Abstract
Autophagy is a highly conserved, degradative process in eukaryotic cells. The rapamycin-sensitive Tor kinase complex 1 (TORC1) has a major role in regulating induction of autophagy; however, the regulatory mechanisms are not fully understood. Here, we find that the protein kinase A (PKA) and Sch9 signaling pathways regulate autophagy cooperatively in yeast. Autophagy is induced in cells when PKA and Sch9 are simultaneously inactivated. Mutant alleles of these kinases bearing a mutation that confers sensitivity to the ATP-analogue inhibitor C3-1′-naphthyl-methyl PP1 revealed that autophagy was induced independently of effects on Tor kinase. The PKA–Sch9-mediated autophagy depends on the autophagy-related 1 kinase complex, which is also essential for TORC1-regulated autophagy, the transcription factors Msn2/4, and the Rim15 kinase. The present results suggest that autophagy is controlled by the signals from at least three partly separate nutrient-sensing pathways that include PKA, Sch9, and TORC1.
INTRODUCTION
Eukaryotic cells respond, and physiologically adapt, to changes in their intracellular and extracellular environment. Autophagy is a major lysosomal/vacuolar degradative pathway for bulk proteins and damaged and/or unnecessary organelles. When cells are starved for some nutrients, a double-membrane vesicle, termed an autophagosome, is formed to sequester cytoplasm. Once completed, the autophagosome subsequently fuses with the lysosome/vacuole, resulting in the breakdown of the contents. Finally, the resulting macromolecules are released back into the cytosol and reused for the synthesis of new proteins that are required for cells to survive during these conditions (Yorimitsu and Klionsky, 2005; Mizushima and Klionsky, 2007). This process is widely conserved in eukaryotes from yeast to mammals, and it is involved in a variety of cellular physiological events such as development, proliferation, remodeling, death, and aging (Levine and Klionsky, 2004; Meijer and Codogno, 2006; Shintani and Klionsky, 2004a). Some yeasts also use a biosynthetic autophagy-related process termed the cytoplasm-to-vacuole targeting (Cvt) pathway that is used for the selective transport of the resident hydrolases aminopeptidase I (Ape1) and α-mannosidase (Yorimitsu and Klionsky, 2005). These hydrolases can also be transported into the vacuole as selective cargos by autophagy.
Although the mechanism of autophagy regulation is not fully understood, the target of rapamycin (Tor) signaling pathway has a major role in controlling induction (Noda and Ohsumi, 1998). Tor is a protein kinase that regulates cellular growth in response to nutrient availability. Tor forms two functionally distinct protein complexes, Tor complex 1 and 2 (TORC1 and TORC2; Loewith et al., 2002). TORC1 is particularly sensitive to the immunosuppressive drug rapamycin. It is also inactivated in nutrient-depleted conditions. Inactivation of TORC1 causes various cellular responses, including induction of autophagy (Schmelzle et al., 2004).
In yeast, ∼30 proteins are identified as functioning specifically in autophagy-related pathways. Most of theses autophagy-related (Atg) proteins localize at a perivacuolar site, termed the phagophore assembly site (PAS), where they are thought to function in the formation of the autophagosome. Of the Atg proteins, Atg1, Atg13, and Atg17 are candidates to receive the signal from TORC1 (Kamada et al., 2000; Kabeya et al., 2005). Rapamycin treatment or shifting to nitrogen starvation, which inactivates TORC1, rapidly alters the phosphorylation state of Atg13 from a hyperphosphorylated to a hypophosphorylated form. This conversion apparently facilitates the interaction of Atg13 with Atg1 and Atg17. Atg1 is a protein kinase, and its activity is stimulated by formation of a complex with Atg13 and Atg17 during autophagy, although the role of Atg1 kinase activity still remains unclear (Nair and Klionsky, 2005).
In addition to TORC1, the RAS/cAMP-dependent protein kinase A (PKA) signaling pathway also regulates autophagy from yeast to mammals (Budovskaya et al., 2004; Furuta et al., 2004; Schmelzle et al., 2004; Mavrakis et al., 2006). The RAS/PKA pathway plays an important role in regulation of growth in response to extracellular nutrients. In rich nutrient conditions in yeast, two redundant small GTPases, Ras1 and Ras2, are activated and they stimulate adenylate cyclase to produce cAMP. PKA consists of three redundant catalytic subunits, Tpk1, Tpk2, and Tpk3, and a regulatory subunit, Bcy1. Binding to cAMP allows the dissociation of Bcy1 from the catalytic subunits and activation of PKA (Thevelein and de Winde, 1999). The RAS/PKA pathway is also associated with some TORC1-regulated responses other than autophagy (Schmelzle et al., 2004).
Constitutive activation of PKA is effective in preventing induction of autophagy by rapamycin or nutrient-depletion, indicating that PKA functions as another negative regulator for autophagy (Budovskaya et al., 2004; Schmelzle et al., 2004). This idea is supported by the observation that inactivation of PKA with a dominant-negative Ras2G22A mutant can induce autophagy in nutrient-rich conditions without rapamycin (Budovskaya et al., 2004). However, the induction of autophagy in this situation is less efficient and slower than that seen with inactivation of TORC1. In Saccharomyces cerevisiae, PKA phosphorylation sites are present in Atg1, Atg13, and Atg18. It is still unclear how or whether the phosphorylation of these Atg proteins by PKA is functionally linked to autophagy. Atg1 is mislocalized in cells expressing a hyperactive Ras mutant, Ras2G19V, whereas an Atg1 mutant lacking PKA phosphorylation sites is properly localized at the PAS in the presence of this mutant (Budovskaya et al., 2005). However, this altered Atg1 did not display constitutive autophagy activity in the presence of the Ras2G19V mutant. Therefore, mislocalization of Atg1 does not by itself account for the defect in autophagy that results from the hyperactive Ras allele.
The protein kinase Sch9 is a homologue of mammalian protein kinase B (PKB)/Akt or p70S6 kinase, both of which are also involved in regulation of growth in response to nutrients. The relationship between the RAS/PKA and Sch9 signaling pathways is unclear, but they may function in parallel in several physiological pathways. SCH9 was originally identified as a multicopy suppressor of RAS/PKA signaling defects. Conversely, activation of PKA suppresses the slow growth phenotype seen with deletion of SCH9 (Toda et al., 1988). In addition, several lines of evidence also indicate that the three kinases PKA, Sch9, and TORC1 are involved in at least some of the same cellular processes. For example, these kinases may converge in the regulation of a protein kinase, Rim15, which is required for entry into stationary phase through activation of G0-related traits (Pedruzzi et al., 2003). In addition, loss of any of the signaling pathways regulated by these kinases extends yeast longevity, which is dependent on Msn2/4 and Rim15 (Fabrizio et al., 2001; Kaeberlein et al., 2005).
Here, we report that autophagy is stimulated in nutrient-rich conditions without inactivation of TORC1, by inactivation of both PKA and Sch9 by using inhibitor-sensitive alleles of SCH9 and TPK1, TPK2, and TPK3. The Atg1 kinase complex is essential for PKA/Sch9-mediated autophagy. Moreover, depletion of Msn2/4 and/or Rim15 blocks autophagy that is induced by PKA and Sch9 inactivation. Our results suggest that these three intracellular nutrient-sensory kinases cooperatively regulate the induction of autophagy.
MATERIALS AND METHODS
Strains and Media
The S. cerevisiae strains used in this study are listed in Table 1. Gene deletions were performed by a polymerase chain reaction (PCR)-based procedure. Yeast strains were grown or incubated in YPD, synthetic medium (SMD) or starvation medium (SD-N) as described previously (Cheong et al., 2005). Rapamycin (Fermentek, Jerusalem, Israel) and C3-1′-naphthyl-methyl PP1 (1NM-PP1; a kind gift from Dr. Kevan Shokat, University of California, San Francisco) were used at a final concentration of 0.2 μg/ml and 0.1 μM, respectively.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| TYY164 | W303-1B atg1Δ::kan | This study |
| TYY166 | Y2864 sch9T492Gatg1Δ::kan | This study |
| TYY167 | W303-1B tpk1M164Gtpk2M147Gtpk3M165Gsch9T492Gatg1Δ::kan | This study |
| TYY172 | W303-1B pho13Δ::kan pho8Δ60::HIS3 | This study |
| TYY173 | W303-1B tpk1M164Gtpk2M147Gtpk3M165Gpho13Δ::kan pho8Δ60::HIS3 | This study |
| TYY174 | W303-1B tpk1M164Gtpk2M147Gtpk3M165Gsch9T492Gpho13Δ::kan pho8Δ60::HIS3 | This study |
| TYY175 | W303-1B vac8Δ::kan | This study |
| TYY176 | W303-1B tpk1M164Gtpk2M147Gtpk3M165Gvac8Δ::kan | This study |
| TYY177 | Y2864 sch9T492Gvac8Δ::URA3 | This study |
| TYY178 | W303-1B tpk1M164Gtpk2M147Gtpk3M165Gsch9T492Gvac8Δ::kan | This study |
| TYY179 | W303-1B atg1Δ::kan vac8Δ::HIS3 | This study |
| TYY180 | W303-1B tpk1M164Gtpk2M147Gtpk3M165Gsch9T492Gatg1Δ::kan vac8Δ::URA3 | This study |
| TYY181 | W303-1B pho13Δ::kan pho8Δ60::HIS3 atg1Δ::URA3 | This study |
| TYY182 | W303-1B tpk1M164Gtpk2M147Gtpk3M165Gsch9T492Gpho13Δ::kan pho8Δ60::HIS3 atg1Δ::URA3 | This study |
| TYY183 | W303-1B atg13Δ::HIS3 | This study |
| TYY185 | W303-1B tpk1M164Gtpk2M147Gtpk3M165Gsch9T492Gatg13Δ::HIS3 | This study |
| TYY187 | Y2864 sch9T492Gpho13Δ::kan pho8Δ60::URA3 | This study |
| TYY190 | W303-1B atg17Δ::HIS3 | This study |
| TYY191 | W303-1B tpk1M164Gtpk2M147Gtpk3M165Gsch9T492Gatg17Δ::HIS3 | This study |
| TYY193 | W303-1B tpk1M164Gtpk2M147Gtpk3M165Gsch9T492Gmsn2Δ::LEU2 msn4Δ::HIS3 | This study |
| TYY197 | W303-1B tpk1M164Gtpk2M147Gtpk3M165Gsch9T492Gmsn2Δ::LEU2 msn4Δ::HIS3 rim15Δ::TRP1 | This study |
| TYY199 | W303-1B tpk1M164Gtpk2M147Gtpk3M165Gsch9T492Grim15Δ::HIS3 | This study |
| TYY201 | W303-1B GLN3-myc::HIS3 | This study |
| TYY202 | W303-1B tpk1M164Gtpk2M147Gtpk3M165Gsch9T492GGLN3-myc::HIS3 | This study |
| TYY220 | W303-1B bcy1Δ::LEU2 | This study |
| W303-1B | MATα leu2-3,112 ura3-1 his3-11,15 trp1-1 ade2-1 can1-100 | Thomas and Rothstein (1989) |
| Y2864 | W303-1B gal1::HIS3 | Wang et al. (2004) |
| Y3507 | Y2864 sch9T492G | This study |
| Y3527 | W303-1B tpk1M164Gtpk2M147Gtpk3M165G | This study |
| Y3528 | W303-1B tpk1M164Gtpk2M147Gtpk3M165Gsch9T492G | This study |
Plasmids
Plasmids pCuGFP-AUT7(416), YEp351[APG13], pATG1(415), pATG1K54A(415), and pJU675 (encoding Sch9) have been described previously (Kamada et al., 2000; Abeliovich et al., 2003; Urban et al., 2007; Yen et al., 2007). Plasmid pJU841 encodes a hyperactive Sch9 mutant containing five mutations of T723D, S726D, T737E, S758E, and S765E (Urban et al., 2007). To construct plasmid pNOP1ATG17-PA(314), PCR-amplified ATG17 was introduced into pNOP1-PA(314) by using NcoI and XhoI sites (He et al., 2006). Site-specific mutagenesis to generate mutations of C24R in pATG17-PA(314) and S508A, S515A in pATG1(415) was carried out as described previously (Yen et al., 2007).
Immunoblotting
Yeast cells were grown in SMD medium at 30°C to early log phase, and either 1NM-PP1 or rapamycin was added to each culture to induce autophagy. For starvation conditions, cells were shifted to SD-N medium. At the indicated times, cells were collected, and proteins were precipitated by addition of trichloroacetic acid (TCA). Protein extracts were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) followed by immunoblotting with appropriate antiserum or antibodies.
Assays for Autophagy
For monitoring bulk autophagy, the alkaline phosphatase activity of Pho8Δ60 and processing of green fluorescent protein (GFP)-Atg8 were carried out as described previously (Noda et al., 1995; Shintani and Klionsky, 2004b).
RESULTS
Autophagy Is Induced by Inactivation of PKA and Sch9
Multiple regulatory pathways control cell growth and related processes. For example, PKA and Sch9 have a parallel role in maintaining cell growth (Toda et al., 1988). Although PKA regulates autophagy, a role for Sch9 in autophagy regulation has not been demonstrated in yeast. Thus, to begin to understand the complex regulatory network that controls this essential process, we decided to examine the function of yeast Sch9 and to compare its role with that of PKA. We used strains carrying a mutation in the ATP-binding pocket of Tpk1 (M164G), Tpk2 (M147G), and Tpk3 (M165G), and/or Sch9 (T492G) that confer sensitivity to the cell-permeable ATP-analogue inhibitor 1NM-PP1. In the presence of inhibitor at the concentration used here, only kinases with the appropriate mutation are specifically inhibited (Jorgensen et al., 2004). Hereafter, the strain expressing the triple tpk1, tpk2, and tpk3 mutations is referred to as pka, and the strain expressing the mutant form of Sch9 as sch9; the inhibitor-sensitive quadruple mutant is referred to as pka sch9.
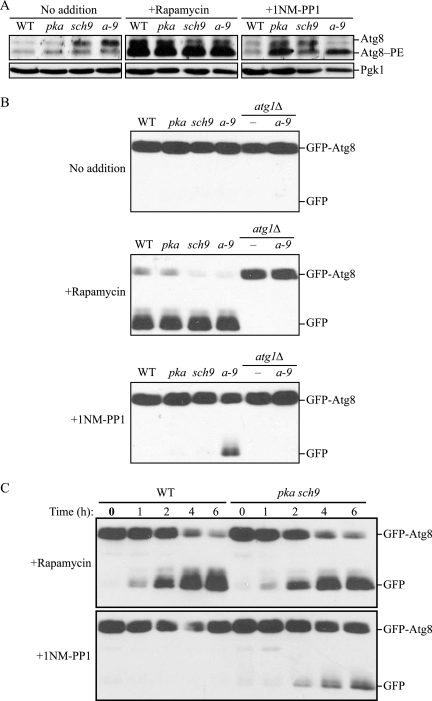
First, we examined the upregulation of Atg8. The expression of ATG8 is up-regulated shortly after induction of autophagy (Huang et al., 2000). Atg8 is an ubiquitin-like protein that is conjugated to phosphatidylethanolamine, generating Atg8–PE. Therefore, we analyzed the synthesis and conjugation of Atg8 as a way to monitor autophagy. Rapamycin treatment resulted in a significant increase in formation of Atg8–PE in wild-type cells as well as in pka, sch9, and pka sch9 cells as expected (Figure 1A). With 1NM-PP1 treatment, Atg8–PE formation was stimulated in pka, sch9 and pka sch9 cells, but not in the wild-type strain. These data suggest that inactivation of PKA and Sch9 induces autophagy.
Figure 1.
Inactivation of PKA and Sch9 induces autophagy. (A) PKA-Sch9 inactivation stimulates Atg8 expression and lipidation. Wild-type (W303-1B), pka (Y3527), sch9 (Y3507), and pka sch9 (a-9; Y3528) cells were grown for 6 h in SMD with or without rapamycin or 1NM-PP1. Proteins were precipitated with TCA and resolved by SDS-PAGE followed by immunoblotting with anti-Ape1 and anti-Pgk1 antiserum (as a loading control). Atg8 and Atg8 conjugated with phosphatidylethanolamine (Atg8–PE) were separated by 12% SDS-PAGE in the presence of 6 M urea followed by immunoblotting with anti-Atg8 or anti-Pgk1 antiserum. (B) GFP-Atg8 processing is enhanced by inactivation of PKA and Sch9. Protein extracts from wild-type, pka, sch9, pka sch9 (a-9), atg1Δ, and atg1Δ pka sch9 (TYY167) cells expressing GFP-Atg8 were analyzed as described in A by using anti-GFP antibodies. (C) Kinetics of GFP-Atg8 processing. Wild-type and pka sch9 cells expressing GFP-Atg8 grown to early log-phase were incubated in SMD containing rapamycin or 1NM-PP1. At the indicated times, TCA-precipitated proteins were subjected to immunoblotting, as described in B.
To verify this result, we monitored processing of GFP-Atg8. When autophagy is induced, GFP-Atg8 is transported into the vacuole inside the autophagic body. Atg8 is degraded after lysis of the autophagic body, whereas the GFP moiety is relatively resistant to proteolysis. Accordingly, monitoring free GFP processed from GFP-Atg8 reflects the level of autophagy (Shintani and Klionsky, 2004b). In contrast to monitoring Atg8–PE levels that reflect induction, the GFP-Atg8 processing assay measures autophagic flux; that is, this assay measures complete autophagy including vacuolar delivery and turnover of the cargo. GFP-Atg8 is also delivered to the vacuole inside of the Cvt vesicles that form under vegetative growth conditions, and by autophagosomes during basal autophagy; however, the level of free GFP resulting from either of these mechanisms is quite low and often undetectable (Figure 1B, no addition). In contrast, when treated with rapamycin, GFP-Atg8 was processed in wild-type, pka, sch9, and pka sch9 cells, but not in atg1Δ and atg1Δ pka sch9 cells that are defective in autophagy. This result indicates that all three mutants were able to induce autophagy in response to rapamycin treatment, that is, when Tor was inactivated. In contrast, 1NM-PP1 treatment revealed the presence of free GFP from GFP-Atg8 only in pka sch9 cells. In addition, the level of free GFP was significantly less than that seen after rapamycin treatment. Processing of GFP-Atg8 in the presence of 1NM-PP1 was not detected in atg1Δ pka sch9 cells, indicating that GFP-Atg8 was processed in an autophagy-dependent manner. To extend our analysis, we observed GFP-Atg8 processing over time after drug treatment (Figure 1C). When treated with rapamycin, wild-type and pka sch9 cells both processed GFP-Atg8 with similar kinetics, and free GFP was first detected within 1 h of drug addition. As expected from the steady-state analysis, 1NM-PP1 treatment did not allow GFP-Atg8 processing in wild-type cells, whereas it induced processing during the time course of drug treatment in the pka sch9 mutant; however, the response was slower than seen with rapamycin, with no detection of free GFP during the first hour, and the level of free GFP was considerably lower at comparable time points.
Processing of GFP-Atg8 monitors the delivery of membrane, specifically the autophagosome inner membrane or autophagic body, to the vacuole and subsequent lysis of the corresponding vesicle within the vacuole lumen. We next decided to examine the uptake of bulk cytoplasm, the cargo of the autophagosome. Pho8Δ60, a truncated variant of the vacuolar alkaline phosphatase Pho8, lacks the N-terminal transmembrane region that normally allows entry into the endoplasmic reticulum (ER), resulting in accumulation of the mutant protein in the cytosol (Noda et al., 1995). Cytosolic Pho8Δ60 is sequestered as a nonspecific cargo within autophagosomes upon induction of autophagy and delivered into the vacuole, where it is processed into an enzymatically active form due to removal of a C-terminal propeptide. Therefore, the alkaline phosphatase activity of Pho8Δ60 reflects the magnitude of autophagic cargo delivery.
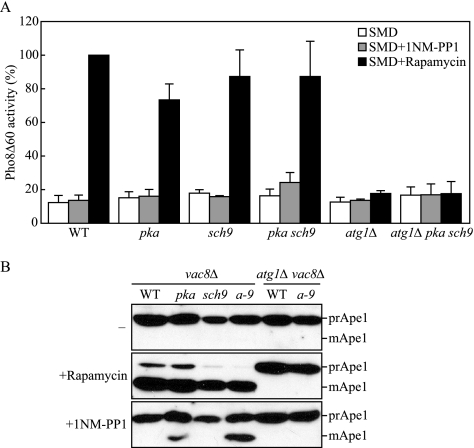
When autophagy was induced by rapamycin treatment, wild-type, pka, sch9, and pka sch9 cells showed a significant increase in Pho8Δ60 activity, whereas atg1Δ and atg1Δ pka sch9 cells defective in autophagy showed only the basal level of activity (Figure 2A). When treated with 1NM-PP1, only the pka sch9 cells showed any increase in the activity of Pho8Δ60, compared with the activity of the cells without treatment. The level of activity was quite low relative to that seen after rapamycin treatment, but this increase was reproducible. In addition, deletion of ATG1 prevented the induction of Pho8Δ60 activity due to 1NM-PP1 treatment in pka sch9 cells, indicating that the induced activity was due to an autophagic process. These results were consistent with the result of GFP-Atg8 processing; that is, only the pka sch9 double mutant displayed an increase in autophagy after inhibition with 1NM-PP1.
Figure 2.
Nonspecific and specific autophagy are induced with inactivation of PKA and Sch9. (A) Wild-type (TYY172), pka (TYY173), sch9 (TYY187), pka sch9 (TYY174) atg1Δ (TYY181), and atg1Δ pka sch9 (TYY182) cells expressing Pho8Δ60, a marker for nonspecific autophagy, were grown for 6 h in SMD with or without rapamycin or 1NM-PP1. The Pho8Δ60 activity was measured as described in Materials and Methods, and it was normalized to the activity of the wild-type cells with rapamycin treatment, which was set to 100%. Error bars indicate the SD of at least three independent experiments. (B) Protein extracts from vac8Δ (TYY175), vac8Δ pka (TYY176), vac8Δ sch9 (TYY177), vac8Δ pka sch9 (a-9; TYY178) vac8Δ atg1Δ (TYY179), and vac8Δ atg1Δ pka sch9 (TYY180) cells were analyzed by immunoblotting, as described in Figure 1, by using antiserum to Ape1.
We undertook one additional assay to confirm the previous results. In this case, we took advantage of the precursor Ape1 (prApe1) accumulation phenotype seen in the vac8Δ mutant. Vac8 is needed for the Cvt pathway, but it is not essential for autophagy; prApe1 that accumulates in the vac8Δ mutant in vegetative conditions is rapidly processed to the mature form when autophagy is induced by starvation (Cheong et al., 2005). Thus, maturation of prApe1 in the vac8Δ background is another assay to monitor autophagic induction. We found that vac8Δ cells accumulated only prApe1 in vegetative conditions regardless of the PKA or Sch9 phenotype (Figure 2B). Rapamycin treatment allowed the essentially complete maturation of prApe1 in all strains, except when ATG1 was additionally deleted, indicating that prApe1 processing occurred by the normal Atg protein-dependent mechanism. Treatment with 1NM-PP1 had no effect on the wild-type strain, but it caused maturation of prApe1 in the pka strain, and to a greater extent in the pka sch9 double mutant background; there was no processing of prApe1 in the sch9 vac8Δ strain, or in the triple mutant strain that was also deleted for ATG1. Together, we concluded that simultaneous inactivation of PKA and Sch9 can trigger autophagy, and as a corollary, that autophagy is negatively regulated cooperatively by the PKA and Sch9 signaling pathways.
PKA-Sch9 Regulation of Autophagy Depends on the Atg1 Kinase Complex
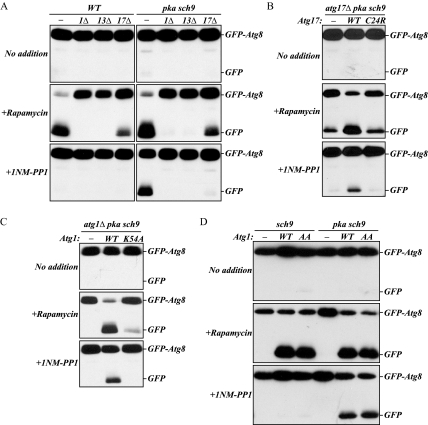
As noted above, autophagy was not induced in atg1Δ cells by inactivation of PKA and Sch9. Atg1 kinase activity is proposed to be involved in the induction of autophagy. Formation of a ternary complex among Atg1, Atg13, and Atg17 allows Atg1 kinase to be fully activated (Kamada et al., 2000; Kabeya et al., 2005). Thus, we examined whether the Atg1–Atg13–Atg17 kinase complex is involved in PKA-Sch9 regulation of autophagy. With rapamycin treatment, GFP-Atg8 processing was completely blocked in atg13Δ cells, similar to the result seen with atg1Δ cells, and partially blocked in atg17Δ cells as observed previously (Kamada et al., 2000; Cheong et al., 2005; Kabeya et al., 2005), in both the wild-type and pka sch9 background (Figure 3A). Similarly, atg1Δ, atg13Δ, and atg17Δ cells with pka sch9 mutations did not process GFP-Atg8 after 1NM-PP1 treatment. These results indicated that induction of autophagy by inactivation of PKA and Sch9 requires the Atg1–Atg13–Atg17 complex.
Figure 3.
Atg1 kinase activity is required for PKA-Sch9 regulation of autophagy. (A) Inactivation of PKA and Sch9 does not induce autophagy without the Atg1–Atg13–Atg17 kinase complex. Protein extracts from wild-type and pka pkb strains, and these strains harboring deletions in ATG1, ATG13, or ATG17 and expressing GFP-Atg8 were subjected to immunoblotting, as described in Figure 1. An Atg17 mutant defective in association with Atg13 (B) and a kinase-defective Atg1 mutant (C) block autophagy when PKA and Sch9 are inactivated. Protein extracts from the atg17Δ pka sch9 (TYY191) (B) or atg1Δ pka sch9 (TYY167) (C) cells harboring the empty vector, and a plasmid expressing wild-type Atg17 or the Atg17C24R mutant (B) or wild-type Atg1 or the Atg1K54A mutant (C) were subjected to immunoblotting, as described in A. (D) atg1Δ sch9 (TYY166) and atg1Δ pka sch9 cells harboring the empty vector, or a plasmid expressing wild-type Atg1 or the Atg1S508,515A mutant (AA) were grown and TCA-precipitated proteins were subjected to immunoblotting, as described in A.
The requirement for Atg17 was confirmed using the Atg17C24R mutant, in which cysteine at position 24 is substituted with arginine. Atg17C24R is not associated with the Atg1–Atg13 complex because of its inability to interact with Atg13, and cells expressing this mutant show only partial autophagy activity (Kabeya et al., 2005). We found that in the pka sch9 background after rapamycin treatment, GFP-Atg8 was processed less efficiently in atg17Δ cells expressing the Atg17C24R mutant, similar to cells with empty vector and at much lower levels than with cells expressing wild-type Atg17 (Figure 3B). Unlike wild-type Atg17, the Atg17C24R mutant also blocked GFP-Atg8 processing in pka sch9 atg17Δ cells treated with 1NM-PP1. This result suggested that the proper interaction between Atg13 and Atg17 is also required to induce autophagy resulting from inactivation of PKA and Sch9.
Furthermore, we continued the analysis by examining whether Atg1 kinase activity is required for autophagy to be induced by inactivation of PKA and Sch9 by using the kinase-defective Atg1 mutant Atg1K54A. The Atg1K54A mutant is largely defective for autophagy (Kamada et al., 2000). We found that GFP-Atg8 processing was essentially blocked in atg1Δ pka sch9 cells expressing the Atg1K54A mutant when treated with either rapamycin or 1NM-PP1 (Figure 3C), indicating that Atg1 kinase activity is also important for induction of PKA–Sch9-mediated autophagy. Together, these results indicated that PKA-Sch9 regulation of autophagy requires the kinase activity of the Atg1–Atg13–Atg17 complex, similar to starvation- or rapamycin-induced autophagy.
Finally, we investigated the role of PKA-dependent phosphorylation of Atg1 in the regulation of autophagy induction. Atg1 has two sites, Ser508 and Ser515, which are phosphorylated by PKA. When both of these serine residues are substituted by alanine, the resulting Atg1S508,515A mutant loses phosphorylation by PKA, but it still retained kinase activity at the same level as the wild-type protein (Budovskaya et al., 2005; our unpublished data). In addition, the Atg1S508,515A mutant efficiently localizes at the PAS in the presence of hyperactive Ras2G19V, whereas wild-type Atg1 is mislocalized when this mutant form of Ras2 is expressed. However, the phosphorylation-defective Atg1 mutant does not gain the ability to induce autophagy that is inhibited by the hyperactive Ras2G19V allele (Budovskaya et al., 2005). One possible explanation for this result could be the presence of active Sch9, because we found that autophagy was efficiently induced only after the inactivation of both PKA and Sch9.
To test this possibility, we examined processing of GFP-Atg8 in sch9-inactivated cells expressing the Atg1S508,515A mutant (Figure 3D). With rapamycin treatment, both wild-type and Atg1S508,515A mutant cells showed a similar level of processed free GFP in both sch9 and pka sch9 cells. 1NM-PP1 treatment allowed pka sch9 cells to process GFP-Atg8 with either the wild-type or mutant Atg1, similar to our previous results (Figure 1B). In contrast, when sch9 cells were treated with 1NM-PP1, neither the wild-type nor the mutant Atg1S508,515A cells showed GFP-Atg8 processing. These results indicated that loss of PKA-mediated phosphorylation of Atg1 did not induce autophagy even with inactivation of Sch9.
Tor Kinase Is Active during Autophagy When PKA and Sch9 Are Inactivated
The rapamycin-sensitive TORC1 has a major role in regulating autophagy. In nutrient-rich conditions, active TORC1 kinase maintains several proteins in a phosphorylated state. The direct or indirect substrates of TORC1 include the transcription factor Gln3 and Atg13. Once TORC1 is inactivated under nutrient starvation conditions or with rapamycin treatment, these proteins are dephosphorylated. Therefore, the activity of TORC1 can be examined by monitoring the phosphorylation status of these proteins. To test the possibility that inactivation of PKA and Sch9 can induce autophagy by modulating the TORC1 activity, we examined the phosphorylation of Gln3 and Atg13.
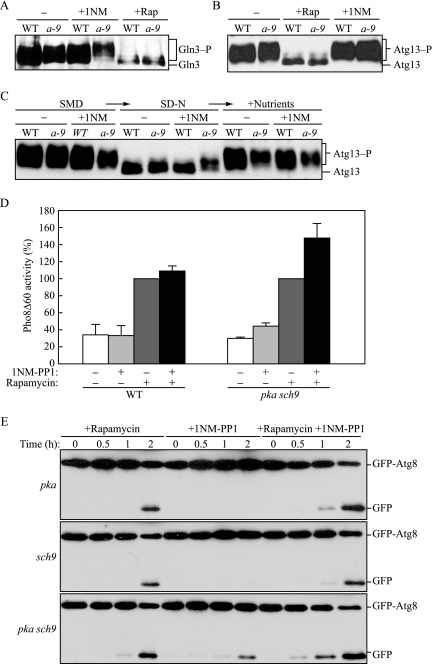
When TORC1 is inactivated, Gln3 is dephosphorylated, which allows it to enter the nucleus and function as an active transcription factor (Beck and Hall, 1999). The phosphorylated and dephosphorylated forms of Gln3 have a different mobility on SDS-PAGE gels, allowing them to be detected separately by immunoblotting. Rapamycin treatment generated a form of Gln3 with faster mobility, corresponding to dephosphorylation, in both wild-type and pka sch9 cells (Figure 4A). In contrast, with 1NM-PP1 treatment, Gln3 still showed the slower mobility characteristic of the phosphorylated form in both wild-type and pka sch9 cells. This result suggested that inactivation of PKA and Sch9 does not affect phosphorylation of Gln3.
Figure 4.
Tor kinase acts in parallel with PKA and Sch9. (A) Gln3 is phosphorylated when PKA and Sch9 are inactivated. Wild-type (TYY201), and pka sch9 (a-9; TYY202) cells expressing Gln3-myc were grown for 2 h in SMD with or without rapamycin or 1NM-PP1. TCA-precipitated proteins were subjected to immunoblotting with anti-myc antibody, as described in Figure 1. (B) Inactivation of PKA and Sch9 does not induce dephosphorylation of Atg13. Wild-type (W303-1B) and pka sch9 (a-9; Y3528) cells harboring a plasmid containing ATG13 (YEp351[APG13]) were grown for 2 h in SMD with or without rapamycin or 1NM-PP1. TCA-precipitated proteins were subjected to immunoblotting with anti-Atg13 antiserum, as described in A. (C) Inactivation of PKA and Sch9 partially affects dephosphorylation of Atg13 that occurs in −N conditions. Wild-type and pka sch9 (a-9) cells harboring the Atg13 plasmid were grown for 2 h in SMD with or without 1NM-PP1, shifted to SD-N for 30 min, and then YNB, amino acids, and vitamins were added to cultures to the same concentrations as those in SMD. At each time point, TCA-precipitated proteins were subjected to immunoblotting, as described in B. (D) Nonspecific autophagy is elevated with inactivation of both the Tor and PKA-Sch9 signaling pathways. Wild-type (TYY172) and pka sch9 (TYY174) cells expressing Pho8Δ60 were grown for 4 h in SMD with or without either or both rapamycin and 1NM-PP1 as indicated. The Pho8Δ60 activity was measured as described in Materials and Methods, and it was normalized to the activity of the cells treated with rapamycin, which was set at 100%. Error bars indicate the SD of at least three independent experiments. (E) pka sch9 (Y3528) cells expressing GFP-Atg8 grown at early log phase were incubated in SMD containing rapamycin and/or 1NM-PP1. At the indicated times, TCA-precipitated proteins were subjected to immunoblotting, as described in Figure 1.
To further analyze TORC1 activity, we examined the phosphorylation status of Atg13, which is hyperphosphorylated in a manner that is dependent on TORC1 activity (Kamada et al., 2000). The dephosphorylated forms of Atg13 resulting from rapamycin treatment showed faster mobility by SDS-PAGE than the hyperphosphorylated forms without treatment (Figure 4B), in agreement with previous results. As observed with Gln3, 1NM-PP1 treatment did not affect the mobility of Atg13 in pka sch9 cells or in the wild-type strain, comparable to the result without any treatment, suggesting that inactivation of PKA and Sch9 did not affect the phosphorylation state of this protein. These results suggested that TORC1 is active during autophagy that is induced by inactivation of PKA and Sch9.
Next, we shifted cells treated with 1NM-PP1 into SD-N medium, and we incubated them for 30 min to test whether the unknown phosphatase that dephosphorylates Atg13 was inactivated when PKA and Sch9 were inhibited (Figure 4C). When shifted to medium lacking nitrogen, Atg13 was no longer detected as the high-molecular-weight species in the wild-type cells with or without 1NM-PP1 treatment, indicating that it was dephosphorylated. Without 1NM-PP1, pka sch9 cells showed the same mobility shift of dephosphorylated Atg13 as the wild-type strain. However, in the presence of 1NM-PP1 the mobility of Atg13 was altered to an intermediate position, suggesting that Atg13 was not fully dephosphorylated. Finally, we examined the ability of Atg13 to be rephosphorylated. After 30-min incubation in SD-N, we added amino acids and yeast nitrogen base to the cultures and incubated them for an additional 30 min. Atg13 was highly phosphorylated in both wild-type and pka sch9 cells with or without 1NM-PP1 treatment. This result indicated that phosphatase activity affecting Atg13 is at most partially affected by PKA-Sch9 inactivation and that there is essentially no defect in rephosphorylation of this protein after a return to vegetative conditions.
PKA-Sch9 Regulate Autophagy in Parallel to the TORC1 Signaling Pathway
Constitutive activation of the RAS/PKA signaling pathway can suppress autophagy induced by rapamycin or starvation. It is possible that the RAS/PKA signaling pathway regulates autophagy downstream of TORC1 or in a parallel pathway. To differentiate between these possibilities, we treated pka sch9 cells with either or both rapamycin and 1NM-PP1 and compared the level of autophagy that was induced. If TORC1 and PKA-Sch9 are involved in autophagy in parallel pathways, we would predict that induction with both rapamycin and 1NM-PP1 would occur at a greater level than that seen with either treatment alone. In contrast, if TORC1 is upstream of PKA-Sch9, or vice versa, we would not expect to see an additive effect.
Wild-type cells did not display any change in Pho8Δ60 activity when treated with 1NM-PP1, whereas the pka sch9 strain showed a small increase (Figure 4D), similar to the result shown in Figure 2A. There was no significant difference in Pho8Δ60-dependent alkaline phosphatase activity when wild-type cells were treated with rapamycin compared with treatment with both rapamycin and 1NM-PP1 (Figure 4D). In contrast, double treatment with rapamycin and 1NM-PP1 resulted in ∼1.5-fold higher activity of Pho8Δ60 in pka sch9 cells than rapamycin treatment alone. This result suggested that rapamycin and 1NM-PP1 have an additive effect on autophagy. To further test this possibility, we observed GFP-Atg8 processing in pka sch9 cells treated with rapamycin and 1NM-PP1 (Figure 4E). Similar to the result seen with the Pho8Δ60 assay, rapamycin treatment resulted in a greater level of free GFP from GFP-Atg8 than seen with 1NM-PP1, and the combination of rapamycin and 1NM-PP1 resulted in a further increase in processing; the latter was particularly evident by the earlier appearance of free GFP seen in cells treated with both drugs. Together, these data suggest that PKA-Sch9 and TORC1 regulate autophagy, at least in part, in parallel pathways.
Msn2/Msn4 and Rim15 Are Involved in PKA-Sch9 Regulation of Autophagy
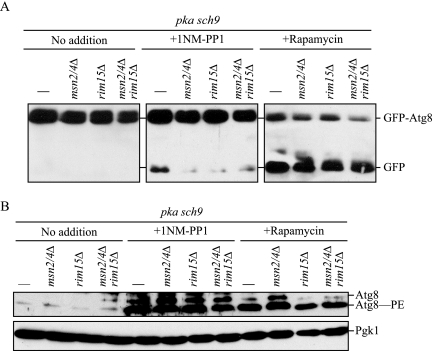
PKA and TORC1 suppress translocation of the redundant transcription factors Msn2 and Msn4 into the nucleus, where they actively transcribe several stress-response genes (Beck and Hall, 1999; Schmelzle et al., 2004). In addition, the activation and the nuclear localization of the protein kinase Rim15, which regulates the transcription of genes under stress conditions, is also regulated by PKA, Sch9, and TORC1; Rim15 is itself activated in the nucleus (Pedruzzi et al., 2003). We investigated whether these factors are required for PKA-Sch9 inhibition-mediated autophagy by the GFP-Atg8 processing assay (Figure 5A). When either or both Msn2/4 and Rim15 were deleted in pka sch9 cells, rapamycin treatment allowed GFP-Atg8 processing comparable with that seen in the pka sch9 background strain. In contrast, with 1NM-PP1 treatment, GFP-Atg8 processing was essentially blocked in pka sch9 cells with either single or double deletion of Msn2/4 and Rim15, being only slightly higher than in the control cells with no drug addition. These results indicated that Msn2/4 and Rim15 are required for the induction of autophagy that occurs upon inhibition of PKA-Sch9, but they are dispensable for TORC1-dependent rapamycin-induced autophagy.
Figure 5.
Msn2/4 and Rim15 are involved in autophagic flux, but not induction, resulting from inactivation of PKA and Sch9, but not from inactivation of the Tor signaling pathway. (A) Autophagic flux assessed by GFP-Atg8 processing was defective in the absence of Msn2/4 and/or Rim15. Protein extracts from pka sch9 (Y3528), msn2/4Δ pka sch9 (TYY193), rim15Δ pka sch9 (TYY197), and msn2/4Δ rim15Δ pka sch9 (TYY199) cells expressing GFP-Atg8 were subjected to immunoblotting, as in described in Figure 1. (B) Depletion of Msn2/4 and Rim15 does not affect enhancement of Atg8 expression or lipidation resulting from inactivation of PKA and Sch9. The cells were grown, and TCA-precipitated proteins were subjected to immunoblotting, as described in A.
We observed up-regulation of expression and lipidation of Atg8 in pka sch9 cells in the presence of 1NM-PP1 similar to the result seen with rapamycin (Figure 1A). Accordingly, we examined Atg8 in Msn2/4- and/or Rim15-depleted pka sch9 cells. When treated with rapamycin or 1NM-PP1, all mutant cells showed significant up-regulation of Atg8/Atg8–PE comparable to that of the pka sch9 background strain (Figure 5B), indicating that the defect in autophagy induction seen by inactivation of PKA-Sch9 without Msn2/4 and Rim15 is not due to an inability to up-regulate the expression of Atg8/Atg8–PE.
Activation of PKA and Sch9 Represses Autophagy
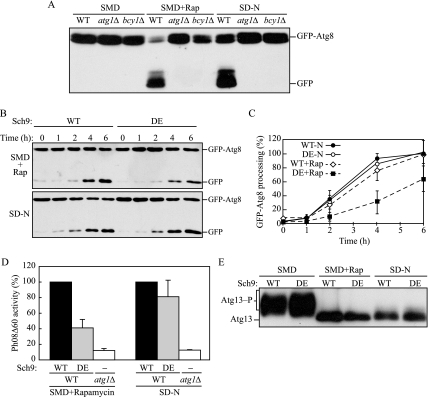
We determined that the simultaneous inactivation of PKA and Sch9 induces autophagy, and we next decided to examine whether activation of these kinases resulted in repression. We examined autophagy in bcy1Δ cells by monitoring GFP-Atg8 processing (Figure 6A). Bcy1 is a regulatory subunit of PKA and its depletion allows PKA to be constitutively active (Toda et al., 1987). No GFP-Atg8 was processed in either autophagy-inducing condition, rapamycin treatment or nitrogen starvation, in bcy1Δ cells, suggesting that PKA suppresses autophagy and can override inactivation of TORC1. This finding is consistent with the observation that the hyperactive Ras2V19G mutant is dominant negative and represses autophagy under otherwise inducing conditions (Budovskaya et al., 2004; Schmelzle et al., 2004).
Figure 6.
Constitutively active PKA and Sch9 suppress autophagy. (A) Constitutively active PKA suppresses autophagy. Wild-type (W303-1B), atg1Δ (TYY164), and bcy1Δ (TYY220) cells expressing GFP-Atg8 were grown for 4 h in SMD with or without rapamycin, or in SD-N. TCA-precipitated proteins were subjected to immunoblotting, as described in Figure 1. (B) A hyperactive Sch9 mutant delays processing of GFP-Atg8 by rapamycin treatment, but not under starvation conditions. Wild-type cells expressing GFP-Atg8 with a plasmid carrying wild type (WT) or hyperactive mutant Sch9 (DE) were grown in SMD containing rapamycin, or in SD-N. At the indicated times, TCA-precipitated proteins were subjected to immunoblotting, as described in A. (C) Band intensities of GFP-Atg8 and free GFP were quantified and normalized to those of wild-type cells treated with rapamycin or under starvation conditions for 6 h. Error bars indicate the SD of at least three independent experiments. DE, hyperactive Sch9. (D) The hyperactive Sch9 mutant blocks bulk autophagy by rapamycin treatment, but not under starvation conditions. Wild-type (TYY172) cells with a plasmid expressing WT or DE, or atg1Δ (TYY181) cells with the empty vector were grown for 4 h in SMD containing rapamycin, or in SD-N. The Pho8Δ60 activity was measured as described in Materials and Methods and normalized to the activity of the wild-type cells, which was set at 100%. Error bars indicate the SD of at least three independent experiments. (E) Hyperactive Sch9 does not affect dephosphorylation of Atg13 in response to rapamycin or nitrogen starvation. Wild-type (W303-1B) cells harboring the Atg13 plasmid along with a plasmid carrying WT or DE Sch9 were grown for 30 min in SMD with or without rapamycin, or in SD-N. TCA-precipitated proteins were subjected to immunoblotting, as described in Figure 4.
In addition, we checked whether enforced activation of Sch9 affects autophagy by using a Sch9DDEEE mutant, in which five residues at the phosphorylation sites are substituted with acidic residues (T723D, S263D, T737E, S758E, and S765E), resulting in a hyperactive Sch9 kinase (Urban et al., 2007). First, we tested GFP-Atg8 processing in cells with a plasmid carrying a wild-type or hyperactive mutant Sch9 (Figure 6B). Unlike the situation seen in bcy1Δ cells expressing hyperactive PKA, when treated with rapamycin, hyperactive Sch9 cells showed only a partial defect in GFP-Atg8 processing. When quantified, the processing with the hyperactive mutant was ∼60% that of the wild-type strain after 6-h treatment with rapamycin (Figure 6C). Furthermore, there was no significant difference in the processing between the wild-type and hyperactive mutant under starvation conditions (Figure 6, B and C). To extend this analysis, we performed the Pho8Δ60 assay with cells expressing a wild-type or hyperactive mutant Sch9 (Figure 6D). Rapamycin treatment of the hyperactive mutant showed ∼40% of the Pho8Δ60 activity compared with that of the wild-type strain, although under starvation conditions, the activity with the mutant was comparable with that of wild type, which was consistent with the results of GFP-Atg8 processing.
These results suggested that Sch9 is also a negative regulatory factor for autophagy. However, in contrast to PKA, hyperactive Sch9 was not able to completely suppress autophagy induction in rapamycin or SD-N conditions. To gain further insight, we examined the effect of hyperactive Sch9 on the phosphorylation status of Atg13 (Figure 6E). In nutrient-rich conditions without rapamycin, Atg13 was phosphorylated in the presence of hyperactive Sch9 at similar levels as seen with wild-type Sch9. Similarly, either nitrogen starvation or rapamycin treatment allowed Atg13 to be dephosphorylated to a similar extent with both wild-type and hyperactive Sch9. These results indicated that activation of Sch9 does not affect phosphorylation and dephosphorylation of Atg13, which is consistent with the result that activation of the Ras/PKA pathway does not affect phosphorylation and dephosphorylation of Atg13 (Budovskaya et al., 2004).
DISCUSSION
Autophagy is a degradative process induced primarily in response to nutrient starvation. There are several nutrient sensory kinases in yeast, including PKA, Sch9, and Tor. TORC1 plays a major role in regulation of autophagy, and our present data demonstrate that PKA and Sch9 signaling pathways cooperatively regulate induction of autophagy. Loss of all three TPK1, TPK2, and TPK3 genes causes lethality (Smith et al., 1998), and deletion of SCH9 is synthetically lethal with reduced activity of the RAS/PKA pathway (Lorenz et al., 2000). To avoid these issues, we used strains carrying a mutation in the ATP-binding pocket of three Tpk proteins and/or Sch9 and controlled the activity of these kinases with the ATP-competitive inhibitor 1NM-PP1. Inactivation of PKA and Sch9 shows phenomena observed upon induction of autophagy, such as induction of Atg8 synthesis and increase in Atg8–PE formation (Figure 1A). Accordingly, we examined whether the complete autophagic process is driven using three different sets of assays; processing of GFP-Atg8 (Figure 1, B and C), Pho8Δ60 activity (Figure 2A), and maturation of prApe1 in a vac8Δ background (Figure 2B). All three assays provide evidence for a significant level of autophagic activity when PKA and Sch9 are inactivated simultaneously.
The dominant-negative Ras2G22A mutant is not very effective for induction of autophagy; it takes more than 60 h after induction of the Ras2G22A mutant phenotype to reach autophagic activity comparable to that seen with wild-type cells under starvation conditions (Budovskaya et al., 2004). One explanation for this result can be seen with our observation that Sch9 remains active as a negative regulator. We also observed that treatment of pka vac8Δ cells with 1NM-PP1 showed a slight maturation of prApe1 compared to pka sch9 vac8Δ cells (Figure 2B). The results from this sensitive assay also raise the possibility that the contribution of PKA and Sch9 for regulation of autophagy is not equivalent. This is supported by our observation that activation of PKA by BCY1 deletion completely inhibits autophagy induced in rapamycin and starvation, whereas hyperactive Sch9 partially blocks rapamycin-induced autophagy but it has no effect on starvation-induced autophagy (Figure 6). Although PKA and Sch9 regulate autophagy redundantly, PKA might have a predominant effect.
To understand how PKA and Sch9 function for induction of autophagy, we checked the activity of TORC1 kinase by examining the phosphorylation state of two TORC1 substrate proteins, Gln3 and Atg13 (Beck and Hall, 1999; Kamada et al., 2000). Both proteins in pka sch9 cells with 1NM-PP1 treatment were as highly phosphorylated as those without treatment (Figure 4, A and B), indicating that TORC1 remains active during inactivation of PKA and Sch9. Therefore, we conclude that induction of autophagy can result from inactivation of PKA and Sch9 and that it does not require simultaneous inactivation of TORC1.
Activity of the Atg1 kinase complex is indispensable for starvation-induced autophagy (Cheong et al., 2005; Kabeya et al., 2005). We found that PKA-Sch9 inhibition-induced autophagy also depends on these components (Figure 3, A–C). A high level of Atg1 kinase activity requires efficient association of Atg1 with Atg17 and dephosphorylated Atg13. Although the role of Atg1 kinase activity is still inconclusive, it may be required for regulation of the magnitude of autophagy but not for its initiation (Nair and Klionsky, 2005). Consistent with this hypothesis, our results showed that the autophagy activity induced by inactivation of PKA and Sch9, which did not cause dephosphorylation of Atg13, was lower than that induced by inactivation of TORC1 with rapamycin.
We observed additive stimulation of autophagy by simultaneous inactivation of PKA-Sch9 and TORC1 (Figure 4, D and E). This result agrees with the idea that all three kinases regulate autophagy at least partly independently of each other. Furthermore, this observation fits with the view that PKA and Sch9 function redundantly and that PKA, Sch9, and TORC1 also function together in the same pathways (Fabrizio et al., 2001; Pedruzzi et al., 2003; Kaeberlein et al., 2005; Zurita-Martinez and Cardenas, 2005; Chen and Powers, 2006). In contrast, it has been shown recently that Sch9 acts as one of the direct targets of TORC1 to regulate TORC1-dependent cellular processes (Urban et al., 2007). It is also suggested that TORC1 signals through the PKA pathway to control its targets (Schmelzle et al., 2004). It is not known whether PKA acts as a direct effector and substrate of TORC1 similar to Sch9. Thus, the possibility remains that autophagy is regulated by PKA and Sch9 acting solely downstream of TORC1.
We found that Msn2/4 and Rim15 were required for induction of autophagy by inactivation of PKA and Sch9. In contrast, none of these factors is required for autophagy induced by inactivation of TORC1 (Figure 5A). These results suggest that inactivation of PKA and Sch9 specifically changes the expression pattern of genes involved in autophagy through Msn2/4 and Rim15. However, up-regulation of Atg8 was independent of Msn2/4 and Rim15 (Figure 5B). We further analyzed expression of genes in pka sch9 cells by RNA microarry (our unpublished data). We found that several ATG genes were up-regulated in the presence of 1NM-PP1. However, most of these genes, including ATG8, were up-regulated independent of Msn2/4 and Rim15. It is possible that deletion of MSN2/4 and RIM15 affects expression of genes yet unidentified that are required for autophagy. Further study will be needed to identify the factor(s) needed for induction of autophagy through Msn2/4 and Rim15.
In summary, we demonstrate that inactivation of PKA and Sch9 is sufficient to trigger autophagy, suggesting that these kinases are cooperatively involved in negative regulation of autophagy similar to TORC1. Our results also propose in part a parallel, functional connection of PKA, Sch9, and TORC1 to regulate autophagy. Further study to identify the downstream regulator(s) of PKA and Sch9, which involves Msn2/4 and Rim15, will provide greater insight into the mechanism used for the regulation of autophagy.
ACKNOWLEDGMENTS
This work is supported by National Institutes of Health grants GM-53396 (to D.J.K.) and GM-76562 (to J.R.B.).
Abbreviations used:
- 1NM-PP1
C3-1′-naphthyl-methyl PP1
- Atg
autophagy-related
- Atg8–PE
Atg8 conjugated to phosphatidylethanolamine
- PAS
phagophore assembly site
- PKA
protein kinase A
- prApe1
precursor aminopeptidase I
- TORC1
Tor complex 1.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-05-0485) on August 15, 2007.
REFERENCES
- Abeliovich H., Zhang C., Dunn W. A., Jr., Shokat K. M., Klionsky D. J. Chemical genetic analysis of Apg1 reveals a non-kinase role in the induction of autophagy. Mol. Biol. Cell. 2003;14:477–490. doi: 10.1091/mbc.E02-07-0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck T., Hall M. N. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 1999;402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- Budovskaya Y. V., Stephan J. S., Deminoff S. J., Herman P. K. An evolutionary proteomics approach identifies substrates of the cAMP-dependent protein kinase. Proc. Natl. Acad. Sci. USA. 2005;102:13933–13938. doi: 10.1073/pnas.0501046102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budovskaya Y. V., Stephan J. S., Reggiori F., Klionsky D. J., Herman P. K. The Ras/cAMP-dependent protein kinase signaling pathway regulates an early step of the autophagy process in Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:20663–20671. doi: 10.1074/jbc.M400272200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. C., Powers T. Coordinate regulation of multiple and distinct biosynthetic pathways by TOR and PKA kinases in S. cerevisiae. Curr. Genet. 2006;49:281–293. doi: 10.1007/s00294-005-0055-9. [DOI] [PubMed] [Google Scholar]
- Cheong H., Yorimitsu T., Reggiori F., Legakis J. E., Wang C.-W., Klionsky D. J. Atg17 regulates the magnitude of the autophagic response. Mol. Biol. Cell. 2005;16:3438–3453. doi: 10.1091/mbc.E04-10-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P., Pozza F., Pletcher S. D., Gendron C. M., Longo V. D. Regulation of longevity and stress resistance by Sch9 in yeast. Science. 2001;292:288–290. doi: 10.1126/science.1059497. [DOI] [PubMed] [Google Scholar]
- Furuta S., Hidaka E., Ogata A., Yokota S., Kamata T. Ras is involved in the negative control of autophagy through the class I PI3-kinase. Oncogene. 2004;23:3898–3904. doi: 10.1038/sj.onc.1207539. [DOI] [PubMed] [Google Scholar]
- He C., Song H., Yorimitsu T., Monastyrska I., Yen W.-L., Legakis J. E., Klionsky D. J. Recruitment of Atg9 to the preautophagosomal structure by Atg11 is essential for selective autophagy in budding yeast. J. Cell Biol. 2006;175:925–935. doi: 10.1083/jcb.200606084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W.-P., Scott S. V., Kim J., Klionsky D. J. The itinerary of a vesicle component, Aut7p/Cvt5p, terminates in the yeast vacuole via the autophagy/Cvt pathways. J. Biol. Chem. 2000;275:5845–5851. doi: 10.1074/jbc.275.8.5845. [DOI] [PubMed] [Google Scholar]
- Jorgensen P., Rupes I., Sharom J. R., Schneper L., Broach J. R., Tyers M. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 2004;18:2491–2505. doi: 10.1101/gad.1228804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y., Kamada Y., Baba M., Takikawa H., Sasaki M., Ohsumi Y. Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol. Biol. Cell. 2005;16:2544–2553. doi: 10.1091/mbc.E04-08-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M., et al. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- Kamada Y., Funakoshi T., Shintani T., Nagano K., Ohsumi M., Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Klionsky D. J. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- Loewith R., Jacinto E., Wullschleger S., Lorberg A., Crespo J. L., Bonenfant D., Oppliger W., Jenoe P., Hall M. N. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell. 2002;10:457–468. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- Lorenz M. C., Pan X., Harashima T., Cardenas M. E., Xue Y., Hirsch J. P., Heitman J. The G protein-coupled receptor Gpr1 is a nutrient sensor that regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Genetics. 2000;154:609–622. doi: 10.1093/genetics/154.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrakis M., Lippincott-Schwartz J., Stratakis C. A., Bossis I. Depletion of type IA regulatory subunit (RIα) of protein kinase A (PKA) in mammalian cells and tissues activates mTOR and causes autophagic deficiency. Hum. Mol. Genet. 2006;15:2962–2971. doi: 10.1093/hmg/ddl239. [DOI] [PubMed] [Google Scholar]
- Meijer A. J., Codogno P. Signalling and autophagy regulation in health, aging and disease. Mol. Aspects Med. 2006;27:411–425. doi: 10.1016/j.mam.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Klionsky D. J. Protein turnover via autophagy: implications for metabolism. Annu. Rev. Nutr. 2007;27:19–39. doi: 10.1146/annurev.nutr.27.061406.093749. [DOI] [PubMed] [Google Scholar]
- Nair U., Klionsky D. J. Molecular mechanisms and regulation of specific and nonspecific autophagy pathways in yeast. J. Biol. Chem. 2005;280:41785–41788. doi: 10.1074/jbc.R500016200. [DOI] [PubMed] [Google Scholar]
- Noda T., Matsuura A., Wada Y., Ohsumi Y. Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 1995;210:126–132. doi: 10.1006/bbrc.1995.1636. [DOI] [PubMed] [Google Scholar]
- Noda T., Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J. Biol. Chem. 1998;273:3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- Pedruzzi I., Dubouloz F., Cameroni E., Wanke V., Roosen J., Winderickx J., De Virgilio C. TOR and PKA signaling pathways converge on the protein kinase Rim15 to control entry into G0. Mol. Cell. 2003;12:1607–1613. doi: 10.1016/s1097-2765(03)00485-4. [DOI] [PubMed] [Google Scholar]
- Schmelzle T., Beck T., Martin D. E., Hall M. N. Activation of the RAS/cyclic AMP pathway suppresses a TOR deficiency in yeast. Mol. Cell. Biol. 2004;24:338–351. doi: 10.1128/MCB.24.1.338-351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T., Klionsky D. J. Autophagy in health and disease: a double-edged sword. Science. 2004a;306:990–995. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T., Klionsky D. J. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J. Biol. Chem. 2004b;279:29889–29894. doi: 10.1074/jbc.M404399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A., Ward M. P., Garrett S. Yeast PKA represses Msn2p/Msn4p-dependent gene expression to regulate growth, stress response and glycogen accumulation. EMBO J. 1998;17:3556–3564. doi: 10.1093/emboj/17.13.3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevelein J. M., de Winde J. H. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 1999;33:904–918. doi: 10.1046/j.1365-2958.1999.01538.x. [DOI] [PubMed] [Google Scholar]
- Thomas B. J., Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- Toda T., Cameron S., Sass P., Wigler M. SCH9, a gene of Saccharomyces cerevisiae that encodes a protein distinct from, but functionally and structurally related to, cAMP-dependent protein kinase catalytic subunits. Genes Dev. 1988;2:517–527. doi: 10.1101/gad.2.5.517. [DOI] [PubMed] [Google Scholar]
- Toda T., Cameron S., Sass P., Zoller M., Scott J. D., McMullen B., Hurwitz M., Krebs E. G., Wigler M. Cloning and characterization of BCY1, a locus encoding a regulatory subunit of the cyclic AMP-dependent protein kinase in Saccharomyces cerevisiae. Mol. Cell. Biol. 1987;7:1371–1377. doi: 10.1128/mcb.7.4.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban J., et al. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol. Cell. 2007;26:663–674. doi: 10.1016/j.molcel.2007.04.020. [DOI] [PubMed] [Google Scholar]
- Wang Y., Pierce M., Schneper L., Guldal C. G., Zhang X., Tavazoie S., Broach J. R. Ras and Gpa2 mediate one branch of a redundant glucose signaling pathway in yeast. PLoS Biol. 2004;2:E128. doi: 10.1371/journal.pbio.0020128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen W.-L., Legakis J. E., Nair U., Klionsky D. J. Atg27 is required for autophagy-dependent cycling of Atg9. Mol. Biol. Cell. 2007;18:581–593. doi: 10.1091/mbc.E06-07-0612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T., Klionsky D. J. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12:1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurita-Martinez S. A., Cardenas M. E. Tor and cyclic AMP-protein kinase A: two parallel pathways regulating expression of genes required for cell growth. Eukaryot. Cell. 2005;4:63–71. doi: 10.1128/EC.4.1.63-71.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]