Abstract
Utp8p is an essential nucleolar component of the nuclear tRNA export machinery in Saccharomyces cerevisiae. It is thought to act at a step between tRNA maturation/aminoacylation and translocation of the tRNA across the nuclear pore complex. To understand the function of Utp8p in nuclear tRNA export, a comprehensive affinity purification analysis was conducted to identify proteins that interact with Utp8p in vivo. In addition to finding proteins that have been shown previously to copurify with Utp8p, a number of new interactions were identified. These interactions include aminoacyl-tRNA synthetases, the RanGTPase Gsp1p, and nuclear tRNA export receptors such as Los1p and Msn5p. Characterization of the interaction of Utp8p with a subset of the newly identified proteins suggests that Utp8p most likely transfer tRNAs to the nuclear tRNA export receptors by using a channeling mechanism.
INTRODUCTION
Transcription of eukaryotic tRNA genes by RNA polymerase III is highly regulated (White, 2004a,b, 2005; Ernens et al., 2006), and it results in the production of pre-tRNAs. The pre-tRNAs are then converted into mature tRNAs by a multistep process. This process involves removal of the 5′ and 3′ extensions, base modification, addition of the nucleotides C, C, and A to their 3′ ends, and in a small percentage of the transcripts, removal of an intron (for review, see Hopper and Phizicky, 2003). For intronless pre-tRNAs, the maturation process occurs primarily in the nucleus. Maturation of intron-containing pre-tRNAs also occurs in the nucleus. However, the intron-containing tRNAs lacking the extensions are exported to the cytoplasm for removal of the intron, and then they are imported back to the nucleus for reasons that are not fully understood (Yoshihisa et al., 2003; Shaheen and Hopper, 2005; Takano et al., 2005). Mature tRNAs are subsequently subjected to a quality assurance step to ensure that they are functional before export to the cytoplasm.
Studies first conducted in Xenopus laevis (Lund and Dahlberg, 1998) and later in Saccharomyces cerevisiae (Sarkar et al., 1999; Grosshans et al., 2000a; Azad et al., 2001) led to the suggestion that aminoacylation of tRNAs in the nucleus is used to select fully matured and functional tRNAs for export to the cytoplasm. This proof-reading step seems to occur in the nucleolus (Steiner-Mosonyi and Mangroo, 2004). However, nuclear tRNA aminoacylation is not the only mechanism used to inspect the functionality of tRNA, because it is not absolutely required for tRNA export in both X.laevis and S.cerevisiae (Arts et al., 1998b; Azad et al., 2001). Consequently, nuclear tRNA export is thought to occur by two pathways referred to as aminoacylation dependent and aminoacylation independent. However, findings reported suggest that the nuclear tRNA aminoacylation-dependent pathway is primarily responsible for nuclear export of mature tRNAs in S.cerevisiae (Steiner-Mosonyi and Mangroo, 2004).
Translocation of the tRNAs across the nuclear pore complex (NPC) in S.cerevisiae requires Los1p and Msn5p (Hellmuth et al., 1998; Takano et al., 2005). This step in mammalian cells is facilitated by exportin-t and exportin-5 (Kutay et al., 1998; Arts et al., 1998a; Bohnsack et al., 2002; Calado et al., 2002), the orthologues of Los1p and Msn5p, respectively. These proteins are members of the β-karyopherin family of nucleocytoplasmic transport factors that bind the tRNA cargo directly in a RanGTP-dependent manner (Arts et al., 1998a; Hellmuth et al., 1998; Kutay et al., 1998; Bohnsack et al., 2002; Calado et al., 2002). Los1p, Msn5p, and exportin-5 are thought to be receptors of the nuclear aminoacylation-dependent export pathway (Bohnsack et al., 2002; Calado et al., 2002; Steiner-Mosonyi and Mangroo, 2004). However, Los1p may also facilitate export of nonaminoacylated tRNAs based on the finding that exportin-t is able to transport a nonaminoacylated tRNA to the cytoplasm (Arts et al., 1998b). The ATP (CTP):nucleotidyltransferase (Cca1p), an essential enzyme that prepares tRNAs for aminoacylation in the nucleus, cytoplasm, and mitochondrion by adding the nucleotides C, C, and A to the 3′ ends of tRNAs, is also involved in nuclear export of some tRNAs in S.cerevisiae (Feng and Hopper, 2002). Cca1p is thought to function as a tRNA export receptor or an adaptor in a nuclear aminoacylation-independent pathway that permits export of tRNAs obtained from intronless pre-tRNAs (Feng and Hopper, 2002). Furthermore, evidence reported suggests that an unidentified nuclear aminoacylation-independent pathway facilitates nuclear export of tRNAs derived from intron-containing pre-tRNAs (Steiner-Mosonyi et al., 2003).
Cex1p is a cytoplasmic component of the nuclear tRNA export machinery of S.cerevisiae (McGuire and Mangroo, 2007). Cex1p binds tRNA saturably, and it associates with the NPC by interacting with Nup116p. Cex1p was shown to copurify with Los1p and Msn5p, the eukaryotic elongation factor eEF-1A, which delivers aminoacylated tRNAs to the ribosome, and the RanGTPase Gsp1p, but not with Cca1p. Depletion of Cex1p and eEF-1A or Los1p significantly reduced the efficiency of nuclear tRNA export. Cex1p interacted with Los1p but not with eEF-1A in vitro. These findings led to the suggestion that Cex1p is a component of the nuclear aminoacylation-dependent tRNA export pathway, which is responsible for collecting aminoacyl-tRNAs from the nuclear export receptors at the cytoplasmic side of the NPC and transferring them to eEF-1A by using a channeling mechanism (McGuire and Mangroo, 2007).
Utp8p was identified previously using a yeast tRNA three-hybrid interaction method and an in vivo nuclear tRNA export assay to identify proteins that participate in nuclear tRNA export in S.cerevisiae (Steiner-Mosonyi et al., 2003). Utp8p is essential and loss of its function blocks nuclear export of tRNAs derived from intronless and intron-containing pre-tRNAs but not tRNA maturation or nuclear tRNA aminoacylation. Retention of the tRNAs in a Utp8p-depleted strain has been shown to occur in the nucleolus (Steiner-Mosonyi and Mangroo, 2004). Overexpression of Utp8p also increased the efficiency of nuclear export of nonaminoacylated tRNATyr (Steiner-Mosonyi et al., 2003). Utp8p is located in the nucleolus and binds tRNA directly. However, Utp8p does not function as a nuclear tRNA export receptor, because it does not shuttle between the nucleus and cytoplasm. These findings indicated that Utp8p is required for tRNA export by both the aminoacylation-dependent and -independent pathways (Steiner-Mosonyi et al., 2003). Although the function of Utp8p in nuclear tRNA export is not understood, it is thought to act at a step between tRNA maturation/aminoacylation and translocation of the tRNA across the NPC (Steiner-Mosonyi et al., 2003).
Protein binding studies established that Utp8p interacts directly with the tyrosyl-tRNA synthetase in the nucleolus, and with Los1p and Msn5p, suggesting that Utp8p may collect tRNAs from the aminoacyl-tRNA synthetases, and transfer them to the nuclear tRNA export receptors by using a channeling mechanism. The RanGTPase Gsp1p was found to interact directly with Utp8p. Furthermore, Utp8p containing tRNA, Los1p, and Gsp1p formed a complex. In contrast, Gsp1p was not detected in the complex consisting of Los1p and Utp8p lacking tRNA. A model consistent with these data and those reported previously (Hellmuth et al., 1998) suggest that in vivo Utp8p binds Los1p first, and then Gsp1p is recruited to Los1p; interaction of Gsp1p with Utp8p facilitates transfer of the tRNA from Utp8p to Los1p.
MATERIALS AND METHODS
Strains and Plasmids
The yeast strains used in this study are listed in Table 1. The pGEX-2T-TEV plasmid was obtained from Dr. D. Heinrichs (University of Western Ontario). pET19b, pET23d, and pET23a were purchased from Novagen (San Diego, CA). pRS416-GAL1 was described previously (Steiner-Mosonyi et al., 2003). pYX242-NOP1-dsRED and pRS-TYS1-nls1-myc were obtained from Dr. John Aitchison (Seattle Institute of Systems Biology) and Dr. Anita Hopper (The Pennsylvania State University), respectively. pGEX-4T-Gsp1p was obtained from Dr. U. Stochaj (McGill University). The construction of pET19b-UTP8 was described previously (Steiner-Mosonyi et al., 2003). pTU708 containing the C-terminal half of green fluorescent protein (GFP) and pTU707 with the N-terminal half of GFP was obtained from Dr. Martin Chalfie (Columbia University) (Zhang et al., 2004). The pFA6a-based plasmid pKT128, containing yEGFP and the SpHIS5 genes, was obtained from Euroscarf (Frankfurt, Germany). The pGEX-2T-TEV-UTP8 plasmid was generated by polymerase chain reaction (PCR) amplification of UTP8 from genomic DNA, and cloning into the BamHI and EcoRI sites in pGEX-2T-TEV. pGEX-2T-TEV-TYS1 was constructed by PCR amplification of the TYS1 open reading frame (ORF) from pET3d-TYS1 provided by Dr. U. RajBhandary (Massachusetts Institute of Technology), and cloning into the BamHI and SmaI sites in pGEX-2T-TEV. pGEX-2T-TEV-LOS1 was made by introducing the LOS1 ORF into the SmaI and BamHI sites in pGEX-2T-TEV. The pET19b-LOS1 plasmid was prepared by PCR amplification of the LOS1 ORF from genomic DNA, and cloning into the XhoI and BamHI sites in pET19b. pET23a-CCA1 was generated by PCR amplification of the CCA1 ORF from genomic DNA, and cloning into the NheI and NotI sites in pET23a. The pET23d-MSN5 plasmid was constructed by PCR amplification of the MSN5 ORF from genomic DNA, and cloning into the NcoI and NotI sites in pET23d. pET19b-GSP1 was constructed by inserting the GSP1 ORF into the NdeI and BamHI sites in pET19b; the GSP1 ORF was prepared by PCR amplification by using pGEX-4T-Gsp1p as the template. Rabbit anti-Los1p was obtained from Dr. E. Hurt (University of Heidelberg), rabbit anti-Gsp1p was obtained from Dr. J. D. Aitchison, Seattle Institute for Systems Biology, rabbit anti-human TyrRS was obtained from Dr. P. Schimmel (Scripps Institute), and mouse anti-GFP and mAB414 were obtained from Roche Applied Science (Indianapolis, IN) and BAbCo (Berkeley, CA), respectively. mAB414, raised against the vertebrate FG Nups, Nup358, Nup214, and Nup153, recognizes the S.cerevisiae Nup159p and Nup1p, the homologues of Nup214 and Nup153, respectively.
Table 1.
List of strains
| Strain | Genotype | Source |
|---|---|---|
| LOS1-TAP | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, LOS1-TAP::HIS3 | Open Biosystems (Huntsville, AL) |
| BY4741 | MATa, his3Δ, 1 leu2Δ0, met15Δ0, ura3Δ0 | Open Biosystems |
| XPO1-TAP | MATa, his3Δ, 1 leu2Δ0, met15Δ0, ura3Δ0, XPO1-TAP::HIS3 | Open Biosystems |
| UTP8-TAP | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, UTP8-TAP::HIS3 | Open Biosystems |
| NUP120 | MATα, ura3-52, his3-Δ200, trp1-1, leu2-3,112, lys2-80 | American Type Culture Collection (Manassas, VA) |
| nup120 | MATα, ura3-52, his3-Δ200, trp1-1, leu2-3,112, lys2-8, nup120::URA3 | Aitchison et al. (1995) |
| UTP13-TAP | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, UTP13-TAP::HIS3 | Open Biosystems |
| TYS1-TAP | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, TYS1-TAP::HIS3 | Open Biosystems |
| POM152-TAP | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, POM152-TAP::HIS3 | Open Biosystems |
| NUP170-TAP | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, NUP170-TAP::HIS3 | Open Biosystems |
| NUP116-TAP | MATa, NUP116::TAP-K.I. URA3, ade2, arg4, leu2-3,112, trp1-289, ura3-52 | Euroscarf |
| VAS1-TAP | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, VAS1-TAP::HIS3 | Open Biosystems |
| UTP18-TAP | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, UTP18-TAP::HIS3 | Open Biosystems |
| UTP21-TAP | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, UTP21-TAP::HIS3 | Open Biosystems |
| UTP22-TAP | MAT,a his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, UTP22-TAP::HIS3 | Open Biosystems |
| CCA1-TAP | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, CCA1-TAP::HIS3 | Open Biosystems |
| MSN5-TAP | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, MSN5-TAP::HIS3 | Open Biosystems |
| DF5 | MATa/MATα, ura3-52/ura3-52, his3-Δ200/his3Δ200, trp1-1/trp1-1, leu2-3,112/leu2-3,112, lys2-801/lys2-80 | American Type Culture Collection |
| utp8 | derivative of BY4743, utp8::KANR, pCEN-URA-GAL1-UTP8, MATa | Steiner-Mosonyi et al. (2003) |
| UTP8-GFP | derivative of DF5, MATa, UTP8::GFP::SpHIS5 | This study |
| nup120 UTP8-GFP | MATα, ura3-52, his3-Δ200, trp1-1, leu2-3,112, lys2-80, UTP8::GFP::SpHIS5, nup120::URA3 | This study |
| nup120 NUP2-GFP | MATα, ura3-52, his3-Δ200, trp1-1, leu2-3,112, lys2-80, NUP2::GFP::SpHIS5, nup120::URA3 | Rout et al. (2000) |
| NUP2-GFP | MATα, ura3-52, his3-Δ200, trp1-1, leu2-3,112, lys2-80,NUP2::GFP::SpHIS5 | Rout et al. (2000) |
| CSE1-TAP | MATa, CSE1::TAP-K.I. URA3, his3, leu2, met15, ura3 | Open Biosystems |
| UTP8-cgfp | derivative of DF5, UTP8::cgfp::SpHIS5 | This study |
| nup120 UTP8-cgfp | MATα, ura3-52, his3-Δ200, trp1-1, leu2-3,112, lys2-80, UTP8::cgfp::SpHIS5, nup120::URA3 | This study |
Overexpression and Purification of His-tagged Proteins
Escherichia coli BL21 (DE3) Codon Plus RIL (Novagen) with pET19b-LOS1, pET19b-UTP8, pET23a-CCA1, pET19b-GSP1, or pET23d-MSN5 was grown in 1 l of 2YT broth containing 100 μg/ml ampicillin and 34 μg/ml chloramphenicol at 37°C to an A600 of 0.4. The culture was transferred to 15°C, and expression of the His-tagged protein was induced with 200 μM isopropyl β-d-thiogalactoside (IPTG) for 16 h. The cells were harvested by centrifugation, and then they were resuspended in 30 ml of binding buffer (20 mM NaH2PO4, pH 7.5, containing 500 mM NaCl, 5 mM imidazole, and a mixture of protease inhibitors (Complete EDTA-free; Roche Applied Science). The cells were lysed by two passes through a French pressure cell at 10,000 psi. The clarified lysate was applied onto a 1-ml HisTrap HP column (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom). The column was washed with 20 ml of tRNA wash buffer (20 mM NaH2PO4, pH 7.5, containing 500 mM KCl, and 5 mM imidazole), and with 20 ml of 20 mM NaH2PO4, pH 7.5, buffer containing 500 mM NaCl and 50 mM imidazole. The proteins were eluted using a gradient of increasing imidazole concentration to 500 mM. Fractions containing the protein of interest were pooled and dialyzed against 20 mM Tris-HCl, pH 7.5, buffer containing 100 mM NaCl, and they were applied onto a pre-equilibrated HiTrap Q HP column (GE Healthcare) (for Cca1p, a HiTrap S HP column was used, and all steps were done at pH 7.0). The column was washed with 20 mM Tris-HCl, pH 7.5, buffer containing 100 mM NaCl, and proteins were eluted using a gradient of increasing NaCl concentration to 500 mM. Fractions containing purified protein were pooled and dialyzed against IPP150 buffer (25 mM Tris-HCl, pH 7.5, containing 150 mM NaCl, 20% glycerol [wt/vol], and 0.1% Nonidet P-40 [vol/vol]). The protein was concentrated and stored at −80°C.
Overexpression and Purification of Glutathione S-Transferase (GST)-tagged Proteins
E.coli BL21 (DE3) Codon Plus RIL containing pGEX-2T-TEV-TYS1 or pGEX-2T-TEV-UTP8 was grown at 37°C in 1 l of 2YT broth containing 100 μg/ml ampicillin and 34 μg/ml chloramphenicol to an A600 of 0.6. The cells were shifted to 15°C, and expression of the GST-tagged protein was induced with 1 mM IPTG for 4–16 h. The cells were harvested by centrifugation, and then they were resuspended in 30 ml of binding buffer (10 mM Na2HPO4 and 1.8 mM KH2PO4, pH 7.3, containing 140 mM NaCl, 2.7 mM KCl, 1 mM dithiothreitol [DTT], and a mixture of protease inhibitors [Complete EDTA-free, Roche Applied Science]). The cells were lysed by two passes through a French pressure cell at 10,000 psi, and the clarified lysates were applied onto a 1-ml GSTrap HP column (GE Healthcare). The column was washed with 15 ml of binding buffer, and the GST-fusion proteins were eluted with 10 ml of 50 mM Tris-HCl, pH 8.0, buffer containing 10 mM reduced glutathione and 1 mM DTT. The eluates were dialyzed against 20 mM Tris-HCl, pH 7.5, buffer containing 100 mM NaCl, and then they were applied onto a HiTrap Q HP column. The column was washed with 20 mM Tris-HCl, pH 7.5, buffer containing 100 mM NaCl, and proteins were eluted using a NaCl concentration gradient to 500 mM. Fractions containing purified GST-Tys1p or GST-Utp8p were pooled and dialyzed against IPP150 buffer (25 mM Tris-HCl, pH 7.5, containing 150 mM NaCl, 0.1% Nonidet P-40 [vol/vol], and 20% glycerol [wt/vol]). The proteins were concentrated and stored at −80°C. The A260/A280 ratio confirmed that no tRNA was bound to the proteins.
Tandem Affinity Purification (TAP) and Western Blot or Mass Spectrometric Analysis
The TAP-tagged strain was grown in 2 or 8 l of YPD medium to an A600 of 2.0 at 30°C. The cells were harvested by centrifugation, resuspended in 50 ml of Nonidet P-40 buffer (15 mM Na2HPO4 and 10 mM NaH2PO4, pH 7.2, containing 2% Nonidet P-40 (vol/vol), 150 mM NaCl, 2 mM EDTA, 50 mM NaF, 0.1 mM Na3VO4, and protease inhibitors), and lysed at 20,000 psi by using Emulsiflex-C3 high-pressure homogenizer (Avestin, Ottawa, ON, Canada). The lysate was clarified by ultracentrifugation at 142,000 × g for 1.25 h at 4°C, and then it was subjected to affinity purification using IgG-Sepharose (GE Healthcare), or tandem affinity purification by using IgG-Sepharose and calmodulin-Sepharose (Stratagene, La Jolla, CA) as described previously (Rigaut et al., 1999; Puig et al., 2001; Krogan et al., 2004). The proteins in the final eluate were precipitated using 25% trichloroacetic acid, and then they were washed with ice-cold acetone containing 0.05 N HCl, followed by pure acetone. The protein precipitate was dried at room temperature, and it was solubilized in LDS sample buffer (Invitrogen, Carlsbad, CA). The copurifying proteins were separated on 4–12% Novex Bis-Tris gels (Invitrogen) and subjected to liquid chromatography quadrupole time-of-flight tandem mass spectrometry (LCQToF MS-MS) analyses (McGill University Proteomics Facility, Montreal, QC, Canada), or Western blot analyses using rabbit anti-Utp8p, rabbit anti-Los1p, rabbit anti-human TyrRS and rabbit anti-Gsp1p, and the enhanced chemiluminescence detection system (GE Healthcare).
In Vitro Protein Binding
GST-Tys1p, GST-Utp8p, or GST alone in IPP150 buffer containing protease inhibitor and DTT was bound to glutathione (GT)-Sepharose for 2 h in the presence or absence of a large excess of tRNA (20 μM for TyrRS, 6 μM for Utp8p, and 6 μM for Los1p) at 4°C. The resin was washed with IPP150 buffer, and then it was incubated with an equivalent or a twofold molar excess of the interacting proteins (Utp8p, Cca1p, Msn5p, Los1p, or Gsp1p-GTP) for 2 h at 4°C. The resins were washed with IPP150 buffer containing protease inhibitors and DTT, and then they were incubated with the TEV protease (Invitrogen) at 4°C to release the proteins from bound GST. The eluates were subjected to electrophoresis using a 4–12% Novex Bis-Tris gel, and Western blot analysis was performed to detect the proteins. Cca1p and Msn5p were detected using an antibody that recognizes the His-tag. Gsp1p was loaded with GTP in the presence of MgCl2 as described previously (Lee and Aitchison, 1999).
Detection of the Interaction between Utp8p and Tys1p In Vivo by Using Split Green Fluorescent Protein
The plasmid for C-terminal tagging of proteins with the C-terminal half of GFP (cgfp) was constructed by PCR amplification of the cgfp gene from the pTU708 plasmid by using the primers ACGGAGCGAGAGGGTTAATTAA GGGTGGAAGCGGTAAGAAT and ACAGAAGGCGCGCCTCAGTTGTAC AGTTCATCCATGC. This sequence was cloned into the PacI and AscI sites in pKT128, replacing the yEGFP sequence. The cgfp sequence was immediately downstream and in frame with the linker sequence encoding GDGAG, which is necessary to improve the mobility between the cgfp tag and the tagged protein. Tagging of chromosomal Utp8p gene with the cgfp sequence is accomplished by PCR amplification of the cassette by using the primer F5 (GGTGACGGTGCTGGTTTA, preceded by a UTP8-specific 40-mer sequence immediately upstream of the stop codon) and R3 (TCGATGAATTCGA GCTCG, preceded by a UTP8-specific 40-mer sequence complementary to the sequence immediately downstream of the stop codon). The PCR product, 1900 base pairs, was used to transform diploid DF5. Integration of the PCR product into the chromosome was detected by growth of the strain on complete synthetic medium containing dextrose and lacking His (CSD-His). Haploids were isolated by sporulation and tetrad dissection. nup120 UTP8-cgfp was prepared by mating nup120 and UTP8-cgfp followed by sporulation and tetrad dissection.
The plasmid for expression of the N-terminal half of GFP (ngfp) fused to the N-terminal end of a protein was constructed by PCR amplification of the ngfp gene from the pTU707 plasmid by using the primers AGCACGG AGACGGAGTCTAGACCATGGCTAGCAAAGGAGAAGAACTC and AC AGAAGGATCCAGCACCGTCACCGCCAGAGCCAGAGCCACC, and the product was cloned into the BamHI and XbaI sites of pRS416-GAL1, producing pRS416-GAL1-ngfp. The downstream primer introduces a linker sequence identical to that used in the cgfp vector to improve mobility of the tag. pRS416-GAL1-ngfp-TYS1 and pRS416-GAL1-ngfp-TYS1-nls1 was made by cloning the TYS1 and TYS1-nls1 ORF into the BamHI and XhoI sites in pRS416-GAL1-ngfp; the TYS1 and TYS1-nls1 ORFs were amplified from S.cerevisiae genomic DNA and pRS-TYS1-nls1-myc, respectively, by using the primers TCTAATTGACGGATC CATGTCCTCTGCTGCCACG and ACGGATTAAGCTCGAGTTACAATTTGG TTTCCTCTAGTTTCG. These constructs were used to transform the UTP8-cgfp strain. Expression of the N-terminal GFP-tagged protein was induced by growth of the transformants in CS medium containing 2% galactose and 2% raffinose and lacking His and Ura.
RESULTS
Affinity Purification Identified New Utp8p-Interactors
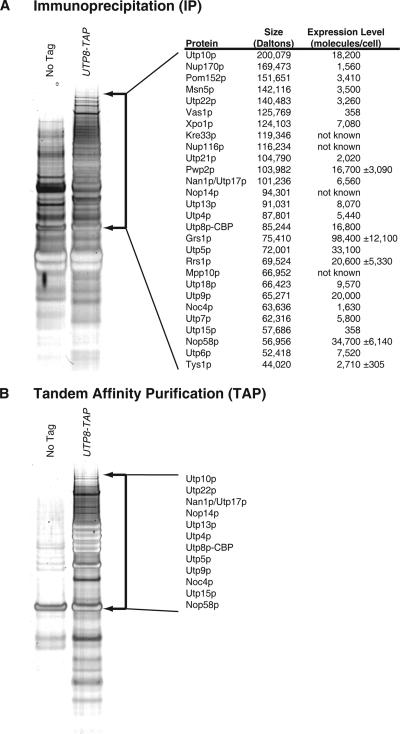
To understand the role of Utp8p in nuclear tRNA export in S.cerevisiae, affinity purification using total cell extract prepared from a strain with the chromosomal UTP8 gene tagged at the 3′ end with a fusion gene encoding a protein A (ProtA)-tobacco etch virus (TEV)-calmodulin binding peptide (CBP) (TAP) tag was used to identify proteins that interact with Utp8p in vivo. To enrich for proteins that may interact weakly with Utp8p and/or of low abundance, total cell extract was subjected to a single purification step by using IgG-Sepharose (immunoprecipitation [IP]) followed by release of Utp8-CBP from ProtA bound to the column by using the TEV protease. To identify proteins that purify nonspecifically, total cell extract from a non-TAP–tagged isogenic strain (BY4741) was also subjected to IP. In addition, extracts prepared from the non-TAP–tagged and UTP8-TAP strains were subjected to TAP by using IgG-Sepharose and calmodulin-Sepharose. The eluate after IP (Figure 1A) or TAP (TAP eluate) (Figure 1B) was subjected to polyacrylamide gel electrophoresis (PAGE) on a 4–12% polyacrylamide gel, and the proteins were detected by silver staining. The gel of each lane was sliced into 1.5-mm sections, and the gel slices were analyzed by LCQToF MS-MS. Proteins below 25 kDa were not subjected to mass spectrometry. The analysis was performed on two independent extracts from each strain. Proteins present in the IP or TAP eluate from the non-TAP–tagged and UTP8-TAP strains were removed from the protein data set obtained from the Utp8-TAP sample.
Figure 1.
Identification of proteins that copurify with Utp8p by LCQToF MS-MS analysis. Total cell lysate prepared from a UTP8-TAP or non-TAP–tagged strain was subjected to affinity chromatography by using IgG-Sepharose (IP; A) or IgG-Sepharose and calmodulin-Sepharose (TAP; B). The proteins that copurified with Utp8-TAP were separated by SDS-PAGE and detected by silver staining of the gels. The gel in each lane was cut in 1.5-mm sections in the region indicated by arrows, and each gel slice was subjected to LCQTof MS-MS analysis. Proteins present in the IP or TAP eluate from the untagged and UTP8-TAP strains were removed from the protein data set obtained from the Utp8-TAP sample. The most significant hits are listed along with the protein size and expression level in A.
The proteins found to copurify with Utp8p by TAP were Nan1p/Utp17p, Utp10p, Utp15p, Utp4p, Utp5p, Utp22p, Utp9p, Nop14p, Noc4p, Utp13p, and Nop58p, which are essential proteins involved in 18S rRNA biogenesis and maturation, and nuclear export of the pre-40s ribosomal subunit (Figure 1B). All these proteins except Utp13p have been shown previously to copurify with Utp8p by TAP (Dragon et al., 2002; Grandi et al., 2002; Krogan et al., 2004, 2006; Gavin et al., 2006). The proteins identified by TAP also copurify with Utp8p by IP (Figure 1A). In addition, several new interactors of Utp8p, including Utp13p, were identified by the IP strategy (Figure 1A). These interactors include Utp18p, Utp21p, Xpo1p, Nup170p, Pom152p, Nup116p, and Msn5p. Furthermore, several aminoacyl-tRNA synthetases, tyrosyl-tRNA synthetase (Tys1p), valyl-tRNA synthetase (Vas1p), glycyl-tRNA synthetase (Grs1p), and arginyl-tRNA synthetase (Rrs1p) were found to copurify with Utp8p.
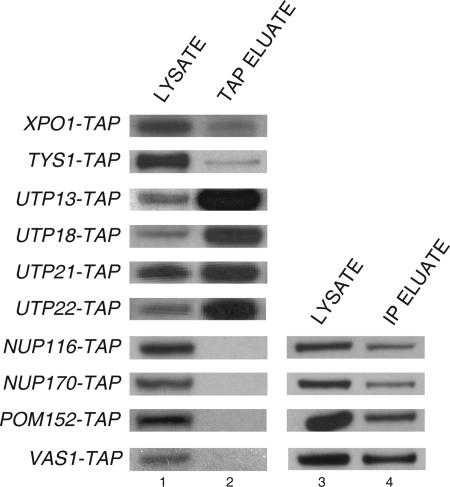
To test the authenticity of the newly identified interactions, TAP was performed with extract prepared from various TAP-tagged strains (Figure 2). Western blot analysis was performed to detect Utp8p in the total cell lysate (lane 1) and TAP eluate (lane 2). The analyses showed that Utp8p copurified with Xpo1p, Tys1p, Utp18p, Utp13p, Utp21p, and Msn5p (see Figure 5 for Msn5p data). Utp8p also copurified with Utp22-TAP, which we and others have found to copurify with Utp8p by TAP (Figures 1 and 2). This finding suggests that these proteins are authentically forming a complex with Utp8p in vivo. Utp8p was not detected in the TAP eluate for Nup116-TAP, Nup170-TAP, Pom152-TAP, and Vas1-TAP, but it was detected in the IP eluate (lane 4). The failure to detect the nuclear pore proteins by TAP is most likely unrelated to their expression level, because Nup170p and Pom152p are relatively abundant proteins (Ghaemmaghami et al., 2003) (Figure 1A). This is consistent with the finding that copurification of Utp8p with Utp15p, which is an extremely low copy number protein (Figure 1A) (Ghaemmaghami et al., 2003), could be easily detected after TAP (Figure 1B). The most likely explanation is that these proteins are interacting weakly with Utp8p or that they associate nonspecifically with Utp8p during chromatography. In contrast to Nup170p and Pom152p, Vas1p is expressed at a low level, and a very small percentage (∼1%) is expected to be in the nucleus based on the amount reported for Tys1p (Azad et al., 2001) (Figure 1A). Thus, this may explain why Vas1p was not found to copurify with Utp8p by TAP.
Figure 2.
Utp8p copurifies with some of the newly identified proteins by TAP and others by IP only. TAP or IP was performed with total cell lysate prepared from the strains indicated. Utp8p in the cell lysate (lanes 1 and 3) and TAP eluate (lane 2) or IP eluate (lane 4) was detected by Western blot analysis using anti-Utp8p.
Figure 5.
Utp8p copurifies with Los1p, Cca1p, and Msn5p, but not with Cse1p, and Los1p only copurifies with Utp8p. Total cell lysate was prepared from LOS1-TAP, MSN5-TAP, CCA1-TAP, CSE1-TAP, UTP8-TAP, UTP21-TAP, UTP4-TAP, or UTP9-TAP, and it was subjected to TAP. Utp8p or Los1p in the cell lysate (lane 1) and TAP eluate (lane 2) was detected by Western blot analysis.
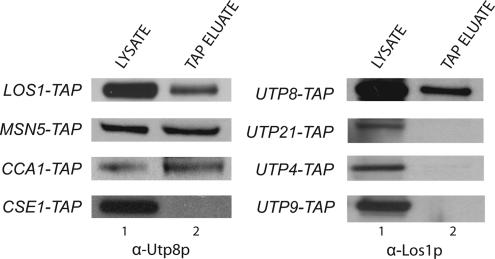
Utp8p Interacts with the Tyrosyl-tRNA Synthetase in the Nucleolus
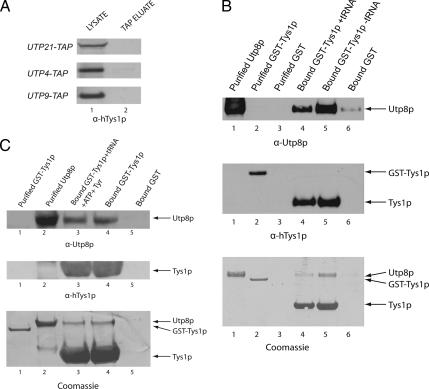
Utp8p was found to copurify with several aminoacyl-tRNA synthetases by IP (Figures 1 and 2). This association between Utp8p and the aminoacyl-tRNA synthetases in vivo seems to be specific, because Tys1p did not copurify with Utp21p, Utp4p, or Utp9p, which are proteins that copurified with Utp8p and that are involved in rRNA biogenesis (Figure 3A). These data suggest that the interaction between Utp8p and the aminoacyl-tRNA synthetases may be related to the function of Utp8p in nuclear tRNA export. We therefore studied Tys1p to gain some insight into how the association of Utp8p with the aminoacyl-tRNA synthetases contributes to its function. In vitro binding was conducted to ascertain whether Utp8p interacts directly with Tys1p or indirectly by binding the tRNA (Figure 3B). GST-Tys1p (410 μg of protein) (lane 2, middle and bottom) was bound to GT-Sepharose in the presence (lane 4) or absence (lane 5) of 333 μM total yeast tRNA (contains 20 μM tRNATyr). Substrate-induced intrinsic fluorescence quenching of tryptophan residues indicated that the apparent affinity of the GST–Tys1 fusion protein for tRNATyr under the binding conditions used is 1 μM (data not shown). Kinetic studies indicated that the Km of Tys1p for tRNATyr is 2 μM (Fechter et al., 2000). The resins were rinsed to remove unbound protein and tRNA, and then incubated with a twofold molar excess of Utp8p (960 μg of protein) (lanes 1, 4, and 5, top and bottom). The same amount of Utp8p was also incubated with GST bound to GT-Sepharose (lane 6, top and bottom). After washing the resins, the TEV protease was used to release Tys1p from bound GST, and Western blot analysis or Coomassie Blue staining was used to detect Utp8p and Tys1p in the eluate. Western blot analysis using anti-Utp8p (top) and Coomassie Blue staining (bottom) show that Utp8p interacts with Tys1p lacking (lane 5) or containing tRNA (lane 4). The amount of Utp8p eluted from the GST-bound resin was extremely low (lane 6). The amount of Utp8p bound to Tys1p complexed to tRNA (lane 4) was lower than that bound to Tys1p free of tRNA (lane 5). Western blot analysis using anti-human TyrRS (lane 4, middle) and Coomassie Blue staining (lane 4, bottom) indicate that this is partly due to a lower amount of GST-Tys1p bound to the GT-resin in the presence of tRNA (lane 4) than in the absence of tRNA (lane 5). The stained gel also shows that Utp8p does not interact stoichiometrically with Tys1p, indicating that the two proteins are interacting weakly.
Figure 3.
Utp8p interacts specifically with Tys1p. (A) Tys1p did not copurify with other Utp proteins. Total cell lysate was prepared from UTP21-TAP, UTP4-TAP, or UTP9-TAP and subjected to TAP. Tys1p in the cell lysate (lane 1) and TAP eluate (lane 2) was detected by Western blot analysis. (B) Utp8p interacts with Tys1p in a tRNA-independent manner in vitro. GST-Tys1p (6 nmol; 410 μg of protein) was incubated with GT-Sepharose in the presence or absence of 20 μM tRNATyr. The resin was washed and incubated with 12 nmol (960 μg of protein) of Utp8p. Utp8p (12 nmol) was also incubated with bound GST (6 nmol). The resin was washed, and Tys1p was released using the TEV protease. Western blot analysis was used to detect Utp8p (top) and Tys1p (middle). The proteins were detected directly by Coomassie Blue staining of an SDS-PAG (bottom). (C) The presence of ATP and tyrosine did not change the tRNA independence of Utp8p binding to Tys1p. The experiment was carried out as described in B using 2 mM ATP and 20 μM Tyr. Western blot analysis was used to detect Utp8p (top) and Tys1p (middle). The proteins were detected directly by Coomassie Blue staining of an SDS-PAG (bottom). Extraction of the tRNA bound to Tys1p released from bound GST in independent experiments, and quantification by measurement of the absorbance at 260 nm indicates the presence of ∼40% of the expected amount if all the GST-Tys1p is bound. Typically, ∼40% of the GST-Tys1p binds to the GT-Sepharose resin.
The interaction between Utp8p and Tys1p was also investigated in the presence of tRNATyr, ATP, and tyrosine (Figure 3C). GST-Tys1p (410 μg of protein) (lane 1) was bound to GT-Sepharose in the presence (lane 3) or absence (lane 4) of 20 μM tRNATyr, 2 mM ATP, and 20 μM Tyr. Utp8p (960 μg of protein) (lane 2) was added to bound Tys1p (lanes 3 and 4) or GST (lane 5). Coomassie Blue staining (bottom) and Western blot analysis were used to detect bound Utp8p (top) and Tys1p (middle). The data show that Utp8p interacts with Tys1p in a tRNA-independent manner. The results also indicate that the presence of ATP and tyrosine did not change the tRNA independence of Utp8p binding to Tys1p (compare lanes 3 and 4). Together, the results suggest that in vivo Utp8p associates directly with Tys1p. The data also suggest that copurification of Utp8p with Vas1p, Grs1p, and Rrs1p during IP (Figure 1A) is physiologically relevant.
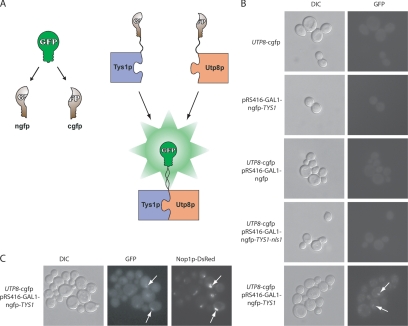
Tys1p has been shown to contain a nuclear localization signal and to be imported into the nucleus (Azad et al., 2001). Therefore, a split GFP system was also used to verify that Utp8p interacts with Tys1p in vivo and to test whether this interaction occurs in the nucleolus (Figure 4A). The split GFP system that we established for S.cerevisiae was adopted from the system developed for use in mammalian cells (Zhang et al., 2004). For this investigation, a haploid strain with the chromosomal UTP8 gene tagged at the 3′ end with the gene encoding the cgfp was isolated by tetrad dissection of a heterozygous diploid strain. The UTP8-cgfp strain was transformed with pRS416-GAL1 without or with the gene for the ngfp fused to Tys1p, a Tys1p mutant defective in nuclear import (TYS1-nls1), or ngfp alone. An isogenic wild-type haploid strain also was transformed with pRS416-GAL1-ngfp-TYS1. Reconstitution of split GFP was detected by direct fluorescence microscopy (Figure 4B). Fluorescence emission was not detected in cells expressing Utp8-cgfp alone (top panel), ngfp-Tys1p alone (second panel), Utp8-cgfp and ngfp (third panel), or in cells expressing Utp8-cgfp and ngfp-Tys1p-nls1 (fourth panel). However, fluorescence emission was observed in cells expressing Utp8-cgfp and ngfp-Tys1p (bottom panel), indicating that Utp8p is interacting with Tys1p; the GFP signal does not seem to be uniformly distributed throughout the cells. Instead, the signal is confined to a specific location, suggesting that this interaction could be occurring in the nucleolus where Utp8p is found at steady state.
Figure 4.
Utp8p associates with Tys1p in the nucleolus. (A) Detection of protein–protein interaction in vivo by reconstitution of split GFP. In the reconstituted split GFP system, a functional GFP is divided into two nonfunctional halves. Refolding of these two halves is accomplished if they are expressed as fusion with proteins that interact in vivo, thus bringing the two halves together. (B) Detection of Utp8p and Tys1p interaction in vivo by using the reconstituted split GFP system. DF5 UTP8-cgfp was grown in CS medium containing 2% raffinose and 2% galactose, and DF5 pRS416-GAL1-ngfp-TYS1, DF5 UTP8-cgfp with pRS416-GAL1-ngfp, DF5 UTP8-cgfp with pRS416-GAL1-ngfp-TYS1-nls1, and DF5 UTP8-cgfp with pRS416-GAL1-ngfp-TYS1 were grown in CS medium containing 2% raffinose and 2% galactose and lacking Ura at 30°C for 3 h. Reconstitution of GFP was detected by direct fluorescence microscopy. (C) Utp8p and Tys1p interaction colocalizes with Nop1p. The UTP8-cgfp strain harboring pRS-URA-GAL1-ngfp-TYS1 and pYX242-LEU-NOP1-dsRED was grown and analyzed as described above. Colocalization of the proteins was determined by overlay analysis of the images.
To ascertain whether Utp8p is interacting with Tys1p in the nucleolus, the nucleolar marker Nop1-dsRED was coexpressed with ngfp-Tys1p in the UTP8-cgfp strain (Figure 4C). Overlay analysis of the reconstituted GFP signal with that of Nop1-dsRED indicates that Utp8p associates with Tys1p in the nucleolus. These data, together with the finding that Utp8p is a tRNA-binding protein, imply that Utp8p interacts directly with Tys1p in the nucleolus to collect the aminoacyl-tRNAs. These results also suggest that Utp8p may be collecting tRNAs from Vas1p, Grs1p, Rrs1p, and the other aminoacyl-tRNA synthetases by interacting with them in the nucleolus. This is consistent with the finding that the function of Utp8p is essential (Giaever et al., 2002; Steiner-Mosonyi et al., 2003). However, why Utp8p copurified with only four aminoacyl-tRNA synthetases is not known. A likely possibility is that the interaction with the other aminoacyl-tRNA synthetases may be too weak to be detected by the affinity purification strategies used.
Utp8p Interacts with Nuclear tRNA Export Receptors
Mass spectrometric and Western blot analyses show that Utp8p associates with Msn5p in vivo (Figures 1A and 5, left). To test whether other proteins that play a role in translocation of tRNAs across the NPC copurify with Utp8p, TAP was performed with extract prepared from LOS1-TAP and CCA1-TAP followed by Western blot analysis to detect Utp8p (Figure 5). Similarly, TAP was conducted with UTP8-TAP followed by Western blot analysis to detect Los1p. Utp8p was detected in total cell lysate (lane 1) prepared from cells expressing Msn5-TAP, Los1-TAP, Cca1-TAP, or Cse1-TAP, which is a protein involved in nuclear export of importin-α (Kunzler and Hurt, 1998; Solsbacher et al., 1998). Utp8p was detected in the TAP eluate (lane 2) obtained from Msn5-TAP, Los1-TAP, and Cca1-TAP. Los1p was also detected in both the total cell lysate from UTP8-TAP (lane 1) and the TAP eluate (lane 2). However, Utp8p was not found in the TAP eluate from Cse1-TAP. This demonstrates that Utp8p did not copurify with Los1p, Msn5p, and Cca1p because of nonspecific interactions with proteins involved in translocating macromolecules across the NPC. Furthermore, TAP analyses indicate that Los1p did not copurify with Utp21p, Utp4p, or Utp9p, suggesting that Utp8p copurified specifically with the nuclear tRNA export proteins (Figure 5).
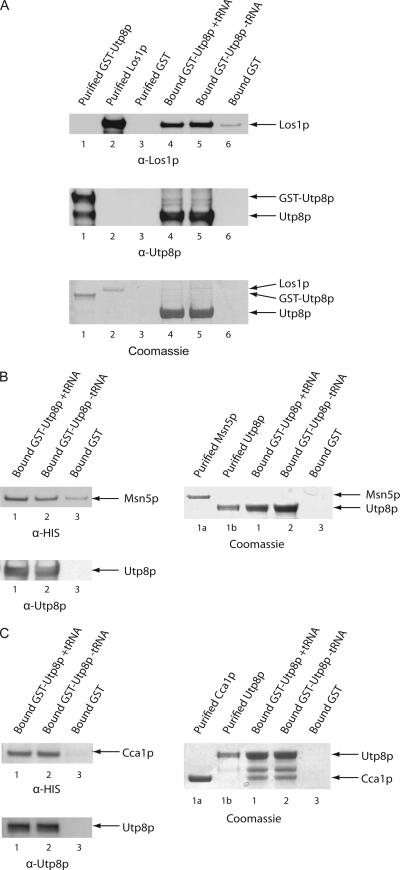
In vitro binding was used to ascertain whether Utp8p interacts directly with Los1p, Msn5p, and Cca1p (Figure 6). GST-Utp8p (480 μg of protein) was bound to GT-Sepharose with or without tRNA (6 μM). Substrate-induced intrinsic fluorescence quenching of tryptophan residues indicated that the affinity of Utp8p for tRNA is 600 nM (data not shown). The resin was washed and incubated with a twofold molar excess of Los1p (A), Msn5p (B), or Cca1p (C). As a control Los1p, Msn5p or Cca1p was incubated with bound GST. Utp8p was released from bound GST by cleavage with TEV, and Western blot analysis was used to detect Los1p (A, top row), Msn5p (B, left, top row), Cca1p (C, left, top row), and Utp8p (A, middle row and B and C, bottom row) in the eluate. Los1p (A, top row) interacts with Utp8p (A, middle row) containing (A, lane 4) or lacking (A, lane 5) tRNA. Furthermore, very little Los1p could be detected in the eluate obtained from the sample containing bound GST (A, lane 6, top row). Coomassie Blue staining of an SDS-PAG shows that a very small amount of Los1p bound to Utp8p, despite using 480 μg (4 nmol) of GST-Utp8p and 8 nmol of Los1p (A, bottom row). Like Los1p, binding of Msn5p (B, left, top row) and Cca1p (C, left, top row) to Utp8p (left, bottom rows) was not dependent on the presence of tRNA (compare lanes 1 and 2). A relatively small amount of Msn5p was in the eluate from the bound GST control (B, left, lane 3, top row), whereas no Cca1p was found (C, left, lane 3, top row). Msn5p could not be detected by Coomassie Blue staining of the SDS-PAG, even though 8 nmol of Msn5p was used (B, right, compare lanes 1a, 1, and 2). In contrast, Cca1p could be detected by Coomassie Blue staining (C, left, compare lanes 1a, 1, and 2). This shows that the Cca1p and Utp8p are not interacting stoichiometrically. These findings indicate that Los1p, Msn5p, and Cca1p interact weakly with Utp8p. The interaction between Cca1p and Utp8p was much stronger than that of Utp8p with Los1p or Msn5p. The data also indicate that the interaction of Utp8p with Cca1p and the nuclear tRNA export receptors is not dependent on tRNA. Collectively, these results suggest that Utp8p may transfer tRNAs to the nuclear tRNA export receptors and Cca1p, which plays a role in translocation of nonaminoacylated tRNAs across the NPC (Feng and Hopper, 2002).
Figure 6.
Utp8p interacts with Los1p, Msn5p, and Cca1p in a tRNA-independent manner in vitro. GST-Utp8p (6 nmol; 480 μg of protein) was incubated with GT-Sepharose with or without 6 μM tRNA. The resin was washed and incubated with a twofold molar excess of Los1p (A), Msn5p (B), or Cca1p (C). Each protein (12 nmol) was also incubated with bound GST (6 nmol). Utp8p was released from bound GST by using the TEV protease. The eluates were subjected to SDS-PAGE, and the proteins were detected by Western blot analysis. Protein binding was also monitored by Coomassie Blue staining of SDS-PAGs. Extraction of the tRNA bound to Utp8p released from bound GST in independent experiments, and quantification by measurement of the absorbance at 260 nm indicates the presence of ∼50% of the expected amount if all the GST-Utp8p is bound. Typically, ∼40–50% of the GST–Utp8p fusion protein binds to the GT-Sepharose resin.
Utp8p, Gsp1p, and Los1p Form a Complex That Is Dependent on tRNA
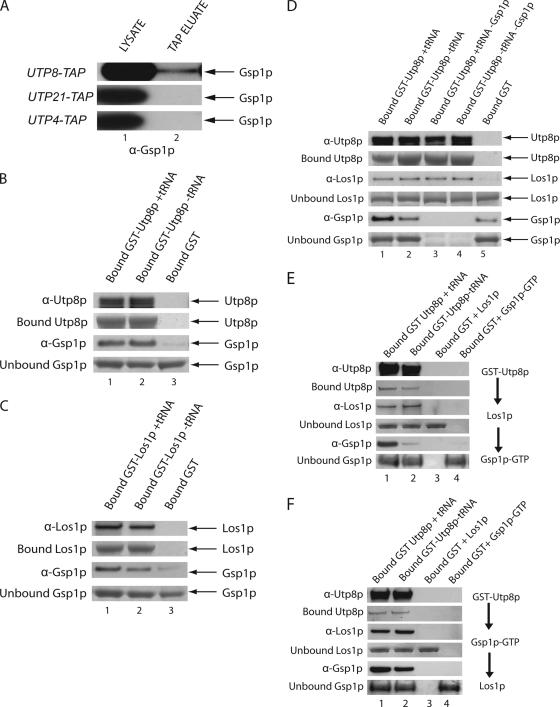
Loading of the tRNA cargo to Los1p has been shown to be dependent on the GTP bound form of Gsp1p, the S.cerevisiae homologue of the mammalian RanGTPase (Hellmuth et al., 1998). Thus, TAP was used to test whether Gsp1p is part of the Utp8p complex in vivo (Figure 7A). Total cell extract was prepared from UTP8-TAP and subjected to TAP, and copurification of Gsp1p was monitored by Western blot analysis. Gsp1p was detected in both the total cell lysate (lane 1) and the TAP eluate (lane 2). TAP analysis also showed that Gsp1p did not copurify with Utp4p or Utp21p (Figure 7A). These findings suggest that Gsp1p copurified specifically with Utp8p and that they are in the same complex in vivo.
Figure 7.
Utp8p interacts with both Gsp1p and Los1p in vitro. (A) Gsp1p copurifies with Utp8p, but not with Utp21p or Utp4p by TAP. Cell lysate from UTP8-TAP, UTP21-TAP, or UTP4-TAP was subjected to TAP, and Gsp1p in the cell lysate (lane 1) and TAP eluate (lane 2) was detected by Western blot analysis. (B) Gsp1p interacts with Utp8p in a tRNA-independent manner. GST-Utp8p (3 nmol; 240 μg of protein) bound to GT-Sepharose in the presence or absence of 6 μM tRNA was incubated with a twofold molar excess of Gsp1p-GTP. The same amount of Gsp1p-GTP was also incubated with bound GST (3 nmol). Utp8p was released from bound GST by cleavage with the TEV protease, and the eluates were subjected to SDS-PAGE. Utp8p (top row) and Gsp1p (third row) were detected by Western blot analysis. The extent of GST-Utp8p binding to the GT-resin was ascertained by Coomassie Blue staining of a SDS-PAGE (second row). The amount of Gsp1p in the wash eluate was determined by Coomassie Blue staining of an SDS-PAG (fourth row). (C) Gsp1p interacts with Los1p in a tRNA-dependent manner. GST-Los1p (3 nmol; 360 μg of protein) was incubated with GT-Sepharose in the presence or absence of 6 μM tRNA and with a twofold molar excess of Gsp1p-GTP. The same amount of Gsp1p-GTP was also incubated with bound GST (3 nmol). Los1p was released from bound GST by using the TEV protease. Los1p (top row) and Gsp1p (third row) were detected by Western blot analysis. The amount of bound GST-Los1p (second row) was detected by Coomassie Blue staining of an SDS-PAG. Unbound Gsp1p in the wash eluate (fourth row) was detected by Coomassie Blue staining of an SDS-PAG. (D) Gsp1p forms a complex with Los1p and Utp8p in a tRNA-dependent manner. GST-Utp8p (3 nmol; 240 μg of protein) was incubated with GT-Sepharose in the presence or absence of 6 μM tRNA. The resin was washed and incubated with an equimolar amount of Los1p and Gsp1p-GTP. Bound GST (3 nmol) was incubated with the same amount of Gsp1p-GTP and Los1p. Utp8p was released from bound GST by using the TEV protease, and Western blot analysis was used to detect Utp8p (top row), Los1p (third row), and Gsp1p (fifth row). The amount of bound GST-Utp8p (second row), and unbound Los1p (fourth row) and Gsp1p (sixth row), was ascertained by Coomassie Blue staining of SDS-PAGs. (E) Gsp1p forms a complex with Los1p and Utp8p in a tRNA-dependent manner when Los1p was incubated with the Utp8p—tRNA complex first. GST-Utp8p (3 nmol; 240 μg of protein) was incubated with GT-Sepharose in the presence or absence of 6 μM tRNA. The resin was washed and incubated with an equimolar amount of Los1p. The resin was washed and incubated with 3 nmol of Gsp1p-GTP. Bound GST (3 nmol) was incubated with the same amount of Gsp1p-GTP or Los1p. Utp8p was released from bound GST by using the TEV protease, and Western blot analysis was used to detect Utp8p (top row), Los1p (third row), and Gsp1p (fifth row). The amount of bound GST-Utp8p (second row), and unbound Los1p (fourth row), and Gsp1p (sixth row) was ascertained by Coomassie Blue staining of SDS-PAGs. (F) Gsp1p forms a complex with Los1p and Utp8p in a tRNA-independent manner when it is incubated with the Utp8p-tRNA complex first. The analysis was conducted as described in E, except Gsp1p was added to Utp8p before the addition of Los1p. Extraction of the tRNA bound to Utp8p released from bound GST in independent experiments, and quantification by measurement of the absorbance at 260 nm indicates the presence of ∼50% of the expected amount if all the GST-Utp8p is bound. Typically, ∼40–50% of the GST—Utp8p fusion protein binds to the GT-Sepharose resin.
In vitro binding was used to determine whether Gsp1p interacts with Utp8p (Figure 7B). GST-Utp8p (240 μg of protein) bound to GT-Sepharose in the presence or absence of molar excesses of tRNA was incubated with a twofold molar excess of Gsp1p-GTP. Similarly, Gsp1p-GTP was incubated with GST bound to GT-Sepharose. The columns were washed, and Utp8p was released from bound GST by using the TEV protease. Utp8p (top row) and Gsp1p (third row) in the eluate were detected by Western blot analysis. The amount of GST-Utp8p bound to the GT-resin was also monitored by Coomassie Blue staining of an SDS-PAG (second row). The amount of Gsp1p in the wash eluate was determined by Coomassie Blue staining of an SDS-PAG (fourth row). Similar amounts of Gsp1p were found bound to GST-Utp8p in the presence (lane 1) or absence of tRNA (lanes 2), but minimally to bound GST (lane 3). Coomassie Blue staining of the blot detected Utp8p (second row) but not Gsp1p (data not shown), indicating that the two proteins are interacting weakly with each other, which is consistent with the amount of Gsp1p in the wash eluate (fourth row, compare lanes 1 and 2 with lane 3). The finding that Utp8p interacts with Gsp1p is interesting, because Utp8p does not possess a typical Ran-binding domain motif found in known Ran-binding proteins.
Utp8p interacts directly with Los1p and Gsp1p in a tRNA-independent manner, and Los1p has been shown to interact with Gsp1p in a tRNA-dependent manner in vitro by using an assay based on Rna1p activation of the GTPase activity of unbound Gsp1p-GTP (Hellmuth et al., 1998). These findings together suggest that Los1p, Utp8p, and Gsp1p may form a complex in vivo. To test this possibility, it was necessary to verify that Los1p interacts with Gsp1p in the protein-binding assay (Figure 7C). GST-Los1p and a twofold molar excess of Gsp1p-GTP were incubated with GT-Sepharose in the presence or absence of molar excesses of tRNA. Los1p was released from bound GST by using the TEV protease, and Western blot analysis was used to detect Los1p (top row) and Gsp1p (third row) in the eluate. The amount of GST-Los1p bound to the GT-resin was ascertained by Coomassie Blue staining of an SDS-PAG (second row). Gsp1p interacted with Los1p in the absence of tRNA (lane 2) and presence of tRNA (lane 1). The amount of bound Gsp1p is significantly higher than that observed with the GST control (lane 3). Analyses of the blot by densitometry indicate that the amount of Gsp1p bound to Los1p in the presence of tRNA is approximately twofold higher than that in the absence of tRNA. The Los1p–Gsp1p interaction observed is very weak, because the majority of Gsp1p was detected in the wash eluate by Coomassie Blue staining of an SDS-PAG (fourth row).
To ascertain whether Utp8p, Gsp1p, and Los1p form a complex in vitro, GST-Utp8p (240 μg) was prebound to GT-Sepharose with or without tRNA, and then it was incubated with equimolar quantities of Los1p and Gsp1p-GTP (Figure 7D). Utp8p was released from bound GST by using the TEV protease, and Western blot analysis was used to detect Utp8p (top row), Los1p (third row), and Gsp1p (fifth row) in the eluate (Figure 7D). The amount of GST-Utp8p bound to GT-Sepharose was monitored by Coomassie Blue staining of an SDS-PAG (second row). As already shown in Figure 6A, Los1p bound to Utp8p in a tRNA-independent manner (third row, lanes 1–4), and it did not interact with GST (lane 5). The strength of this interaction between Utp8p and Los1p was not determined by the presence or absence of Gsp1p, because the amount of bound Los1p did not increase when Gsp1p was present (third row, compare lanes 1 and 3). As before, the interaction between Utp8p and Los1p is weak, because Coomassie Blue staining of the blot did not detect Los1p (data not shown), and the majority of Los1p was found in the wash eluate (fourth row). Gsp1p was detected in the complex consisting of Los1p and Utp8p containing tRNA (fifth row, lane 1); this amount of Gsp1p is higher than that detected in the complex consisting of Los1p and Utp8p lacking tRNA (lane 2). In both cases the amount of Gsp1p is higher than that bound to GST alone (lane 5). Densitometric analyses of the blot indicate that the amount of Gsp1p bound in the presence of tRNA is approximately fourfold higher than in the absence of tRNA. Thus, formation of a Gsp1p–Los1p–Utp8p complex is tRNA dependent. Similarly, the interaction of Gsp1p is weak, because it could not be detected by Coomassie Blue staining of the blot (data not shown), and the majority of Gsp1p was found in the wash eluate (sixth row). Nonetheless, the data show that Utp8p, Los1p, Gsp1p, and tRNA form a quaternary complex in vitro.
Sequential binding analysis was conducted to understand the tRNA dependence of Gsp1p interaction with Utp8p and Los1p (Figure 7, E and F). GST-Utp8p was bound to GT-Sepharose in the presence (lane 1) or absence (lane 2) of 10 μM tRNA. The resins were washed and incubated with a twofold molar excess of Los1p (Figure 7E) or Gsp1p-GTP (Figure 7F). Unbound proteins were removed and incubated with Gsp1p-GTP (Figure 7E) or Los1p (Figure 7F). Bound GST was incubated with Los1p (lane 3) or Gsp1p-GTP (lane 4). Utp8p was released from bound GST by using TEV, and Western blot analysis was used to detect Utp8p (first row), Los1p (third row), and Gsp1p (fifth row) in the eluate. Bound Utp8p (second row) and unbound Los1p (fourth row) and Gsp1p (sixth row) were detected by Coomassie Blue staining of SDS-PAGs. Interaction of Gsp1p with Utp8p and Los1p was highly dependent on tRNA when it was added after Los1p was incubated with Utp8p (Figure 7E, compare lanes 1 and 2), but not when it was incubated with Utp8p before addition of Los1p (Figure 7F, compare lanes 1 and 2). Together, the results suggest that Los1p may be responsible for the tRNA-dependent binding of Gsp1p to the Utp8p–Los1p complex.
Utp8p Seems to Be Located in the Nucleoplasm of a nup120 Mutant Strain
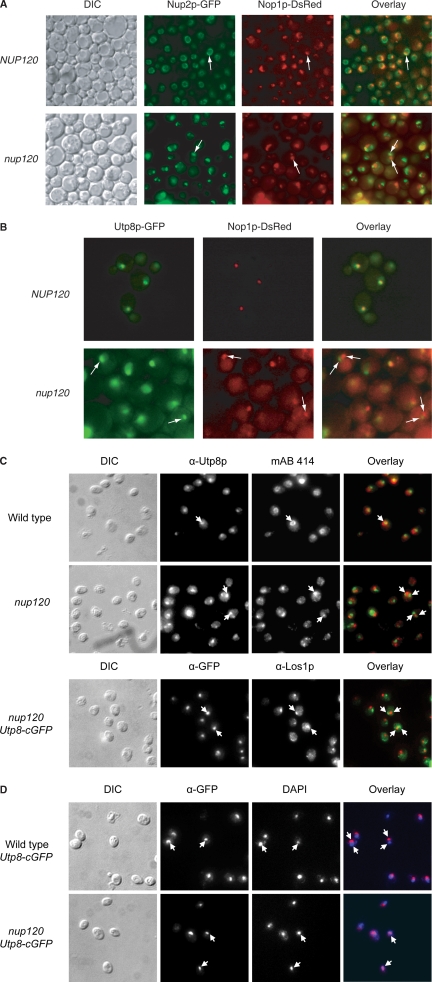
Los1p is found primarily at the NPC, and Utp8p is only detected in the nucleolus at steady state (Simos et al., 1996b; Steiner-Mosonyi et al., 2003). Furthermore, Msn5p is present in the nucleoplasm and at the NPC (Yoshida and Blobel, 2001; Huh et al., 2003). The different location of Utp8p and the two nuclear tRNA export receptors suggests that Utp8p may be transporting the tRNAs out of the nucleolus to deliver them to the tRNA export receptors. Thus, it is anticipated that if Utp8p is transporting tRNAs out of the nucleolus to deliver them to the export receptors, such as Los1p, it should associate with the NPC. In wild-type cells at steady state, Utp8p is not detected at the NPC by direct fluorescence microscopy (Figure 8B). Therefore, a nucleoporin mutant nup120 strain was used to ascertain whether Utp8p concentrates at the NPC. Loss of the function of Nup120p produces a defect in which components of the NPC cluster on the nucleoplasmic side of the nuclear envelope (Aitchison et al., 1995). The clustered NPC has been shown to locate away from the nucleolus (Aitchison et al., 1995). Clustering of the NPC does not block nuclear import, but it affects nuclear export processes, including tRNA export (Aitchison et al., 1995; Sarkar and Hopper, 1998). In addition, nup120 has been used successfully to trap nuclear proteins involved in nuclear export at the NPC, and a cytoplasmic protein involved in nuclear import (Aitchison et al., 1995). To detect the location of Utp8p in the nup120 strain, the chromosomal UTP8 gene was tagged at the 3′ end with GFP. The nucleolus was identified by monitoring the nucleolar protein Nop1p fused to dsRED, and clustering of the NPC in nup120 was monitored using Nup2-GFP.
Figure 8.
Utp8p seems to be located in the nucleoplasm in a nup120 mutant strain. The NUP120 NUP2-GFP, nup120 NUP2-GFP, NUP120 UTP8-GFP, and nup120 UTP8-GFP strains transformed with pYX242-NOP1-DsRED were grown at 23°C for 3 h, and the cellular location of Nup2p, Nop1p, and Utp8p was visualized by direct fluorescence microscopy. Overlay analyses were used to ascertain whether Utp8p colocalizes with Nop1p in the wild-type (A) and mutant Nup120p strains (B). (C) Utp8p does not colocalize with the NPC or Los1p in nup120. Colocalization of Utp8p with Nup159p and Nup1p was investigated by immunofluorescence by using rabbit anti-Utp8p and mAB414, which is a monoclonal antibody made against the vertebrate homologues of Nup159p and Nup1p. The NUP120 UTP8-cgfp and nup120 UTP8-cgfp strains were used to determine whether Utp8p colocalizes with Los1p using rabbit anti-Los1p and a mouse anti-GFP. (D) Utp8p colocalizes with DAPI in nup120. Mouse anti-GFP was used to detect Utp8p-cgfp in the NUP120 UTP8-cgfp and nup120 UTP8-cgfp strains, and DNA was observed by staining with DAPI.
In NUP120, Nup2-GFP is located throughout the nuclear envelope, and Nop1-dsRED is observed as a concentrated signal (Figure 8A). Overlay of the two images shows the position of the nucleolus relative to the nuclear envelope. Nup2-GFP is found clustered in the nuclear envelope of nup120, and the signal for Nop1-dsRED remained unaffected in many of the nup120 cells. Overlay analysis shows that the clustered NPC is located away from the nucleolus; and in some cells, they are found opposite each other. Furthermore, localization of Nop1p in the nucleus verifies that the nup120 strain is not defective in nuclear protein import (Aitchison et al., 1995). Utp8-GFP colocalizes with Nop1-dsRED in NUP120, confirming that Utp8p is located in the nucleolus at steady state (Figure 8B). In contrast, Utp8-GFP does not colocalize with Nop1-dsRED in nup120, indicating that Utp8p is not in the nucleolus.
To test whether Utp8p is at the NPC, immunofluorescence was used to determine whether Utp8p colocalizes with the FG repeat Nups Nup159p and Nup1p in the nup120 mutant strain. Colocalization of Utp8-cGFP with Los1p in nup120 was also investigated by immunofluorescence. In the wild-type strain the FG repeat Nups are located throughout the nuclear envelope and Utp8p is detected as a concentrated signal (Figure 8C). The signals for the Nups and Los1p are concentrated in nup120. However, overlay analyses show that Utp8p did not colocalize with the Nups or Los1p in the clustered NPC.
Colocalization of Utp8p with 4,6-diamidino-2-phenylindole (DAPI) was used to determine whether Utp8p is in the nucleoplasm of nup120 (Figure 8D). The immunofluorescence signal for Utp8-cGFP does not overlap with that of DAPI in the wild-type cells. In contrast, the Utp8p signal overlaps with that of DAPI in many nup120 cells. This finding suggests that Utp8p is in the nucleoplasm of nup120. Altogether, the data suggest that Utp8p may leave the nucleolus to deliver the tRNA to the nuclear tRNA export receptors and Cca1p.
DISCUSSION
Utp8p is an essential nucleolar component of the nuclear tRNA export machinery in S.cerevisiae, which acts at a step between tRNA maturation/aminoacylation and translocation of the tRNA across the NPC. Utp8p was proposed to function as an intranuclear factor that delivers aminoacylated and nonaminoacylated tRNAs to the appropriate nuclear tRNA export pathway (Steiner-Mosonyi et al., 2003). In this study, we found by TAP that Utp8p associates with Los1p, Msn5p, Cca1p, and several aminoacyl-tRNA synthetases, including Tys1p in vivo (Figures 1, 2, and 5). In vitro binding studies also show that Utp8p interacts directly with Tys1p (Figure 3), Los1p (Figure 6A), and Msn5p (Figure 6B). By using a split GFP system, it was determined in vivo that Utp8p interacts with Tys1p in the nucleolus (Figure 4C). Furthermore, Utp8p seems to concentrate in the nucleoplasm of a nup120 strain, which is defective in nuclear export but not nuclear import (Figure 8D). These data together with the steady-state location of Los1p at the NPC and Msn5p in the nucleoplasm and NPC strongly suggest that Utp8p may function as an intranuclear tRNA carrier that transports aminoacyl-tRNAs out of the nucleolus, delivering them to the nuclear tRNA export receptors. This conclusion is also supported by the following evidence: 1) nuclear tRNA aminoacylation has been shown to occur in the nucleolus (Steiner-Mosonyi and Mangroo, 2004), 2) tRNAs are primarily exported from the nucleus in the aminoacylated form (Steiner-Mosonyi and Mangroo, 2004), and 3) Los1p and Msn5p are thought to facilitate nuclear export of aminoacyl-tRNAs (Bohnsack et al., 2002; Calado et al., 2002; Steiner-Mosonyi and Mangroo, 2004). In addition, the data imply that Utp8p may collect the tRNAs from the aminoacyl-tRNA synthetases and transfer them to the export receptors by using a channeling mechanism. This is consistent with reports showing that a channeling strategy is used in mRNA and ribosome biogenesis and export, tRNA maturation, delivery of cytoplasmic tRNAs to aminoacyl-tRNA synthetases, and transfer of aminoacyl-tRNAs from aminoacyl-tRNA synthetases to ribosomes (Stapulionis and Deutscher, 1995; Simos et al., 1996a; Yoo and Wolin, 1997; Wolin and Matera, 1999; Grosshans et al., 2000a,b; Deinert et al., 2001; Milkereit et al., 2001, 2003; Strasser et al., 2002). Utp8p also interacts directly with Cca1p (Figure 6C), which is in agreement with evidence suggesting that Cca1p plays a role in facilitating translocation of nonaminoacylated tRNAs across the NPC (Feng and Hopper, 2002). This finding further supports the previous suggestion that Utp8p also delivers nonaminoacylated tRNAs to the export receptors of the aminoacylation-independent tRNA export pathway (Steiner-Mosonyi et al., 2003). The reason why the nucleolus is the starting point for nuclear tRNA export by the aminoacylation-dependent and -independent pathways in S.cerevisiae is not known. A plausible explanation is that the final tRNA maturation step, which is presently not known, is located in this compartment.
The GTPase Ran/Gsp1p plays an essential role in both nuclear import and export processes facilitated by β-karyopherins (Gorlich and Kutay, 1999). For nuclear export, interaction of the export receptor with the GTP bound form of Ran/Gsp1p facilitates binding of the cargo to the receptor. The resulting ternary complex moves across the NPC and once in the cytoplasm, the GTPase activity of Ran/Gsp1p is activated by the RanGTPase-activating protein RanGAP/Rna1p. Hydrolysis of GTP to GDP by Ran/Gsp1p facilitates dissociation of the export receptor–cargo–Ran/Gsp1p complex. Los1p has been shown to copurify with Gsp1p by affinity purification and to interact with Gsp1p-GTP in a tRNA-dependent manner in vitro (Hellmuth et al., 1998). This finding led to the suggestion that loading of Los1p and Msn5p with tRNA is dependent on Gsp1p-GTP in vivo. This has also been shown for exportin-t and exportin-5 (Arts et al., 1998a; Kutay et al., 1998; Bohnsack et al., 2002; Calado et al., 2002). Gsp1p also copurified with Utp8p by TAP. Furthermore, TAP resulted in the co-isolation of Utp8p and Los1p, or Msn5p (Figure 5). In vitro binding analyses established that Utp8p interacts directly with Los1p, Msn5p, and Gsp1p (Figures 6, A and B, and 7B). Together, the data suggest that Utp8p may associate with the export receptor–Gsp1p complex in vivo. Therefore, in vitro binding studies were conducted to test whether Utp8p, Los1p, and Gsp1p form a complex. The analyses indicate that Gsp1p forms a complex with Utp8p and Los1p, but only when Utp8p contained tRNA (Figure 7D). This suggests that Gsp1p, Utp8p, tRNA, and Los1p could be forming a quaternary complex in vivo. The tRNA dependence of Gsp1p binding is most likely related to Los1p instead of Utp8p based on the following observations: 1) it has been shown that Gsp1p associates with Los1p in the presence of tRNA (Hellmuth et al., 1998); 2) in vitro protein binding studies also found that the Gsp1p–Los1p interaction is dependent on tRNA (Figure 7C); 3) we have found that the interaction between Utp8p and Gsp1p is not dependent on tRNA (Figure 7B); and 4) Gsp1p interacted with Los1p and Utp8p in a highly tRNA-dependent manner when Los1p was incubated with the Utp8p–tRNA complex first (Figure 7E), but not when it was added to the Utp8p–tRNA complex before addition of Los1p (Figure 7F). In addition, the finding that Gsp1p interacts directly with Utp8p (Figure 7B) as well as Los1p (Hellmuth et al., 1998) (Figure 7C) suggests that the Gsp1p–Utp8p–tRNA–Los1p complex is formed by Utp8p interacting with Los1p, and Gsp1p interacting with both proteins. Although the significance of this interaction network is not fully understood, it is possible that this is needed to transfer the tRNA from Utp8p to the export receptor. A model that is consistent with the data from this work and studies reported previously (Hellmuth et al., 1998), suggests that Utp8p loaded with tRNA binds Los1p first to indicate the presence of tRNA; Gsp1p is subsequently recruited to Los1p, allowing Los1p to interact with the tRNA. Gsp1p then interacts with Utp8p to facilitate release of the tRNA. However, further studies are required to fully understand the mechanism of Utp8p-mediated loading of Los1p and Msn5p and the significance of the interaction of Gsp1p with Utp8p. Nevertheless, it is possible that the model proposed for Los1p loading may be applicable to other Ran/Gsp1p-regulated nuclear export processes, such as ribosome export, which is complex and involves a large number of proteins.
The in vitro binding data indicate that Utp8p interacts directly with Los1p, Msn5p, Cca1p (Figure 6), and Tys1p (Figure 3) in a tRNA-independent manner. This commonality of the function of Utp8p might mean that in vivo it has to bind aminoacyl-tRNA synthetases to collect the tRNA, and the tRNA export proteins to transfer the tRNA. However, why Utp8p remained associated with the Tys1p–tRNA complex, and with Cca1p, Los1p, and Msn5p when it contained tRNA is not understood. A possible explanation is that in vivo dissociation of Utp8p from the export proteins, and the Utp8p–tRNA complex from the aminoacyl-tRNA synthetases, is mediated by proteins that have not been identified; by analogy to ribosome export, these proteins may also be needed to traffic Utp8p between the nucleolus and the location of the nuclear export proteins. Thus, it is possible that the intranuclear phase of nuclear tRNA export may be more complex than anticipated and that it involves a mechanism analogous to that used to move preribosomes from the nucleolus to the nucleoplasm for export to the cytoplasm (Tschochner and Hurt, 2003). Future studies will address the issue of how Utp8p dissociates from the aminoacyl-tRNA synthetases and the export receptors, and the mechanism by which Utp8p shuttles between the nucleolus and the nuclear tRNA export proteins.
Biogenesis and export of ribosome to the cytoplasm is a complex process involving a large number of proteins (Tschochner and Hurt, 2003; Granneman and Baserga, 2005; Houseley et al., 2006). Ribosomal proteins made in the cytoplasm are imported into the nucleolus and associate with the 35S pre-rRNA to form pre-90S ribosomes. The 35S pre-rRNA of the preribosome is processed to produce the pre-large and pre-small ribosomal subunits. The pre-rRNA of each subunit undergoes additional processing and the ribosomal subunits are exported to cytoplasm. For production of the 40S small ribosomal subunit, the preribosomal subunit containing the 20S pre-rRNA is exported by Xpo1p to the cytoplasm (Moy and Silver, 1999), where the 20S rRNA is processed to the mature 18S rRNA, giving rise to functional subunits. A large protein complex known as the SSU processome has been shown to be involved in processing of pre-18S rRNA (Dragon et al., 2002). More recently, Utp8p, Nan1p/Utp17p, Utp10p, Utp15p, Utp4p, Utp5p, and Utp9p of the SSU processome have been shown to form a subcomplex that regulates transcription of rDNA (Gallagher et al., 2004; Krogan et al., 2004). Our TAP analysis also showed that these Utp proteins copurified with Utp8-TAP. We have shown previously that depletion of Utp8p for 24 h did not affect synthesis of 25S or 18S rRNA, whereas nuclear tRNA export was affected within 6 h of depletion of Utp8p (Steiner-Mosonyi et al., 2003). However, other studies have shown that depletion of Utp8p over a 48-h period affects transcription of rDNA. Although the major function of Utp8p is in nuclear tRNA export, it is evident that it also plays a role in rDNA transcription. In contrast to Utp8p, we found that depletion of Utp4p, Utp5p, Utp10p, or Utp15p of the subcomplex did not affect nuclear tRNA export, which supports their function in rDNA transcription (data not shown). The significance of the involvement of Utp8p but not the others in both nuclear tRNA export and rDNA transcription is not understood. A possible explanation is that Utp8p is also playing a regulatory role that coordinates nuclear tRNA export and biogenesis of ribosome. This communication between the two pathways may be important for controlling the rate of protein synthesis based on the demand for protein synthesis, which is dictated by a number of signals such as nutrient availability. Xpo1p may also be associated with the function of Utp8p in regulating biogenesis of ribosomes, because Xpo1p copurified with Utp8p by TAP and loss of the function of Xpo1p did not result in a nuclear tRNA export defect (data not shown) (Feng and Hopper, 2002). However, further studies are required to understand the implication of the function of Utp8p in both nuclear tRNA export and ribosome biogenesis.
Several S.cerevisiae genome-scale protein–protein interaction studies using TAP have been reported in the past several years. However, these studies did not detect an interaction between Utp8p, Los1p, Msn5p, Gsp1p, Cca1p, Utp21p, Utp18p, and several aminoacyl-tRNA synthetases and nucleoporins (Krogan et al., 2004, 2006; Gavin et al., 2006). Like the studies reported we also could not detect an interaction between Utp8p and these proteins by TAP followed by LCQToF MS-MS. However, we did find these interactions by performing IP and LCQToF MS-MS analysis, and by TAP followed by Western blot analysis. In vitro binding analyses indicate that Utp8p interacts weakly with Tys1p, Los1p, Cca1p, Msn5p, and Gsp1p, which is probably a reflection of the dynamic nature of the tRNA export process. This finding suggests that the major factor contributing to the lack of detection of these interactions by TAP and mass spectrometry is most likely the poor yield of the prey proteins. Thus, it is very probable that the interaction network established for each protein of S.cerevisiae is incomplete. An alternative strategy that can be used to identify proteins that interact transiently in vivo is reconstitution of split GFP. This strategy has been used to detect protein–protein interaction in mammalian cells and bacteria but not previously in S.cerevisiae (Ozawa et al., 2001, 2003, 2005; Ozawa and Umezawa, 2001; Jeong et al., 2006). We established this split GFP system for S.cerevisiae, and we showed that it can easily detect the weak interaction between Utp8p and Tys1p in vivo (Figure 4). Furthermore, it was possible to identify the cellular location of the interaction at steady state. Thus, the expansion of this strategy to other proteins in the S.cerevisiae Genome Database would add greatly to the available protein–protein interaction data both by increasing the number of interactions detectable and by pinpointing the location of the interaction within cells. Such data would not only provide a more complete interaction network of each protein, it would also facilitate a large-scale transition from the assignment of proteins to functional pathways to the assignment of more precise functions to individual proteins.
ACKNOWLEDGMENTS
This work was supported by an operating grant from the Canadian Institutes of Health Research. B.R.S. and J.B.P. are recipients of Ontario Graduate Scholarships. We thank Drs. D. Heinrichs, J. Aitchison, U. Stochaj, E. Hurt, P. Schimmel, M. Chalfie, U. RajBhandary, A. Hopper, and S. Wente for generous gifts of plasmids and antibodies. We also thank the anonymous reviewers for constructive and insightful suggestions.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-11-1016) on July 18, 2007.
REFERENCES
- Aitchison J. D., Blobel G., Rout M. P. Nup120p: a yeast nucleoporin required for NPC distribution and mRNA transport. J. Cell Biol. 1995;131:1659–1675. doi: 10.1083/jcb.131.6.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arts G. J., Fornerod M., Mattaj I. W. Identification of a nuclear export receptor for tRNA. Curr. Biol. 1998a;8:305–314. doi: 10.1016/s0960-9822(98)70130-7. [DOI] [PubMed] [Google Scholar]
- Arts G. J., Kuersten S., Romby P., Ehresmann B., Mattaj I. W. The role of exportin-t in selective nuclear export of mature tRNAs. EMBO J. 1998b;17:7430–7441. doi: 10.1093/emboj/17.24.7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad A. K., Stanford D. R., Sarkar S., Hopper A. K. Role of nuclear pools of aminoacyl-tRNA synthetases in tRNA nuclear export. Mol. Biol. Cell. 2001;12:1381–1392. doi: 10.1091/mbc.12.5.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack M. T., Regener K., Schwappach B., Saffrich R., Paraskeva E., Hartmann E., Gorlich D. Exp5 exports eEF1A via tRNA from nuclei and synergizes with other transport pathways to confine translation to the cytoplasm. EMBO J. 2002;21:6205–6215. doi: 10.1093/emboj/cdf613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calado A., Treichel N., Muller E. C., Otto A., Kutay U. Exportin-5-mediated nuclear export of eukaryotic elongation factor 1A and tRNA. EMBO J. 2002;21:6216–6224. doi: 10.1093/emboj/cdf620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deinert K., Fasiolo F., Hurt E. C., Simos G. Arc1p organizes the yeast aminoacyl-tRNA synthetase complex and stabilizes its interaction with the cognate tRNAs. J. Biol. Chem. 2001;276:6000–6008. doi: 10.1074/jbc.M008682200. [DOI] [PubMed] [Google Scholar]
- Dragon F., et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002;417:967–970. doi: 10.1038/nature00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernens I., Goodfellow S. J., Innes F., Kenneth N. S., Derblay L. E., White R. J., Scott P. H. Hypoxic stress suppresses RNA polymerase III recruitment and tRNA gene transcription in cardiomyocytes. Nucleic Acids Res. 2006;34:286–294. doi: 10.1093/nar/gkj402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fechter P., Rudinger-Thirion J., Theobald-Dietrich A., Giege R. Identity of tRNA for yeast tyrosyl-tRNA synthetase: tyrosylation is more sensitive to identity nucleotides than to structural features. Biochemistry. 2000;39:1725–1733. doi: 10.1021/bi992276t. [DOI] [PubMed] [Google Scholar]
- Feng W., Hopper A. K. A Los1p-independent pathway for nuclear export of intronless tRNAs in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2002;99:5412–5417. doi: 10.1073/pnas.082682699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher J.E.G., Dunbar D. A., Granneman S., Mitchell B. M., Osheim Y., Beyer A. L., Baserga S. J. RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes Dev. 2004;18:2506–2517. doi: 10.1101/gad.1226604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin A. C., et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., Weissman J. S. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Giaever G., et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- Gorlich D., Kutay U. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- Grandi P., et al. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell. 2002;10:105–115. doi: 10.1016/s1097-2765(02)00579-8. [DOI] [PubMed] [Google Scholar]
- Granneman S., Baserga S. J. Crosstalk in gene expression: coupling and co-regulation of rDNA transcription, pre-ribosome assembly and pre–rRNA processing. Curr. Opin. Cell Biol. 2005;17:281–286. doi: 10.1016/j.ceb.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Grosshans H., Hurt E., Simos G. An aminoacylation-dependent nuclear tRNA export pathway in yeast. Genes Dev. 2000a;14:830–840. [PMC free article] [PubMed] [Google Scholar]
- Grosshans H., Simos G., Hurt E. Review: transport of tRNA out of the nucleus-direct channeling to the ribosome? J. Struct. Biol. 2000b;129:288–294. doi: 10.1006/jsbi.2000.4226. [DOI] [PubMed] [Google Scholar]
- Hellmuth K., Lau D. M., Bischoff F. R., Kunzler M., Hurt E., Simos G. Yeast Los1p has properties of an exportin-like nucleocytoplasmic transport factor for tRNA. Mol. Cell Biol. 1998;18:6374–6386. doi: 10.1128/mcb.18.11.6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper A. K., Phizicky E. M. tRNA transfers to the limelight. Genes Dev. 2003;17:162–180. doi: 10.1101/gad.1049103. [DOI] [PubMed] [Google Scholar]
- Houseley J., LaCava J., Tollervey D. RNA-quality control by the exosome. Nat. Rev. Mol. Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Jeong J., Kim S. K., Ahn J., Park K., Jeong E. J., Kim M., Chung B. H. Monitoring of conformational change in maltose binding protein using split green fluorescent protein. Biochem. Biophys. Res. Commun. 2006;339:647–651. doi: 10.1016/j.bbrc.2005.11.056. [DOI] [PubMed] [Google Scholar]
- Krogan N. J., et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Krogan N. J., et al. High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell. 2004;13:225–239. doi: 10.1016/s1097-2765(04)00003-6. [DOI] [PubMed] [Google Scholar]
- Kunzler M., Hurt E. C. Cse1p functions as the nuclear export receptor for importin alpha in yeast. FEBS Lett. 1998;433:185–190. doi: 10.1016/s0014-5793(98)00892-8. [DOI] [PubMed] [Google Scholar]
- Kutay U., Lipowsky G., Izaurralde E., Bischoff F. R., Schwarzmaier P., Hartmann E., Gorlich D. Identification of a tRNA-specific nuclear export receptor. Mol. Cell. 1998;1:359–369. doi: 10.1016/s1097-2765(00)80036-2. [DOI] [PubMed] [Google Scholar]
- Lee D. C., Aitchison J. D. Kap104p-mediated nuclear import. Nuclear localization signals in mRNA-binding proteins and the role of Ran and Rna. J. Biol. Chem. 1999;274:29031–29037. doi: 10.1074/jbc.274.41.29031. [DOI] [PubMed] [Google Scholar]
- Lund E., Dahlberg J. E. Proofreading and aminoacylation of tRNAs before export from the nucleus. Science. 1998;282:2082–2085. doi: 10.1126/science.282.5396.2082. [DOI] [PubMed] [Google Scholar]
- McGuire A. T., Mangroo D. Cex1p is a novel cytoplasmic component of the Saccharomyces cerevisiae nuclear tRNA export machinery. EMBO J. 2007;26:288–300. doi: 10.1038/sj.emboj.7601493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milkereit P., Gadal O., Podtelejnikov A., Trumtel S., Gas N., Petfalski E., Tollervey D., Mann M., Hurt E., Tschochner H. Maturation and intranuclear transport of pre-ribosomes requires Noc proteins. Cell. 2001;105:499–509. doi: 10.1016/s0092-8674(01)00358-0. [DOI] [PubMed] [Google Scholar]
- Milkereit P., Strauss D., Bassler J., Gadal O., Kuhn H., Schutz S., Gas N., Lechner J., Hurt E., Tschochner H. A Noc complex specifically involved in the formation and nuclear export of ribosomal 40 S subunits. J. Biol. Chem. 2003;278:4072–4081. doi: 10.1074/jbc.M208898200. [DOI] [PubMed] [Google Scholar]
- Moy T. I., Silver P. A. Nuclear export of the small ribosomal subunit requires the Ran-GTPase cycle and certain nucleoporins. Genes Dev. 1999;13:2118–2133. doi: 10.1101/gad.13.16.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa T., Nishitani K., Sako Y., Umezawa Y. A high-throughput screening of genes that encode proteins transported into the endoplasmic reticulum in mammalian cells. Nucleic Acids Res. 2005;33:e34. doi: 10.1093/nar/gni032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa T., Sako Y., Sato M., Kitamura T., Umezawa Y. A genetic approach to identifying mitochondrial proteins. Nat. Biotechnol. 2003;21:287–293. doi: 10.1038/nbt791. [DOI] [PubMed] [Google Scholar]
- Ozawa T., Takeuchi M., Kaihara A., Sato M., Umezawa Y. Protein splicing-based reconstitution of split green fluorescent protein for monitoring protein-protein interactions in bacteria: improved sensitivity and reduced screening time. Anal. Chem. 2001;73:5866–5874. doi: 10.1021/ac010717k. [DOI] [PubMed] [Google Scholar]
- Ozawa T., Umezawa Y. Detection of protein-protein interactions in vivo based on protein splicing. Curr. Opin. Chem. Biol. 2001;5:578–583. doi: 10.1016/s1367-5931(00)00244-1. [DOI] [PubMed] [Google Scholar]
- Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E., Wilm M., Seraphin B. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- Rout M. P., Aitchison J. D., Suprapto A., Hjertaas K., Zhao Y., Chait B. T. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S., Azad A. K., Hopper A. K. Nuclear tRNA aminoacylation and its role in nuclear export of endogenous tRNAs in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 1999;96:14366–14371. doi: 10.1073/pnas.96.25.14366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S., Hopper A. K. tRNA nuclear export in saccharomyces cerevisiae: in situ hybridization analysis. Mol. Biol. Cell. 1998;9:3041–3055. doi: 10.1091/mbc.9.11.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen H. H., Hopper A. K. Retrograde movement of tRNAs from the cytoplasm to the nucleus in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2005;102:11290–11295. doi: 10.1073/pnas.0503836102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simos G., Segref A., Fasiolo F., Hellmuth K., Shevchenko A., Mann M., Hurt E. C. The yeast protein Arc1p binds to tRNA and functions as a cofactor for the methionyl- and glutamyl-tRNA synthetases. EMBO J. 1996a;15:5437–5448. [PMC free article] [PubMed] [Google Scholar]
- Simos G., Tekotte H., Grosjean H., Segref A., Sharma K., Tollervey D., Hurt E. C. Nuclear pore proteins are involved in the biogenesis of functional tRNA. EMBO J. 1996b;15:2270–2284. [PMC free article] [PubMed] [Google Scholar]
- Solsbacher J., Maurer P., Bischoff F. R., Schlenstedt G. Cse1p is involved in export of yeast importin alpha from the nucleus. Mol. Cell Biol. 1998;18:6805–6815. doi: 10.1128/mcb.18.11.6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapulionis R., Deutscher M. P. A channeled tRNA cycle during mammalian protein synthesis. Proc. Natl. Acad. Sci. USA. 1995;92:7158–7161. doi: 10.1073/pnas.92.16.7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner-Mosonyi M., Leslie D. M., Dehghani H., Aitchison J. D., Mangroo D. Utp8p is an essential intranuclear component of the nuclear tRNA export machinery of Saccharomyces cerevisiae. J. Biol. Chem. 2003;278:32236–32245. doi: 10.1074/jbc.M302779200. [DOI] [PubMed] [Google Scholar]
- Steiner-Mosonyi M., Mangroo D. The nuclear tRNA aminoacylation-dependent pathway may be the principal route used to export tRNA from the nucleus in Saccharomyces cerevisiae. Biochem. J. 2004;378:809–816. doi: 10.1042/BJ20031306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser K., et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002;417:304–308. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- Takano A., Endo T., Yoshihisa T. tRNA actively shuttles between the nucleus and cytosol in yeast. Science. 2005;309:140–142. doi: 10.1126/science.1113346. [DOI] [PubMed] [Google Scholar]
- Tschochner H., Hurt E. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 2003;13:255–263. doi: 10.1016/s0962-8924(03)00054-0. [DOI] [PubMed] [Google Scholar]
- White R. J. RNA polymerase III transcription–a battleground for tumour suppressors and oncogenes. Eur. J. Cancer. 2004a;40:21–27. doi: 10.1016/j.ejca.2003.09.027. [DOI] [PubMed] [Google Scholar]
- White R. J. RNA polymerase III transcription and cancer. Oncogene. 2004b;23:3208–3216. doi: 10.1038/sj.onc.1207547. [DOI] [PubMed] [Google Scholar]
- White R. J. RNA polymerases I and III, growth control and cancer. Nat. Rev. Mol. Cell Biol. 2005;6:69–78. doi: 10.1038/nrm1551. [DOI] [PubMed] [Google Scholar]
- Wolin S. L., Matera A. G. The trials and travels of tRNA. Genes Dev. 1999;13:1–10. doi: 10.1101/gad.13.1.1. [DOI] [PubMed] [Google Scholar]
- Yoo C. J., Wolin S. L. The yeast La protein is required for the 3′ endonucleolytic cleavage that matures tRNA precursors. Cell. 1997;89:393–402. doi: 10.1016/s0092-8674(00)80220-2. [DOI] [PubMed] [Google Scholar]
- Yoshida K., Blobel G. The karyopherin Kap142p/Msn5p mediates nuclear import and nuclear export of different cargo proteins. J. Cell Biol. 2001;152:729–739. doi: 10.1083/jcb.152.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihisa T., Yunoki-Esaki K., Ohshima C., Tanaka N., Endo T. Possibility of cytoplasmic pre-tRNA splicing: the yeast tRNA splicing endonuclease mainly localizes on the mitochondria. Mol. Biol. Cell. 2003;14:3266–3279. doi: 10.1091/mbc.E02-11-0757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. F., Ma C., Chalfie M. Combinatorial marking of cells and organelles with reconstituted fluorescent proteins. Cell. 2004;119:137–144. doi: 10.1016/j.cell.2004.09.012. [DOI] [PubMed] [Google Scholar]