line (line 7249) also showed expression in SMCs, although not as strong. The other two lines were negative for staining. Although two founder lines did not express the 3×ICR-TK LacZ transgene, it seems unlikely that the cell-restricted activity observed in lines 7240 and 7249 was due solely to locus-dependent activation associated with the site of transgene insertion. Rather, results support that the 227-bp intronic sequence can direct transcription at least in some SMCs in vivo when coupled with a minimal TK promoter.
Figure 8.
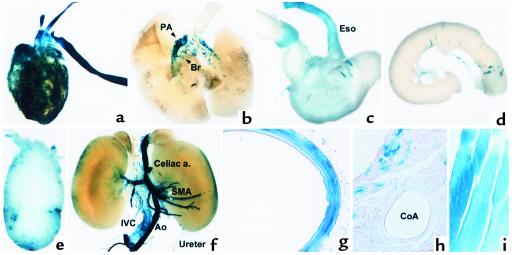
Transgene expression of the intronic CArG region minimal TK-LacZ. Various tissues of 4-week-old transgenic mice of the 3×ICR-TK LacZ line 7240 were stained for β-galactosidase activity. (a and b) Anterior view of the heart and lung. (c) The esophagus, stomach, and duodenum. (d) A part of the small intestine. (e) The bladder. (f) Anterior view of abdominal organs and great blood vessels. (g–i) Histological examination of the thoracic aorta (g), cardiac muscle and coronary artery (h), and intercostal muscle (i) of the 3×ICR-TK LacZ transgenic mouse. PA, pulmonary artery; SMA, superior mesenteric artery; CoA, coronary artery.
In line 7240, reporter expression was particularly prominent in the large arteries, including the aorta, carotid, and pulmonary arteries (Figure 8, a, b, f, g). Reporter expression was also strong in intermediate-size arteries (Figure 8f). Transgene expression in smaller arteries was relatively weaker than that in large arteries, and not all the smaller arteries were stained positive. Reporter expression was also observed in large veins, including the vena cava (Figure 8f). While the expression in vascular SMCs was very strong, transgene expression was very weak in visceral SMCs. Only a few cells were stained positive in the stomach, intestine, and bladder (Figure 8, c–e). Interestingly, strong reporter expression was also observed in the heart and skeletal muscle. In the heart, while cardiac muscle cells were stained positive for β-galactosidase expression, no transgene expression was observed in SMCs in coronary vessels (Figure 8h). Various skeletal muscle cells also expressed the transgene (Figure 8i). The data provide evidence that the conserved region containing the intronic CArG is capable of driving transcription in subsets of SMCs in vivo but lacks the complete SMC specificity seen with the endogenous SM-MHC gene and the –4200/+11600 LacZ transgene.
Discussion
Multiple CArG elements are required for in vivo transcription of SMC-specific genes.
The present studies demonstrate that multiple CArG elements play a key role in regulation of SM-MHC in vivo. Results of the present studies also provide the very first evidence, to our knowledge, showing that CArG elements exhibit differential activity in different subtypes of SMC, although previous studies have shown that CArG elements of other SMC marker genes are differentially required in SMCs vs. striated muscle cells in vivo (13–16). Mutational analyses of the SM α-actin CArG elements demonstrated that the intronic CArG was dispensable for expression in embryonic skeletal and cardiac muscle cells, but was required for expression in SMCs. In contrast, CArG B in the 5′-flanking sequence was required for all three muscle types. The more proximal CArG element of the SM22α promoter was found to be required for expression in all tissues in transgenic mice, while the distal CArG element was dispensable for expression in arterial SMCs, but had a minor role in skeletal and cardiac muscle cells (13–15). Results of our analyses of the 3×ICR-TK LacZ transgene showed that the region was sufficient to drive expression in only a subset of SMC subtypes and, unlike the endogenous SM-MHC gene, was also expressed in skeletal and cardiac muscle. Taken together, the results (a) demonstrate the multiplicity of regulatory programs that control expression of SMC-differentiation marker genes in vivo; (b) suggest that each of the multiple CArG elements mediates distinct information for transcriptional regulation in different cell types in vivo; and (c) imply that the spatial and temporal regulation of SMC genes is not governed by a single regulatory region or an enhancer. Indeed, the model that has emerged is that transcription of these SMC marker genes in vivo requires multiple positive and negative transcriptional regulatory modules that mediate responsiveness of SMC genes to changing environmental cues. We will discuss the implications of this multiplicity of transcriptional regulation and how CArG elements and SRF may function within this complex transcriptional regulatory program in the following sections.
SMC-specific transcriptional control requires multiple transcriptional regulatory modules.
A growing body of evidence indicates that spatial and temporal regulation of gene expression requires a complex regulatory system consisting of multiple regulatory modules, each of which contains multiple cis elements (25). For example, the sea urchin Endo 16 gene contains at least 33 transcription factor–binding sites in seven modules within a 2-kb region, and the modules function differentially at various stages of development (26). The results of the transgenic mice of the CArG-mutant constructs indicate that at least two regions (i.e., the 5′-flanking CArG and intronic CArG regions) are required for in vivo transcription of the SM-MHC gene. We are further mapping transcriptional regulatory modules in the SM-MHC gene locus. Preliminary data indicate that the 5′-flanking and first intron contain multiple positive and negative transcriptional regulatory regions and that different SMC subtypes require different subsets of modules (Manabe and Owens, unpublished observations). Why might SM-MHC transcription require such a complex transcriptional regulatory scheme in vivo?
It is evident that vascular SMCs within different vascular beds reside in vastly divergent local environments in vivo. Differences in the physiological role of vascular beds with respect to blood pressure, flow, and tone require very diverse vessel wall structures (27). SMCs are thus undoubtedly exposed to quite different vasoactive/neuronal stimuli and environmental cues from one vascular bed to another. If one considers the differences between vascular and visceral SMCs, the diversity is even more prominent, and it is well established that SMCs are derived from different embryological origins (28). Finally, one must also consider that the functions of SMCs can vary greatly during development and in adult animals due to their key role in matrix deposition and vessel morphogenesis, as well as in vascular repair (28). Due to these many differences, SMCs in vivo need to respond to very diverse inputs (environmental cues) that activate various intracellular signaling pathways and coordinately express necessary genes. It is thus conceivable that even to control the same gene, such as SM-MHC, SMC subpopulations in different environments may need to use distinct sets of regulatory pathways. In other words, the SM-MHC gene regulatory program evolved so that it uses various regulatory pathways to control transcription in heterogenous extra- and intracellular environments. In fact, the differential requirement of the intronic CArG and CArG2 of the SM-MHC gene supports a hypothesis that distinct transcriptional regulatory programs are activated in SMC-subtypes.
Elastic artery-specific activity of the intronic CArG and heterogeneity of SMCs.
The most striking feature of the intronic CArG mutant was that the transgene was completely silent in elastic arteries such as the aorta, whereas expression was easily detectable in the intermediate and small-size arteries directly branched from the aorta. There are at least two possible explanations for the differential requirement of the intronic CArG that are not necessarily mutually exclusive. First, we need to consider the heterogeneity in the embryonic origin of SMCs between the large and smaller arteries. It has been postulated that SMCs have at least three embryonic origins: local mesenchymal cells, neural crest cells, and proepicardial cells (28). In the aorta, the aortic bulb and ascending aorta consist mainly of neural crest–derived SMCs, and the descending aorta consists mainly of mesenchymal-derived SMCs (29). However, in intronic CArG-mutant transgenic mice, SMCs in the aorta were stained negative irrespective of the position of the cells, and no known difference in lineage fits the distribution of the intronic CArG dependency. Therefore, it is unlikely that differences in embryonic origin solely determined the requirement of intronic CArG for transcriptional control.
Second, as discussed above, the heterogeneity in phenotype and function of SMCs in vivo is likely to require multiple transcriptional programs to control the same gene. The differences in the physiological functions of SMCs in elastic versus muscular arteries would also require SMCs to express distinct sets of genes to fulfill their functional roles. It is thus conceivable that the intronic CArG is integrated in a regulatory program that processes environmental cues unique to elastic arteries and controls gene expression important for the function of such vessels. A number of genes, including ion channels, contractile proteins, growth factors/receptors, and transcription factors have been shown to be differentially expressed in vascular beds. For example, a transcription factor, CHF1 (Hrt2/Hey2/HERP1/gridlock), has been shown to be expressed mainly in the aorta (30). It would be of interest to compare the transcriptional regulatory mechanisms of these genes and also to determine the function of differentially expressed transcription factors in control of SMC subtype–selective gene-regulatory programs.
SRF-dependent control of SM-MHC transcription in vivo.
The present studies provide evidence showing binding of SRF to the CArG elements of the endogenous SM-MHC gene in the context of intact chromatin, as opposed to oligonucleotide fragments employed in typical DNA-binding studies. Landerholm et al. recently showed that in an in vitro avian proepicardial cell differentiation system two types of dominant-negative SRF inhibited SMC differentiation and reduced expression of SMC marker genes, including SM α-actin and SM22α (6). These data demonstrate the significant role of SRF in the control of endogenous SMC-differentiation marker genes. A critical question is thus: how can SRF, which is clearly not SMC-specific, regulate SMC-specific gene expression? Various hypotheses have been postulated that are not mutually exclusive. First, although SRF is clearly not cell specific, there are very large differences in the level of SRF expression between different cell types that may contribute, at least in part, to cell-specific SRF-CArG–dependent gene expression (31). Second, the binding affinity of SRF may be regulated in a cell type–dependent manner by interactions with other proteins, such as MHox (32) or by phosphorylation (33). Third, SRF may form SMC-specific multiprotein complexes. Although we did not observe SMC-specific higher-order complexes in EMSA experiments, it is possible that a longer probe might form such a multiprotein complex in EMSAs. Lastly, chromatin remodeling may play a significant role in the regulation of activity of transcriptional regulatory modules. It is now well established that transcription factor binding to cis elements is greatly affected by chromatin structure. It has also been shown that various transcription factors bind histone acetylases and deacetylases and thereby modify chromatin structure (34). The results of ChIP assays (Figure 7) demonstrated that the SM-MHC CArG regions were bound by SRF only in SMCs, although nuclear extracts of L6 myocytes were perfectly capable of binding the CArG elements in EMSAs (Manabe and Owens, unpublished observations). Conversely, SRF bound the skeletal α-actin promoter only in L6 myocytes, but not in SMCs or fibroblasts. These data are potentially extremely important in that they suggest that the transcriptional regulatory regions of the endogenous SM-MHC gene are only active in SMC chromatin. That is, the transcriptional regulatory regions of the SM-MHC gene may be in “closed state” in the non-SM cell lines.
SMC subtype–selective transcription control mechanisms in vascular diseases.
In contrast to the main function of mature SMCs (i.e., contraction), one of the major functions of SMCs in developing blood vessels is to contribute to formation of the vascular wall through cell proliferation and production of ECM components. Such functions are also extremely important during repair of vascular injury and may contribute to postangioplasty restenosis (11). As such, it is likely that a part of the transcriptional regulatory programs that are normally activated in vascular development is reactivated by vascular injury and alters gene expression. It would thus be important to study the functions of the CArG elements during vascular development and in neointimal formation induced by vascular injury.
It is well-known that some vascular beds, including the coronary arteries and aorta, are more prone to atherogenesis (35). Our data provide evidence for SMC subtype–selective transcriptional regulatory mechanisms. It is tempting to speculate that this multiplicity in the transcriptional control mechanisms might in some way be related to differential susceptibility of different vessels to atherosclerosis. The modularity of the SM-MHC transcription program might also allow us to design gene therapy vectors to target specific subsets of SMCs. SMC-selective activity obtained by the intronic CArG region coupled with a minimal TK promoter in transgenic mice suggests that the region could be used as a building block for such vectors. The results of the present studies have revealed the complex nature of transcriptional control of the SM-MHC gene in vivo in SMC subtypes and the role that multiple cis-regulatory modules play in processing divergent environmental cues in vivo. Further studies on the SM-MHC gene regulation should provide additional insights into the complex and dynamic regulatory mechanisms that normally control SMC differentiation and how these processes are altered during phenotypic modulation of SMCs during injury repair and development of vascular diseases.
Acknowledgments
This study was supported by grants from the NIH (RO1 HL-57353, PO1 HL-19242, and RO1 HL-38854 to G.K. Owens and U54 HD-28934 to the University of Virginia Research Histology Core) and by a fellowship grant from the Virginia Affiliate of the American Heart Association (VA-F98255V to I. Manabe). We gratefully acknowledge the expert technical assistance of Diane Raines, Margaret Ober, and Angela Miller.
References
- 1.Owens GK. Regulation of differentiation of vascular smooth muscle cells. Physiol Rev. 1995;75:487–517. doi: 10.1152/physrev.1995.75.3.487. [DOI] [PubMed] [Google Scholar]
- 2.Kuro-o M, et al. Developmentally regulated expression of vascular smooth muscle myosin heavy chain isoforms. J Biol Chem. 1989;264:18272–18275. [PubMed] [Google Scholar]
- 3.Miano JM, Cserjesi P, Ligon KL, Periasamy M, Olson EN. Smooth muscle myosin heavy chain exclusively marks the smooth muscle lineage during mouse embryogenesis. Circ Res. 1994;75:803–812. doi: 10.1161/01.res.75.5.803. [DOI] [PubMed] [Google Scholar]
- 4.Kuro-o M, et al. cDNA cloning of a myosin heavy chain isoform in embryonic smooth muscle and its expression during vascular development and in arteriosclerosis. J Biol Chem. 1991;266:3768–3773. [PubMed] [Google Scholar]
- 5.Aikawa M, et al. Redifferentiation of smooth muscle cells after coronary angioplasty determined via myosin heavy chain expression. Circulation. 1997;96:82–90. doi: 10.1161/01.cir.96.1.82. [DOI] [PubMed] [Google Scholar]
- 6.Landerholm TE, et al. A role for serum response factor in coronary smooth muscle differentiation from proepicardial cells. Development. 1999;126:2053–2062. doi: 10.1242/dev.126.10.2053. [DOI] [PubMed] [Google Scholar]
- 7.Yamagishi H, Olson EN, Srivastava D. The basic helix-loop-helix transcription factor, dHAND, is required for vascular development. J Clin Invest. 2000;105:261–70. doi: 10.1172/JCI8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Madsen CS, Regan CP, Owens GK. Interaction of CArG elements and a GC-rich repressor element in transcriptional regulation of the smooth muscle myosin heavy chain gene in vascular smooth muscle cells. J Biol Chem. 1997;272:29842–29851. doi: 10.1074/jbc.272.47.29842. [DOI] [PubMed] [Google Scholar]
- 9.Watanabe N, et al. BTEB2, a Kruppel-like transcription factor, regulates expression of the SMemb/Nonmuscle myosin heavy chain B (SMemb/NMHC-B) gene. Circ Res. 1999;85:182–191. doi: 10.1161/01.res.85.2.182. [DOI] [PubMed] [Google Scholar]
- 10.Miano JM, Carlson MJ, Spencer JA, Misra RP. Serum response factor-dependent regulation of the smooth muscle calponin gene. J Biol Chem. 2000;275:9814–9822. doi: 10.1074/jbc.275.13.9814. [DOI] [PubMed] [Google Scholar]
- 11.Owens, G. 1996. Role of alterations in the differentiated state of vascular smooth muscle cells in atherogenesis. In Atherosclerosis and coronary artery disease. V. Fuster, R. Ross, and E. Topol, editors. Lippincott-Raven Publishers. Philadelphia, Pennsylvania, USA. 401–420.
- 12.Madsen CS, et al. Smooth muscle-specific expression of the smooth muscle myosin heavy chain gene in transgenic mice requires 5′-flanking and first intronic DNA sequence. Circ Res. 1998;82:908–917. doi: 10.1161/01.res.82.8.908. [DOI] [PubMed] [Google Scholar]
- 13.Moessler H, et al. The SM 22 promoter directs tissue-specific expression in arterial but not in venous or visceral smooth muscle cells in transgenic mice. Development. 1996;122:2415–2425. doi: 10.1242/dev.122.8.2415. [DOI] [PubMed] [Google Scholar]
- 14.Li L, Liu Z, Mercer B, Overbeek P, Olson EN. Evidence for serum response factor-mediated regulatory networks governing SM22alpha transcription in smooth, skeletal, and cardiac muscle cells. Dev Biol. 1997;187:311–321. doi: 10.1006/dbio.1997.8621. [DOI] [PubMed] [Google Scholar]
- 15.Kim S, Ip HS, Lu MM, Clendenin C, Parmacek MS. A serum response factor-dependent transcriptional regulatory program identifies distinct smooth muscle cell sublineages. Mol Cell Biol. 1997;17:2266–2278. doi: 10.1128/mcb.17.4.2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mack CP, Owens GK. Regulation of smooth muscle alpha-actin expression in vivo is dependent on CArG elements within the 5′ and first intron promoter regions. Circ Res. 1999;84:852–861. doi: 10.1161/01.res.84.7.852. [DOI] [PubMed] [Google Scholar]
- 17.Regan CP, Manabe I, Owens GK. Development of a smooth muscle-targeted cre recombinase mouse reveals novel insights regarding smooth muscle myosin heavy chain promoter regulation. Circ Res. 2000;87:363–369. doi: 10.1161/01.res.87.5.363. [DOI] [PubMed] [Google Scholar]
- 18.Watanabe M, et al. Structure and characterization of the 5′-flanking region of the mouse smooth muscle myosin heavy chain (SM1/2) gene. Circ Res. 1996;78:978–989. doi: 10.1161/01.res.78.6.978. [DOI] [PubMed] [Google Scholar]
- 19.Madsen CS, Hershey JC, Hautmann MB, White SL, Owens GK. Expression of the smooth muscle myosin heavy chain gene is regulated by a negative-acting GC-rich element located between two positive-acting serum response factor-binding elements. J Biol Chem. 1997;272:6332–6340. doi: 10.1074/jbc.272.10.6332. [DOI] [PubMed] [Google Scholar]
- 20.Boshart M, Kluppel M, Schmidt A, Schutz G, Luckow B. Reporter constructs with low background activity utilizing the cat gene. Gene. 1992;110:129–130. doi: 10.1016/0378-1119(92)90456-y. [DOI] [PubMed] [Google Scholar]
- 21.Manabe I, et al. Isolation of the embryonic form of smooth muscle myosin heavy chain (SMemb/NMHC-B) gene and characterization of its 5′-flanking region. Biochem Biophys Res Commun. 1997;239:598–605. doi: 10.1006/bbrc.1997.7512. [DOI] [PubMed] [Google Scholar]
- 22.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alberts AS, Geneste O, Treisman R. Activation of SRF-regulated chromosomal templates by Rho-family GTPases requires a signal that also induces H4 hyperacetylation. Cell. 1998;92:475–487. doi: 10.1016/s0092-8674(00)80941-1. [DOI] [PubMed] [Google Scholar]
- 24.Wen SC, et al. Loading of DNA-binding factors to an erythroid enhancer. Mol Cell Biol. 2000;20:1993–2003. doi: 10.1128/mcb.20.6.1993-2003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arnone MI, Davidson EH. The hardwiring of development: organization and function of genomic regulatory systems. Development. 1997;124:1851–1864. doi: 10.1242/dev.124.10.1851. [DOI] [PubMed] [Google Scholar]
- 26.Yuh C-H, Bolouri H, Davidson EH. Genomic cis-regulatory logic: experimental and computational analysis of a sea urchin gene. Science. 1998;279:1896–1902. doi: 10.1126/science.279.5358.1896. [DOI] [PubMed] [Google Scholar]
- 27.Milnor, W.R. 1990. Cardiovascular physiology. Oxford University Press. New York, New York, USA. 290–326.
- 28.Hungerford JE, Little CD. Developmental biology of the vascular smooth muscle cell: building a multilayered vessel wall. J Vasc Res. 1999;36:2–27. doi: 10.1159/000025622. [DOI] [PubMed] [Google Scholar]
- 29.Topouzis S, Majesky MW. Smooth muscle lineage diversity in the chick embryo. Two types of aortic smooth muscle cell differ in growth and receptor-mediated transcriptional responses to transforming growth factor-beta. Dev Biol. 1996;178:430–445. doi: 10.1006/dbio.1996.0229. [DOI] [PubMed] [Google Scholar]
- 30.Chin MT, et al. Cardiovascular basic helix loop helix factor 1, a novel transcriptional repressor expressed preferentially in the developing and adult cardiovascular system. J Biol Chem. 2000;275:6381–6387. doi: 10.1074/jbc.275.9.6381. [DOI] [PubMed] [Google Scholar]
- 31.Belaguli NS, Schildmeyer LA, Schwartz RJ. Organization and myogenic restricted expression of the murine serum response factor gene. A role for autoregulation. J Biol Chem. 1997;272:18222–18231. doi: 10.1074/jbc.272.29.18222. [DOI] [PubMed] [Google Scholar]
- 32.Hautmann MB, Thompson MM, Swartz EA, Olson EN, Owens GK. Angiotensin II-induced stimulation of smooth muscle alpha-actin expression by serum response factor and the homeodomain transcription factor MHox. Circ Res. 1997;81:600–610. doi: 10.1161/01.res.81.4.600. [DOI] [PubMed] [Google Scholar]
- 33.Garat C, et al. Induction of smooth muscle alpha-actin in vascular smooth muscle cells by arginine vasopressin is mediated by c-Jun amino-terminal kinases and p38 mitogen-activated protein kinase. J Biol Chem. 2000;275:22537–22543. doi: 10.1074/jbc.M003000200. [DOI] [PubMed] [Google Scholar]
- 34.Ng HH, Bird A. Histone deacetylases: silencers for hire. Trends Biochem Sci. 2000;25:121–126. doi: 10.1016/s0968-0004(00)01551-6. [DOI] [PubMed] [Google Scholar]
- 35.Ross R. Atherosclerosis: an inflammatory disease. N Engl J Med. 1999;340:115–126. doi: 10.1056/NEJM199901143400207. [DOI] [PubMed] [Google Scholar]