Abstract
To understand the mechanism of cell migration, one needs to know how the parts of the motile machinery of the cell are assembled and how they move with respect to each other. Actin and myosin II are thought to be the major structural and force-generating components of this machinery (Mitchison and Cramer, 1996; Parent, 2004). The movement of myosin II along actin filaments is thought to generate contractile force contributing to cell translocation, but the relative motion of the two proteins has not been investigated. We use fluorescence speckle and conventional fluorescence microscopy, image analysis, and computer tracking techniques to generate comparative velocity and assembly maps of actin and myosin II over the entire cell in a simple model system of persistently migrating fish epidermal keratocytes. The results demonstrate contrasting polarized assembly patterns of the two components, indicate force generation at the lamellipodium–cell body transition zone, and suggest a mechanism of anisotropic network contraction via sliding of myosin II assemblies along divergent actin filaments.
INTRODUCTION
Crawling cell motion involves a cycle of several distinct processes: protrusion at the front of the cell, attachment to the substratum, and forward translocation of the cell body followed or accompanied by detachment and withdrawal of the rear of the cell. Crawling motion is thought to be dependent on the actin–myosin II cytoskeletal system (Mitchison and Cramer, 1996): protrusion is thought to be driven by the assembly of actin network, which is anchored to the substratum through integrin-containing adhesions; forward translocation of the cell body and contraction of the rear are thought to depend on the interaction of the actin network with the motor protein myosin II. Actin assembly during the protrusion at the leading edge of motile cells has recently received the most attention both in experimental (Pantaloni et al., 2001; Pollard and Borisy, 2003; Ridley et al., 2003) and theoretical studies (for review, see Mogilner, 2006). In contrast, the mechanisms involved in the forward translocation of the cell body remain largely unclear: the exact layout and the mode of action of the contractile actin–myosin II machinery are controversial. Qualitative models proposed in the literature include shortening of small contractile units similar to muscle sarcomeres, myosin II-dependent transport along uniformly polarized actin arrays, and a dynamic network contraction mechanism where contraction results from alignment of actin filaments by myosin II assemblies (Cramer, 1999; Verkhovsky et al., 1999a). More sophisticated biophysical models aim to understand the contractile cytoskeletal machinery in quantitative physical terms. In the past, considering the cytoskeleton as a gel made of cross-linked semiflexible polymers has helped understanding its passive visco-elastic properties. More recently, attempts were made to develop biophysical models taking into account intrinsic activity of the cytoskeleton: polarized assembly of actin filaments and their dynamic interaction with motor proteins. Recent studies analyzed the dynamics of polymer motion in the bundles of aligned actin filaments in the presence of myosin motor proteins (Kruse and Julicher 2003) and developed a general approach to describe the cytoskeletal network of actin filaments and motor proteins in terms of active gel (Kruse et al., 2005, 2006). The model of aligned filament bundles may be applicable to the dynamics of stress fibers; the active gel model describes some general properties of the lamellipodium, such as retrograde flow; but neither of the models have been yet directly tested by comparing their quantitative predictions to the experimental cell dynamics.
In a different approach, Rubinstein et al. (2005) attempted to develop a specific model describing the dynamics of a simple motile cell, fish epidermal keratocyte. Fish keratocytes are characterized by a fast and persistent motion and a simple shape and cytoskeletal organization, which remain nearly constant during migration (Lee et al., 1993; Grimm et al., 2003). The steady-state character of keratocyte migration simplifies quantitative description of the cytoskeletal dynamics and imposes constrains on the theoretical models. Thus, keratocytes represent the favorable cell system to match theoretical models to the experimental data. Rubinstein et al. (2005) combined in their model the molecular reactions of protrusion, attachment to the substrate, and retraction of the cell rear with actin turnover. The model captures the overall character of keratocyte motion, but it does not take into account several important aspects of the process, e.g., the cell body motion and the actin retrograde flow in the lamellipodium are neglected, and the actin–myosin II interaction is considered only at the very rear of the cell.
To test the theoretical models and ultimately understand the mechanism of cell migration in quantitative physical terms, the complete data on the motion and assembly of actin and myosin II in moving cells are of critical importance. In particular, to be able to distinguish between the models of the myosin-dependent contraction at the back of the cell, it is necessary to study the relative motion of myosin II with respect to actin. Partial maps of the motion and assembly of actin in the lamellipodia and lamella of migrating cells were obtained using fluorescent speckling microscopy and computer tracking techniques (Ponti et al., 2004; Vallotton et al., 2004, 2005). However, there is no comparable data on the motion of myosin II, and the relative motion of the two components has not been investigated. Here, we track actin and myosin II in migrating fish epidermal keratocytes and generate maps of the motion and assembly of the two components over the entire cell, and we generate the map of the relative motion of myosin II with respect to actin.
MATERIALS AND METHODS
Cell Culture and Microscopy
Fish epidermal keratocytes were cultured as described previously (Verkhovsky et al., 2003). Fluorescence microscopy was performed using a Nikon Eclipse TE300 inverted microscope (Nikon, Tokyo, Japan) with 100× Plan objective (numerical aperture 1.25) and Micromax 1024BFT cooled charge-coupled device (CCD) camera (Roper Scientific, Trenton, NJ) operated with MetaMorph software (Molecular Devices, Sunnyvale, CA) at a rate of 1 frame per 3 s. Z-series images were acquired at a rate of one series every 4 s by using MicroMax 512BFT cooled CCD camera and piezoelectric translator (Princeton Instruments, Princeton, NJ).
Microinjection
Rhodamine-phalloidin (Sigma-Aldrich, St. Louis, MO) was prepared for microinjection as described previously (Vallotton et al., 2005). Alexa568-phalloidin and Alexa488-phalloidin (Invitrogen, Carlsbad, CA) were prepared identically to obtain 2–4 μg/ml phalloidin solution in 15% dimethyl sulfoxide. Tetramethylrhodamine-myosin II was prepared and injected as described previously (Svitkina et al., 1997). For double fluorescence imaging, cells were first injected with Alexa488-phalloidin and then with tetramethylrhodamine-myosin II.
Tracking
Fluorescence images were first treated as described previously (Vallotton et al., 2005) to equalize intensity and increase speckle contrast over entire cell (Figure 1, A and B). Speed measurement was based on a texture-tracking algorithm implemented with MatLab (Mathworks, Natick, MA), briefly as follows. Images were segmented in square regions with dimensions of 30 × 30 pixels2 (1.95 × 1.95 μm2). The regions were partially overlapping with each other (the distance between adjacent regions was 10 pixels). Each region was compared with regions shifted with respect to its original position by a distance of up to 20 pixels in the subsequent image of the time-lapse sequence. A score based on the cross-correlation between regions defined the most probable displacement and thus the velocity map in the substrate coordinates. Tracking approach similar in principle to our tracking algorithm was described recently (Hebert et al., 2005).
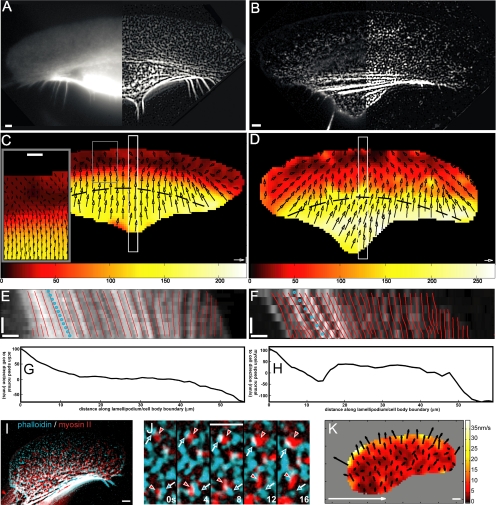
Figure 1.
Velocity maps of actin and myosin II in migrating keratocytes. Distribution of microinjected tetramethylrhodamine B isothiocyanate-phalloidin (A) and rhodamine-myosin II (B); left panels represent raw images; right, processed images. Actin (C) and myosin (D) velocities are shown with arrows, velocity amplitude is color-coded, and inset in C shows zoomed region of velocity inversion at the back of the lamellipodium. Dashed lines in C and D indicate positions where lateral velocity profiles displayed in G and H were measured. Kymographs (E and F) of regions shown with white boxes in C and D are superimposed with simulated kymographs (red lines) created using velocity data. Blue dotted lines represent overall cell velocity. Lateral motion of actin (G) and myosin II (H) in the bundles at the lamellipodium–cell body transition zone: projection of the velocity normal to the cell motion is plotted as a function of the distance along the bundle (indicated with dashed lines in C and D). The direction of the velocity from left to right is represented by positive values and the opposite direction by negative values. Simultaneous localization of actin (cyan) and myosin II (red): overview (I); zoomed region of the lamellipodium (relative displacement highlighted with arrowheads, time indicated in seconds) (J); and map of relative myosin II velocity with respect to actin (K). Horizontal bar, 3 μm; vertical bar, 10 s; white arrow, 100 nm/s.
We did not apply any outlier detection filter for possible false tracking. Smooth variation of the velocity vectors in the map suggested that our tracking procedure in most cases yielded true displacement vectors. Indeed, since the measurements were made in overlapping regions, velocity vectors in the maps were expected to vary smoothly. False tracking would have manifested itself in sharp differences between adjacent velocity vectors, but this was not observed.
The interpolation of the velocity data projected on a line yielded simulated kymographs (Figure 1, E and F). The displacement maps obtained for several pairs of frames were superimposed to obtain time-averaged velocity maps (Figure 1, C and D). Typically, ∼10 frames with the interval between frames of 3–4 s were used for time-averaging. Specifically, for the images represented in Figure 1, C and D, the data were averaged over the total time interval of 36 s (12 frames for Figure 1C and 9 frames for Figure 1D).
The F-actin concentration map was obtained by fixing the same cell that was used for velocity measurement and staining it with saturating amount of rhodamine-phalloidin as described previously (Vallotton et al., 2005) (Figure 4A). To obtain concentration map of cytoskeletal myosin II, we estimated the intensity of diffuse fluorescence due to the soluble myosin species and subtracted it from the images of injected cells (Figure 4D). The minimum of the intensity of the 5- × 5-μm region around each pixel of the image was taken as a measure of diffuse fluorescence in the cell interior. At the cell border, the minimum was taken in 2.5- × 5-μm regions to accommodate the cell outline more precisely. Stretching/compression maps were generated from time-averaged velocity maps (Figure 4, B and E). Normalized polymerization/depolymerization maps were generated from normalized concentration maps and time-averaged velocity maps using the mass conservation principle as described by Vallotton et al. (2004). Stretching/compression and polymerization/depolymerization maps obtained from individual pairs of frames without time-averaging displayed essentially the same patterns as in time-averaged maps, but more noise (data not shown). Actin velocity at different distances from the substratum was measured by applying the above-mentioned tracking protocol to each plane. Relative velocity map of myosin II with respect to actin (Figure 1I) was obtained by subtracting actin velocity map from that of myosin II in the same cell.
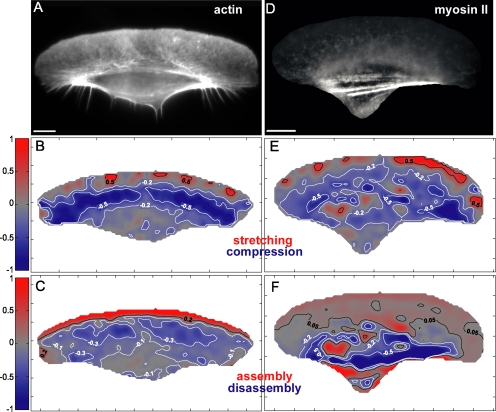
Figure 4.
Stretching/compression and assembly/disassembly maps of actin and myosin II. Maps of concentration (A and D), velocity divergence (B and E) and assembly (C and F) of actin (A–C) and myosin II (D–F) are shown. Negative divergence (blue) reflects compression; in assembly maps, positive values represent assembly, and negative values represent disassembly. Note the blue contraction area in B and E at the lamellipodium–cell body boundary. Bar, 10 μm.
RESULTS
Recently developed feature-based tracking approaches match individual particles (e.g., fluorescence speckles) in sequential images of the time-lapse sequences (Ponti et al., 2004; Vallotton et al., 2004). Because all particles are similar, mismatches are likely to occur (i.e., particle A in the frame i could be falsely identified with particle B in the frame i + 1), but they are subsequently corrected based on information about the overall trend of motion. However, if motion velocity changes significantly within the cell, correction may smoothen or mask velocity changes. Here, we tracked the cytoskeletal motion over the entire cell during a steady-state migration. To capture gradients of velocity, we used matching of small regions of the image, rather than matching individual particles. Because each region has its individual pattern of features, mismatches were not likely, and no correction based on average velocity trend was applied. For actin tracking (Supplemental Video 1), cells were injected with low concentration of fluorescent phalloidin, resulting in fluorescent speckling images (Vallotton et al., 2005) (Figure 1A, left). Myosin II (Supplemental Video 2) is naturally distributed in a form of small discrete clusters (Svitkina et al., 1997), which we tracked using conventional fluorescence images of microinjected rhodamine-myosin II (Figure 1B, left). All images were processed as described previously (Vallotton et al., 2005) to equalize the intensity between the cell body and the lamellipodium and to enhance the contrast of local features throughout the cell (Figure 1, A and B, right). All examined cells displayed similar motion patterns and velocity amplitudes of each of the two labeled proteins.
Maps of Actin and Myosin II Motion in the Lamellipodia and Ventral Part of the Cell Body
We obtained detailed velocity maps in the substrate optical section (encompassing the lamellipodium and the ventral part of the cell body) for six cells for each actin and myosin II. Of these cells, four cells were double-labeled for both proteins. Representative maps are shown in Figure 1. The front part of the lamellipodia exhibited retrograde flow of actin features (Figure 1C), whereas myosin velocity in this region was typically zero or low anterograde velocity (Figure 1D). Only one cell with atypically low overall motion velocity (6 μm/min) displayed retrograde myosin motion at the front of the lamellipodium (data not shown). Velocity of both proteins changed at the back of the lamellipodia (Figure 1, C and D): for actin, we observed change of direction from retrograde to anterograde and an increase in the rate of motion; for myosin, anterograde velocity significantly increased. The boundary where velocity changed formed a line nearly parallel to the leading edge, dividing the lamellipodia approximately in half. Anterograde velocity of both actin and myosin II further increased in the cell body, but it always remained lower than the overall cell velocity. To verify the tracking results, we obtained kymographs by cutting and pasting side-by-side narrow regions from sequential images. Alternatively, velocity profiles obtained from tracking data were used to simulate kymographs for the same regions. Close match between kymographs obtained from images and simulated kymographs confirmed the accuracy of tracking (Figure 1, E and F). Note that kymographs only display motion velocity along a selected direction, whereas the two-dimensional maps provide the information about all directions of motion. Our maps demonstrated that at the lateral wings of the cell, the direction of motion was predominantly lateral toward the cell center, whereas little or no lateral motion was observed in the central part of the cell (Figure 1, C and D). Visual examination of the movies also revealed several instances when myosin particles at the lateral cell wings moved away from the cell center (Supplemental Video 2). To display the lateral motion of actin and myosin II in the bundles at the lamellipodium–cell body transition zone, we plotted the projection of the velocity in the direction normal to the cell motion along the line parallel to the leading edge at approximately half the distance between the front and rear of the cell (Figure 1, G and H). Projection data confirmed predominantly centripetal motion of actin and myosin II at the cell wings and the virtual absence of motion in the central part of the bundle. However, myosin velocity profile along the bundle was generally less smooth than the actin velocity profile, and it displayed short intervals where myosin velocity was directed away from the cell center.
Results of tracking of actin and myosin II in different cells suggested differences in velocity between the two components. In the cells labeled simultaneously for both actin and myosin II (Figure 1I and Supplemental Video 3), the relative motion of the two proteins was not visually apparent, but it could be detected upon careful examination of zoomed images (Figure 1J and Supplemental Video 4). Our tracking procedure allowed reliable detection of the relative motion of the two components. Difference velocity map demonstrated forward motion of myosin II relative to actin throughout the cell (Figure 1K). The relative velocity was low compared with the cell velocity, and it had a graded distribution with maximum at the front of the lamellipodium and minimum at the cell rear. Forward translocation of myosin II with respect to actin in the lamellipodium is consistent with the motion toward the barbed ends of actin filaments driven by myosin motor activity.
To investigate the role of myosin activity in the motility of keratocytes, we treated the cells with blebbistatin, an inhibitor of myosin motor activity. Blebbistatin at 100 μM decreased the average cell velocity by approximately a factor of 2 (measured before the eventual fragmentation of the cells), and it induced, in the majority of the cells, the separation of the lamellipodia from the cell body (26 of 33 cells vs. 2 of 50 untreated cells), and the eventual splitting of the cell into multiple fragments (Figure 2A). Similar effects were described previously for the inhibitors of the activity of myosin light chain kinase (Verkhovsky et al., 1999b). Blebbistatin also induced desorganization of the actin–myosin II bundles at the lamellipodium–cell body boundary (Figure 2, B and C), consistent with the earlier hypothesis (Svitkina et al., 1997) that the alignment of actin and myosin filaments into the bundles was driven by myosin motor activity.
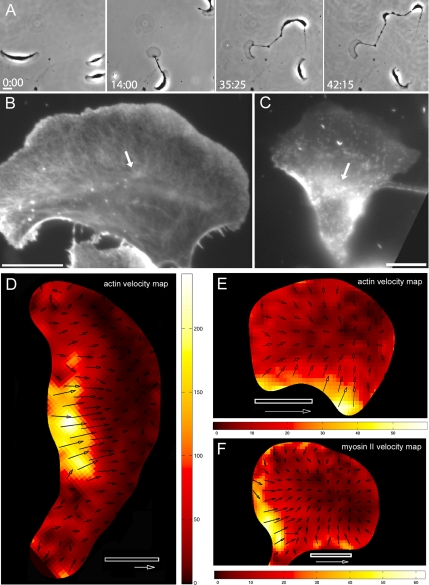
Figure 2.
Effect of blebbistatin on the shape, motility, cytoskeletal structure, and intracellular motion of actin and myosin II in fish epidermal keratocytes. (A) Time-lapse sequence illustrates the progressive fragmentation of the cell treated with 100 μM blebbistatin. (B) Distribution of actin and (C) distribution of microinjected rhodamine-myosin II in blebbistatin-treated keratocytes. Note the loose assemblies (arrows) of actin and myosin II at the lamellipodium–cell body transition zone (compare to distinct bundles in nontreated cells as shown in Figure 4, A and D). Velocity maps of actin in the intact cell treated with blebbistatin (D) and of actin (E) and myosin II (F) in the cell fragments generated as a result of blebbistatin treatment. Bars, 10 μm. Time is indicated in minutes:seconds. Arrows, 100 nm/s.
Motion tracking demonstrated that all blebbistatin-treated cells exhibited significantly altered patterns of motion of actin and myosin II. Even the cells that remained intact and moved relatively fast were clearly distinguished from the control cells by the absence of continuous zone of actin retrograde flow at the front, lack of clearly defined velocity transition zone, relatively small zone of high anterograde velocity, and overall apparently disorganized pattern of motion of actin and myosin II. Representative actin velocity map is shown on Figure 2D; myosin II maps displayed similarly disorganized pattern (data not shown). Cell fragments induced by blebbistatin treatment were characterized by very low intracellular velocities of actin and myosin II and overall centripetal direction of the motion of both proteins. Similar low values and identical orientation of the velocities of actin and myosin II suggested that the relative velocity of the two proteins in blebbistatin-treated cells was also very low. However, direct determination of the relative velocity in the same cell was impossible because of the phototoxicity of blebbistatin under illumination in fluorescein/green fluorescent protein channel (Kolega, 2004). For double-color fluorescent imaging in our setup, one of the probes had to be in the channel where blebbistatin was phototoxic. Illumination of the cells under these conditions resulted in the instantaneous arrest of motion, precluding the measurement of protein velocities.
Dynamics of Actin and Myosin II in the Retraction Fibers
As described above, we have detected the motion of myosin II with respect to actin in keratocyte lamellipodia. However, due to the high density of actin filaments and the variability of their orientation in the lamellipodium, it was impossible to identify individual actin tracks for myosin motion and to determine whether the myosin aggregates were following single actin tracks, multiple tracks or changing tracks. In contrast, retraction fibers at the rear of the cell contained bundles of parallel actin filaments, representing well-defined tracks for myosin motion. We analyzed the motion of actin and myosin II in the retraction fibers to gain insight into the possible patterns of mutual arrangement and relative motion of the two proteins. Retraction fibers were not captured by our tracking procedure because of their small area; consequently, we examined the motion within fibers visually. Phalloidin speckles arose at the tips of the fibers, and they moved toward the cell body (Figure 3A and Supplemental Video 5), suggesting actin assembly at the tips. Although retrograde motion of phase-dense nodules was observed previously in retraction fibers and it was attributed to motor-dependent transport along actin filaments (Cramer and Mitchison, 1997), our present observations indicate retrograde flow within the actin core of the fiber. Actin assembly at the tips of the fibers is consistent with the idea that retraction fibers are a form of filopodia (Svitkina et al., 2003). In contrast to actin, myosin II in the retraction fibers exhibited motion both toward the cell body and away from it (Figure 3, B and C). Similar patterns of motion were reported previously in filopodia for myosin X (Sousa and Cheney, 2005). Motion away from the cell body is consistent with myosin motor polarity and actin filament polarity in retraction fibers with barbed ends toward the tip of the fiber (Svitkina et al., 1997). Therefore, this motion likely represents active translocation of the motor protein along actin filaments. In contrast, motion toward the cell body may be due to the retraction of the entire fiber into the cell body with myosin II somehow entangled in the bundle and moving passively with actin.
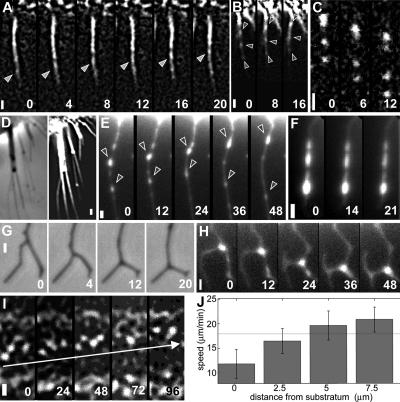
Figure 3.
Actin and myosin dynamics in retraction fibers and on apical surface of the cell body. Movement of actin toward (A) and of myosin II toward (B) and away (C) from the cell body in retraction fibers. (D) Retraction fibers retain actin filaments after cytochalasin D treatment: phase-contrast image on the left and phalloidin labeling on the right. (E–H) Dynamics of myosin II in retraction fibers after treatment with cytochalasin D. (E) Bidirectional movement of myosin II in a single fiber. (F) Lack of motion of myosin features in fibers of the cells treated with blebbistatin and cytochalasin D. Phase-contrast (G) and tetramethylrhodamine–myosin fluorescence microscopy (H) sequences of zipping of two branched fibers. (I) Forward motion of actin with respect to the cell on the apical surface of the cell body (7.5 μm from substrate). (J) Actin velocity in the cell body varies with the distance from the substratum. SD and mean cell speed (horizontal line) are indicated. Leading edge of the cell is up; all images are aligned with respect to the cell. Bars, 1 μm. Time is indicated in seconds.
Another possibility was that the motion away from the cell body is also due to the motion of the entire fiber rather than myosin translocation along it, e.g., passive stretching of the fiber between its substrate-attached tip and moving cell body. To eliminate the effect of the cell body motion, we blocked keratocyte migration with 0.7 μg/ml cytochalasin D. Cells ceased migration and retracted within seconds; actin and myosin in the cell body exhibited chaotic motion, and eventually they collapsed into aggregates (data not shown). However, retraction fibers in cytochalasin-treated cells contained residual actin filaments (as demonstrated by phalloidin staining of fixed cells; Figure 3D), and they remained attached to the substrate. Myosin spots exhibited fast motion along fibers in two directions: predominantly away from the cell body but also toward it (Figure 3E and Supplemental Video 6). Pretreatment for 1 h with 100 μM myosin inhibitor blebbistatin blocked this motion (Figure 3F), supporting the idea that myosin motion depended on its motor activity.
Interestingly, approximately two thirds of the retraction fibers in cytochalasin-treated cells were split into branches; the bifurcation points were not static, but they moved away from the cell, zipping the two branches together (Figure 3G and Supplemental Video 7). As the zipping point moved along the two branches, the unzipped branch portions changed their orientation so that the angle between the two branches increased. Myosin II-positive spots were found at the bifurcations (Figure 3H), suggesting that zipping was powered by myosin aggregates moving along and bringing together actin filaments of the two branches. Thus, analysis of the retraction fiber dynamics provided evidence for myosin motion along divergent actin tracks concomitant with track reorientation.
Motion of Actin at the Apical Part of the Cell Body
We have also tracked actin motion in the cell body at different distances from the substrate. Actin velocity increased with the distance from the substrate, and at the apical surface of the cell body (7.5 μm above the substrate for the cell shown in Figure 3I), actin displayed a velocity higher than the overall cell velocity. Together with the result that actin motion in the substrate plane was slower than the cell motion, this suggests the rotation of the cytoskeleton in the cell body. Cell body rotation was previously documented with membrane markers and internalized particles (Anderson et al., 1996). If the cytoskeleton in the cell body moved forward exclusively by rotation, the velocity in the substrate plane would be zero, and the velocity in the apical plane would be twice the cell velocity. In reality, the cell velocity was more than half of the velocity at the apical plane, indicating that rotation was relatively slow with respect to forward translocation (Figure 3J). Thus, transfer of actin polymer between optical sections due to rotation was insignificant compared with the movement within the substrate plane.
Maps of Assembly/Disassembly and Stretching/Compression of Actin and Myosin II Network
We computed the maps of stretching/compression and the net assembly and disassembly of actin and myosin II network essentially as described by Vallotton et al. (2004). Stretching/compression maps were defined as velocity divergence maps with positive divergence values reflecting stretching and negative values reflecting compression of the network (Figure 4, B and E). Negative divergence zone at the lamellipodium–cell body transition in the divergence maps of actin and myosin II velocity was a manifestation of contraction in this region of the cell (Figure 4, B and E). Assembly/disassembly maps were computed based on the mass conservation principle and on the velocity (Figure 1) and concentration maps (Figure 4, A and D) of the two proteins. For the simplicity, we have used velocity and concentration maps for the substrate optical plane, and we assumed that polymeric forms of actin and myosin II were confined in the substrate optical plane during the cell motion. This assumption was validated by the observations that fluorescence intensities of both actin and myosin II were significantly higher in the substrate plane than in the other optical planes and that the rotation of the cell body was slow compared with the cell motion, rendering the exchange of the polymer between the planes relatively insignificant compared with its motion and turnover within the substrate plane. Resulting maps demonstrated that the net assembly of actin was confined to the front of the lamellipodia, whereas myosin II assembled in a distributed manner with uniform low assembly level throughout the lamellipodium and some focused assembly spots in the cell body (Figure 4, C and F). Disassembly of actin was faster at the back of the lamellipodia than in the cell body, whereas fast disassembly of myosin II occurred in dense fibers at the lamellipodium–cell body transition and in the cell body. No significant assembly of actin was detected in fibers at the transition zone, indicating that these structures arise via network contraction rather than assembly.
DISCUSSION
In this study, we have analyzed the motion and assembly of the two principal components of the cell motile machinery, actin and myosin II, over the entire cell in the process of persistent migration. Resulting two-dimensional maps provide a comprehensive quantitative description of the protein dynamics, and they offer insight into the mode of actin–myosin interaction.
The pattern of actin motion and assembly in keratocytes displays several features that are found in other types of migrating cells and that may be fundamental to actin-dependent motility. In particular, our maps show intense assembly of actin network at the leading edge of the cell, retrograde flow of the network away from the leading edge, anterograde motion in the cell body domain, and a convergence of the retrograde flow from the front with anterograde flow from the back in the middle of the cell.
Actin assembly at the leading edge is a well-established feature of migrating cells, but recent work (Watanabe and Mitchison, 2002; Ponti et al., 2004; Gupton et al., 2005) has also suggested significant sites of actin assembly away from the leading edge. Our maps highlight the leading edge as a dominant site of the net actin assembly in keratocytes. Distribution of actin throughout the cell could be fully accounted for by assembly at the front, motion of the assembled network, and gradual disassembly, which was more intense at the back of the lamellipodium than in the cell body. Faster disassembly rate in the lamellipodium is consistent with the localization of ADF/cofilin in this zone (Svitkina and Borisy, 1999). Note that our method detected only net assembly and disassembly; thus, actin turnover (simultaneous assembly and disassembly) sites away from the leading edge may be unidentified. The assembly/disassembly pattern of myosin II was significantly different from that of actin: myosin II assembled in a distributed manner at the front of the cell and in a few spots within the cell body. The sites of myosin disassembly generally coincided with the sites of high myosin polymer concentration, i.e., in myosin-containing bundles at the lamellipodium–cell body transition zone and in the cell body. The distribution of the sites of myosin assembly and disassembly may be consistent with the hypothesis that myosin turnover is driven by the mass action mechanism (Verkhovsky et al., 1999b): contraction at the back of the lamellipodium and in the cell body domain may result in the increase in the local concentration of myosin polymer, thus favoring the reaction of disassembly, which would bring the polymer concentration back to the chemical equilibrium level. Contraction may also result in depletion of myosin polymer from adjacent areas stimulating the assembly reaction next to disassembly. Despite their contrasting features, the patterns of actin and myosin II assembly were both distinctly polarized along the cell motility direction, thereby contributing to the polarization of the contractile machinery of a migrating cell.
Retrograde actin flow is thought to be a consequence of the essential forces involved in the cell migration: polymerization force pushing against the membrane and contractile force in the actin–myosin network (Cramer, 1997; Verkhovsky et al., 1999a). Retrograde flow has long been recognized as a common feature of many types of migrating cells, but it was only recently detected in keratocytes with kymograph and particle tracking techniques (Jurado et al., 2005; Vallotton et al., 2005). Our tracking approach based on cross-correlation between small areas of the image produced similar results, further supporting the notion of the generality of the retrograde flow phenomenon.
Anterograde flow of actin structures and convergence zone of the retrograde and anterograde flow are also thought to be a characteristic feature of migrating cells (Salmon et al., 2002). However, these features are not inherent to all motile systems. Motile machinery recruited by intracellular pathogens such as Listeria works entirely through treadmilling (i.e., assembly at the front and disassembly at the back), and it does not involve any forward motion of the cytoskeletal polymers (Pantaloni et al., 2001). For keratocytes, forward motion of the cytoskeletal elements was reported in the cell body and at the lamellipodium–cell body boundary (Svitkina et al., 1997; Verkhovsky et al., 1999a; Vallotton et al., 2005). Here, we have for the first time generated complete polymer velocity maps for both the cell body and the lamellipodium. According to these maps, transition from retrograde to anterograde actin flow occurs at approximately half-width of the lamellipodium and the anterograde velocity subsequently increases sharply at the lamellipodium–cell body transition. A steep gradient of velocity in this zone results in the compression of the network as displayed in the stretching/compression map (Figure 4B). Transition of velocity approximately at half-width of the lamellipodium is consistent with the recently proposed polar gel model of the lamellipodium motion (Kruse et al., 2006); in contrast, this feature is not captured by another recent model, a two-dimensional multiscale model of a moving keratocyte (Rubinstein et al., 2005).
Thus, keratocyte actin dynamics could be summarized as follows: the extension of the leading edge of the lamellipodium is associated with the new assembly of actin–myosin network, whereas the movement of the back of the lamellipodium and the bulk of the cell body involves forward translocation of the network. Additionally, faster translocation of the actin structures at the top of the cell body than at the substrate level suggests that at the back of the cell, the actin network lifts up from the substrate and rotates. Rotation of the cell body was demonstrated previously with membrane markers (Anderson et al., 1996), and we now confirm it at the level of the cytoskeleton. Forward motion of the cell body is thus contributed by the two processes: forward translation of the cytoskeletal network and its rotation. This movement is expected to be driven by forces that are distinct from the forces of actin assembly driving front protrusion.
There are conflicting theories about how and where forces for the translocation of the cell body are generated. According to one model, forces result from contraction at the lateral wings of keratocyte (Anderson et al., 1996; Oliver et al., 1999), whereas another hypothesis, the dynamic network contraction model, holds that forces are developed along the entire lamellipodium–cell body transition zone (Svitkina et al., 1997; Verkhovsky et al., 1999a). The distribution of the gradients of velocity and the sites of compression in the cytoskeletal network may help to identify the sites of force production. The change of velocity from retrograde to anterograde and the compression along the lamellipodium–cell body transition zone are consistent with the force generation along this zone as stipulated in the dynamic network contraction model. However, compression of the network may also be driven by the forces developed elsewhere rather then by the internal contractile forces, e.g., a section of the lamellipodial–cell body transition zone may be compressed passively by the forces developed at other locations. The shape of the compression zone may help to distinguish whether the local compression is active or passive. If the center of the cell was driven passively by the forces transmitted from the wings, the central portion of the compression zone would be expected to lag behind the lateral segments. In reality, the compression zone had a convex forward shape, that is, the central part was leading, indicating that motion in the center was driven by local force rather than by force transmitted from the lateral wings.
Localization of myosin II at the lamellipodium–cell body boundary and the effects of the myosin inhibitor blebbistatin suggest that myosin II plays a role in the generation of the force compressing the cytoskeletal network and driving the cell body forward. Indeed, when the myosin motor activity was inhibited by blebbistatin, the actin–myosin bundles at the lamellipodium–cell body boundary became disorganized, the cell velocity decreased, and the cell body could not keep up with the motion of the lamellipodium, resulting in fragmentation of the cells. Disorganization of the velocity transition zone and of the overall pattern of actin and myosin motion in the presence of blebbistatin also suggested that the force production at the lamellipodium–cell body transition was compromised. However, cell body translocation was only partially inhibited by blebbistatin, suggesting that either myosin II activity was not completely blocked by this drug or that there are other redundant mechanisms contributing to the cell body translocation. One of the possibilities is that in the absence of myosin activity the force could be produced by the depolymerization-driven entropic network contraction (Mogilner and Oster, 1996). Regardless, fragmentation of the cell suggested that when myosin activity was attenuated, cell body could not follow front protrusion in a robust and effective way. The role of myosin II in maintaining cell shape and integrity is consistent with previous studies in mammalian cells showing that inhibition of myosin II resulted in formation of long processes and loss of coherent cell movement (Even-Ram et al., 2007). However, movement velocity of mammalian cells increased rather than decreased upon inhibition of myosin II activity (Even-Ram et al., 2007). This difference may reflect differences in substrate adhesion strength between keratocytes and mammalian cells. It was established in theoretical and experimental studies (DiMilla et al., 1991; Gupton and Waterman-Storer, 2006) that the overall speed of the cell is not directly proportional to the strength of myosin-mediated contraction but that it depends of the balance of contractility and adhesion strength and on the optimal organization of the motile machinery where contractility and adhesion are interdependent. Inhibition of contractility in strongly adherent mammalian cells may compromise the force-dependent maturation of focal adhesions, rendering the cells less adherent and more motile. In contrast, motile machinery of weakly adherent keratocytes is optimized for fast migration; thus, perturbation of contractility may compromise the efficiency of motion.
To gain insight on how the motor activity of myosin II contributes to the network compression and cell body translocation, here we have analyzed the relative motion of myosin II with respect to actin for the first time in a migrating cell. Our tracking protocol detected a forward motion of myosin II assemblies relative to actin network with a velocity significantly lower than the cell velocity. The resolution of the light microscopy did not permit visualizing clear events of myosin assemblies moving along distinct actin tracks, but there were several reasons to think that detected relative motion indeed represented active motion of myosin motor proteins along actin filaments. The direction of the motion toward the front of the cell was consistent with the orientation of actin filaments and known motor polarity of the myosin II. Conversely, other mechanisms of motion, such as convective motion or a motion driven by microtubule-dependent motors were unlikely considering the velocity distribution. Observed gradient of relative velocity with the maximum at the front of the lamellipodium was not consistent with convective motion, because the convection would be expected to be attenuated at the sites of the highest filamentous actin density, i.e., at the front of the lamellipodium. Likewise, the motion could not be attributed to the activity of microtubule-dependent motor proteins, because the front of keratocyte lamellipodium lacked microtubules. Finally, blebbistatin inhibited myosin motion along retraction fibers in the stationary cells, suggesting that this motion was dependent on myosin motor activity.
Does the pattern of the relative motion of myosin II with respect to actin provide any clues about the contraction mechanism? If forward motion of the cell originated from myosin-driven transport along uniformly polarized actin filaments (Cramer et al., 1997), one would expect actin to be stationary, and myosin velocity with respect to actin to be comparable with the cell velocity. This was not the case, suggesting that pure transport mechanism was unlikely to contribute to the cell body translocation. If the motion resulted from contraction of small units similar to muscle sarcomeres (Maciver, 1996; Cramer, 1999), one may expect bidirectional shearing of myosin with respect to actin on the scale of individual sarcomeric units, but no relative motion on a scale greater than a sarcomeric unit. We did not detect significant relative motion of actin and myosin II along most of the actomyosin bundle at the lamellipodium–cell body boundary. This may be consistent with sarcomeric units comparable or smaller than the regions used in our tracking protocol, and thereby avoiding detection. However, sarcomeric contraction is unlikely to operate in the direction of cell motion, because the relative velocity of myosin II with respect to actin was nonzero and directed uniformly forward.
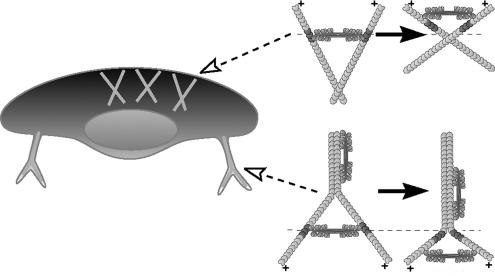
The mechanism that fits the best with the forward motion of both proteins with higher velocity of myosin II is the network contraction mechanism where myosin assemblies slide forward along divergent actin filaments (Svitkina et al., 1997; Verkhovsky et al., 1999a), resulting in filament bending and/or rotation, and compression of the entire network in the forward direction (Figure 5, top). The direction of filament rotation and compression of the network is dependent on the adhesion pattern and distribution of resistance in the network. The barbed ends of the actin filaments in the lamellipodium are immobilized in the dense actin network that is strongly attached to the substrate at the front of the cell, whereas the pointed filament ends at the back of the lamellipodium could be moved easier because of the sparser actin network and weaker substrate attachment in this region. Therefore, the sliding of myosin II along divergent actin filaments in the lamellipodium is expected to rotate filaments around their barbed ends and to realign them parallel to the leading edge. Observed decay of the forward velocity of myosin II with respect to actin at the cell rear may be explained by progressive reorientation of actin filaments parallel to the leading edge, which would eventually eliminate the forward component of myosin II velocity. The movement of individual myosin assemblies along divergent actin filaments could not be directly visualized in the lamellipodium because of the density of the actin network. However, the feasibility of this mechanism is supported by static electron microscopy images (Svitkina et al., 1997) where actin filaments of different orientations were observed to converge in the vicinity of myosin clusters at the lamellipodium–cell body boundary. In addition, our present light microscopy data are suggestive of a similar motion of myosin assemblies along divergent filaments in the retraction fibers of cytochalasin-treated cells. There, myosin aggregates were observed to zip together two branches of the fiber while moving to their distal ends (Figure 5, bottom). Zipping of the retraction fibers could not be considered completely analogous to network rearrangement at the lamellipodium–cell body transition, because the organization of the actin network in two cases was very different (branched network in the lamellipodium and bundled filaments in the retraction fibers), but the fundamental similarity between the two cases was that myosin II activity resulted in the deformation and realignment of actin filaments. The exact mode of deformation in the retraction fibers was presumably different from that in the lamellipodium, because of the different filament attachment pattern. In retraction fibers, both barbed and pointed filament ends were immobilized through attachment to the substrate and the cell body, respectively. Myosin motion therefore resulted in zipping together the proximal portions of filaments and alignment of their distal portions in the perpendicular direction.
Figure 5.
Motion of myosin II along divergent actin filaments. In the lamellipodium (top), this motion results in reorientation of actin filaments and overall compression of actin–myosin II network in the forward direction. In retraction fibers (bottom), a similar process zips two branches of the fiber, concomitantly widening the angle between unzipped fiber portions. Fixed positions along actin filaments are marked to illustrate actin motion.
Thus, the patterns of actin and myosin II motion help to distinguish between different descriptive schemas of the cell translocation mechanism, but the next challenge is to go beyond schematic cartoons. Keratocytes are arguably the most persistent and regular of all migrating cells, and they are likely to be the favorite target of the forthcoming comprehensive biophysical models of cell migration. Our two-dimensional maps of the velocity and assembly of the major components of the motile machinery may serve as a reference data set to test and refine such quantitative models. Future research may combine the tracking of the intracellular motion, force-tracking at the substrate level, and the data on intracellular elasticity to derive the map of internal cytoskeletal forces and correlate it to the distribution and activities of specific molecules.
Supplementary Material
ACKNOWLEDGMENTS
The work was supported by Swiss Science Foundation grant 31-61589 (to A.B.V.). V.M.L. was supported by grant BDF 01. 000006495 “Bourse de formation à l'étranger 2001” from the Institut National de la Santé et de la Recherche Médicale.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-09-0859) on July 18, 2007.
 The online version of this article contains supplemental material MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material MBC Online (http://www.molbiolcell.org).
REFERENCES
- Anderson K. I., Wang Y. L., Small J. V. Coordination of protrusion and translocation of the keratocyte involves rolling of the cell body. J. Cell Biol. 1996;134:1209–1218. doi: 10.1083/jcb.134.5.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer L. P. Molecular mechanism of actin-dependent retograde flow in lamellipodia of motile cells. Front. Biosci. 1997;2:260–270. doi: 10.2741/a189. [DOI] [PubMed] [Google Scholar]
- Cramer L. P. Organization and polarity of actin filament networks in cells: implications for the mechanism of myosin-based cell motility. Biochem. Soc. Symp. 1999;65:173–205. [PubMed] [Google Scholar]
- Cramer L. P., Mitchison T. J. Investigation of the mechanism of retraction of the cell margin and rearward flow of nodules during mitotic cell rounding. Mol. Biol. Cell. 1997;8:109–119. doi: 10.1091/mbc.8.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer L. P., Siebert M., Mitchison T. J. Identification of novel graded polarity actin filament bundles in locomoting heart fibroblasts: implications for the generation of motile force. J. Cell Biol. 1997;136:1287–1305. doi: 10.1083/jcb.136.6.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMilla P. A., Barbee K., Lauffenburger D. A. Mathematical model for the effects of adhesion and mechanics on cell migration speed. Biophys. J. 1991;60:15–37. doi: 10.1016/S0006-3495(91)82027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Even-Ram S., Doyle A. D., Conti M. A., Matsumoto K., Adelstein R. S., Yamada K. M. Myosin IIA regulates cell motility and actomyosin–microtubule crosstalk. Nat. Cell Biol. 2007;9:299–309. doi: 10.1038/ncb1540. [DOI] [PubMed] [Google Scholar]
- Grimm H. P., Verkhovsky A. B., Mogilner A., Meister J. J. Analysis of actin dynamics at the leading edge of crawling cells: implications for the shape of keratocyte lamellipodia. Eur. Biophys. J. 2003;32:563–577. doi: 10.1007/s00249-003-0300-4. [DOI] [PubMed] [Google Scholar]
- Gupton S. L., et al. Cell migration without a lamellipodium: translation of actin dynamics into cell movement mediated by tropomyosin. J. Cell Biol. 2005;168:619–631. doi: 10.1083/jcb.200406063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupton S. L., Waterman-Storer C. M. Spatiotemporal feedback between actomyosin and focal-adhesion system optimizes rapid cell migration. Cell. 2006;125:1361–1374. doi: 10.1016/j.cell.2006.05.029. [DOI] [PubMed] [Google Scholar]
- Hebert B., Costantino S., Wiseman P. W. Spatiotemporal image correlation spectroscopy (STICS) theory, verification, and application to protein velocity mapping in living CHO cells. Biophys. J. 2005;88:3601–3614. doi: 10.1529/biophysj.104.054874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurado C., Haserick J. R., Lee J. Slipping or gripping? Fluorescent speckle microscopy in fish keratocytes reveals two different mechanisms for generating a retrograde flow of actin. Mol. Biol. Cell. 2005;16:507–518. doi: 10.1091/mbc.E04-10-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolega J. Phototoxicity and photoinactivation of blebbistatin in UV and visible light. Biochem. Biophys. Res. Commun. 2004;320:1020–1025. doi: 10.1016/j.bbrc.2004.06.045. [DOI] [PubMed] [Google Scholar]
- Kruse K., Joanny J. F., Julicher F., Prost J., Sekimoto K. Pi-by-no theory of active polar gels: a paradigm for cytoskeletal dynamics. Eur. Phys. J. E. Soft Matter. 2005;16:5–16. doi: 10.1140/epje/e2005-00002-5. [DOI] [PubMed] [Google Scholar]
- Kruse K., Joanny J. F., Julicher F., Prost J. Contractility and retrograde flow in lamellipodium motion. Phys. Biol. 2006;3:130–137. doi: 10.1088/1478-3975/3/2/005. [DOI] [PubMed] [Google Scholar]
- Kruse K., Julicher F. F. Self-organization and mechanical properties of active filament bundles. Phys. Rev. E. Stat. Nonlin. Soft Matter Phys. 2003;67 doi: 10.1103/PhysRevE.67.051913. 051913. [DOI] [PubMed] [Google Scholar]
- Lee J., Ishihara A., Jacobson K. The fish epidermal keratocyte as a model system for the study of cell locomotion. Symp. Soc. Exp. Biol. 1993;47:73–89. [PubMed] [Google Scholar]
- Maciver S. K. Myosin II function in non-muscle cells. Bioessays. 1996;18:179–182. doi: 10.1002/bies.950180304. [DOI] [PubMed] [Google Scholar]
- Mitchison T. J., Cramer L. P. Actin-based cell motility and cell locomotion. Cell. 1996;84:371–379. doi: 10.1016/s0092-8674(00)81281-7. [DOI] [PubMed] [Google Scholar]
- Mogilner A. On the edge: modelling protrusion. Curr. Opin. Cell Biol. 2006;18:32–39. doi: 10.1016/j.ceb.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Mogilner A., Oster G. Cell motility driven by actin polymerisation. Biophys. J. 1996;71:3030–3045. doi: 10.1016/S0006-3495(96)79496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver T., Dembo M., Jacobson K. Separation of propulsive and adhesive traction stresses in locomoting keratocytes. J. Cell Biol. 1999;145:589–604. doi: 10.1083/jcb.145.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantaloni D., Le Clainche C., Carlier M. F. Mechanism of actin-based motility. Science. 2001;292:1502–1506. doi: 10.1126/science.1059975. [DOI] [PubMed] [Google Scholar]
- Parent C. A. Making all the right moves: chemotaxis in neutrophils and Dictyostelium. Curr. Opin. Cell Biol. 2004;16:4–13. doi: 10.1016/j.ceb.2003.11.008. [DOI] [PubMed] [Google Scholar]
- Pollard T. D., Borisy G. G. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- Ponti A., Machacek M., Gupton S. L., Waterman-Storer C. M., Danuser G. Two distinct actin networks drive the protrusion of migrating cells. Science. 2004;305:1782–1786. doi: 10.1126/science.1100533. [DOI] [PubMed] [Google Scholar]
- Ridley A. J., Schwartz M. A., Burridge K., Firtel R. A., Ginsberg M. H., Borisy G., Parsons J. T., Horwitz A. R. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- Rubinstein B., Jacobson K., Mogilner A. Multiscale two-dimensional modeling of a motile simple-shaped cell. Multiscale Model. Simul. 2005;3:413–439. doi: 10.1137/04060370X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon W. C., Adams M. C., Waterman-Storer C. M. Dual-wavelength fluorescent speckle microscopy reveals coupling of microtubule and actin movements in migrating cells. J. Cell Biol. 2002;158:31–37. doi: 10.1083/jcb.200203022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina T. M., Borisy G. G. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J. Cell Biol. 1999;145:1009–1026. doi: 10.1083/jcb.145.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallotton P., Danuser G., Bohnet S., Meister J. J., Verkhovsky A. B. Tracking retrograde flow in keratocytes: news from the front. Mol. Biol. Cell. 2005;16:1223–1231. doi: 10.1091/mbc.E04-07-0615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallotton P., Gupton S. L., Waterman-Storer C. M., Danuser G. Simultaneous mapping of filamentous actin flow and turnover in migrating cells by quantitative fluorescent speckle microscopy. Proc. Natl. Acad. Sci. USA. 2004;101:9660–9665. doi: 10.1073/pnas.0300552101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkhovsky A. B., Chaga O. Y., Schaub S., Svitkina T. M., Meister J. J., Borisy G. G. Orientational order of the lamellipodial actin network as demonstrated in living motile cells. Mol. Biol. Cell. 2003;14:4667–4675. doi: 10.1091/mbc.E02-10-0630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkhovsky A. B., Svitkina T. M., Borisy G. G. Network contraction model for cell translocation and retrograde flow. Biochem. Soc. Symp. 1999a;65:207–222. [PubMed] [Google Scholar]
- Verkhovsky A. B., Svitkina T. M., Borisy G. G. Self-polarization and directional motility of cytoplasm. Curr. Biol. 1999b;9:11–20. doi: 10.1016/s0960-9822(99)80042-6. [DOI] [PubMed] [Google Scholar]
- Sousa A. D., Cheney R. E. Myosin-X: a molecular motor at the cell's fingertips. Trends Cell Biol. 2005;15:533–539. doi: 10.1016/j.tcb.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Svitkina T. M., Bulanova E. A., Chaga O. Y., Vignjevic D. M., Kojima S., Vasiliev J. M., Borisy G. G. Mechanism of filopodia initiation by reorganization of a dendritic network. J. Cell Biol. 2003;160:409–421. doi: 10.1083/jcb.200210174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina T. M., Verkhovsky A. B., McQuade K. M., Borisy G. G. Analysis of the actin-myosin II system in fish epidermal keratocytes: mechanism of cell body translocation. J. Cell Biol. 1997;139:397–415. doi: 10.1083/jcb.139.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N., Mitchison T. J. Single-molecule speckle analysis of actin filament turnover in lamellipodia. Science. 2002;295:1083–1086. doi: 10.1126/science.1067470. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.