Abstract
The mitotic spindle consists of a complex network of proteins that segregates chromosomes in eukaryotes. To strengthen our understanding of the molecular composition, organization, and regulation of the mitotic spindle, we performed a system-wide two-hybrid screen on 94 proteins implicated in spindle function in Saccharomyces cerevisiae. We report 604 predominantly novel interactions that were detected in multiple screens, involving 303 distinct prey proteins. We uncovered a pattern of extensive interactions between spindle proteins reflecting the intricate organization of the spindle. Furthermore, we observed novel connections between kinetochore complexes and chromatin-modifying proteins and used phosphorylation site mutants of NDC80/TID3 to gain insights into possible phospho-regulation mechanisms. We also present analyses of She1p, a novel spindle protein that interacts with the Dam1 kinetochore/spindle complex. The wealth of protein interactions presented here highlights the extent to which mitotic spindle protein functions and regulation are integrated with each other and with other cellular activities.
INTRODUCTION
The faithful inheritance of chromosomes is essential for the propagation of organisms. Central to this process in eukaryotes is the mitotic spindle, an elaborate array of microtubules and associated proteins that positions and segregates chromosomes during cell division. The fundamental nature of this dynamic structure is reflected by the significant number of components that are shared by humans and many simpler organisms including Saccharomyces cerevisiae. The proteins involved in spindle function not only encompass tubulin, motor proteins, and other microtubule-associated proteins, but also the microtubule-organizing centers, kinetochore complexes, chromatin-associated proteins, regulatory kinases and phosphatases, and the anaphase-promoting complex. The dependence of cell division on the mitotic spindle makes its disruption both a cause of diseases and a target for anticancer treatments.
Although our understanding of the mitotic spindle has increased significantly in recent years, our knowledge of the mechanisms that intricately choreograph chromosome segregation remains incomplete. Different models, each consistent with available observations, have been proposed to explain spindle dynamics, chromosome capture by microtubules, force generation on chromosomes, and checkpoint function (Mogilner et al., 2006). Achieving a complete understanding of mitosis at the molecular level would be aided by an in-depth interaction network map of the proteins involved. Such a map would facilitate elucidation of the functions and organization of spindle proteins and of their roles within the greater context of the cellular environment.
Systems approaches such as systematic two-hybrid screens are necessary to reveal the myriad patterns of protein interactions that underlie complex processes. In yeast, genome-wide approaches including systematic tandem affinity purification (Gavin et al., 2006; Krogan et al., 2006), synthetic genetic arrays (Tong et al., 2004), and two-hybrid screens (Uetz et al., 2000; Ito et al., 2001) have provided a wealth of data from which models of functional interactions and pathways can be generated. However, the scale of genome-wide assays necessitates a high level of stringency and uniform experimental conditions to maximize their efficiency. An advantage of a study that focuses on an individual cellular process is that a denser interaction map can be created because of the additional experimental flexibility and customizability. By intensively probing the mitotic machinery in our two-hybrid study, we generated a high-resolution map of protein interactions within the mitotic spindle and with proteins not conventionally considered to be part of the spindle.
MATERIALS AND METHODS
Strains and Growth Conditions
The plasmids and yeast strains used in this study are listed in Supplementary Material 5. Yeast strains were grown in either YP (yeast extract/peptone) or minimal medium supplemented with 2% glucose and appropriate nutrients. Geneticin (G418; GIBCO BRL, Rockville, MD) was used at a concentration of 0.4 mg/ml.
For C-terminal tagging with three tandem green fluorescent proteins (GFPs), the SHE1 open reading frame (ORF) was subcloned into the BamHI site of pYS47 (Sun et al., 2007), using primers oJW131 (CGCGGATCCCAAGATCTAAAGTACACAGATCG) and oJW132 (CGCGGATCCCCGCCAAATAGGTCTATCACT), to generate pJW15. The orientation of the ORF was confirmed by digesting pJW15 with AflIII and AgeI. pJW15 was linearized with AatII and transformed into a wild-type yeast strain. Transformants were selected on minimal medium plates lacking histidine. The diploids were sporulated to isolate haploids expressing She1-3GFP.
The C-terminal tagging of genes with monomeric RFP (mRFP, a generous gift from Roger Tsien, University of California, San Diego, La Jolla, CA) was performed as described (Longtine et al., 1998).
Two-Hybrid Screen
Genome-wide two-hybrid screens were performed as described by the Yeast Resource Center (http://depts.washington.edu/∼yeastrc/). Briefly, each prospective bait gene was amplified from the genomic DNA of DDY1102 by PCR, and a unique restriction site was added at each terminus of the amplified fragment. The genes were then cloned into the vector pOBD2 or pBDC for fusion of the Gal4p-DBD to the N- or C-terminus, respectively (Uetz et al., 2000; Millson et al., 2003). After verification of the cloning by restriction digest, the plasmids were transformed into PJ69-4a for mating with ∼6000 strains hosting the Gal4p-AD–fused genome-wide array and subsequent screening (Hazbun et al., 2003). Each bait was then rescreened against an array of 732 preys that exhibited interactions in the initial screen. Both screens were performed in duplicate. Verification of the Gal4p-AD–fused strains was performed by sequencing 20 strains, indicating that the array strain identities were correctly positioned. Graphical representations of protein interaction networks were created with Cytoscape software (http://www.cytoscape.org) unless otherwise noted. Comparison of the spindle two-hybrid data set with the database of physical interactions hosted by the Saccharomyces Genome Database was also performed with Cytoscape software.
Microscopy
Indirect immunofluorescence microscopy on intact yeast cells was performed as described (Ayscough and Drubin, 1998). The rabbit anti-GFP (Torrey Pines Biolabs, San Diego, CA) and YOL1/34 anti-α-tubulin antibody (Accurate Chemical and Scientific, Westbury, NY) were used at dilutions of 1:2000 and 1:500, respectively. Fluorescein- or rhodamine-conjugated anti-IgG heavy-chain secondary antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA) were used at 1:100 dilution. Fluorescein- or rhodamine-conjugated goat anti-rat secondary antibodies (ICN Biomedicals/Cappel, Cosa Mesa, CA) were used at 1:500 dilution. Images were obtained on a Nikon TE300 microscope (Melville, NY) equipped with an 100×/NA 1.4 objective and an Orca-100 camera (Hamamatsu, Bridgewater, NJ) controlled by Image ProPlus software (Phase-3 Imaging Systems, Milford, MA).
Live cell imaging was performed on log-phase cells grown at 25°C. Cells were adhered to concanavalin A-coated (Sigma, St. Louis, MO) coverslips and sealed into 50 μl of minimal medium with vacuum grease (Dow Corning, Midland, MI) on a glass slide. Fluorescent images were obtained on an Olympus IX81/71 microscope (Melville, NY) using an 100×/NA 1.4 objective and a Orca-ER camera (Hamamatsu) controlled by Metamorph software (Universal Imaging, West Chester, PA). Image processing was performed with Image J software (http://rsb.info.nih.gov/ij/).
RESULTS
Two-Hybrid Overview
We conducted a yeast two-hybrid screen of proteins implicated in mitotic and/or meiotic spindle function in S. cerevisiae. For this screen, we selected 113 proteins that are either components of the spindle, regulate its activity, or are directly required for its wild-type function during mitosis and/or meiosis (Table 1). In addition to proteins that comprise the spindle structurally, we included proteins in mitotic regulatory pathways such as the Cdc14-early-anaphase-release (FEAR) and spindle checkpoint pathways to gain insight into mitotic protein regulation. The genes encoding these proteins were cloned into “bait” vectors containing the DNA-binding domain (DBD) of the Gal4p transcription factor. Of the 113 genes cloned as baits, 19 fusion constructs were either strongly self-activating or lethal when transformed into S. cerevisiae and were not used further (Table 1). The remaining clones were screened in duplicate against an array of ∼6000 “prey” yeast strains expressing individual ORFs fused to the Gal4-activation domain (AD). Pair-wise interactions were scored as multiple hits if they were detected in duplicate or as single hits if they were detected in only one of the two trials. To further saturate our data set and to retest the single hit interactions, we rescreened the spindle baits in duplicate against a mini-array of 732 preys encompassing the majority of prey interactants from the initial screen. The data set was filtered for dubious ORFs, transposon and viral genes, and common false positives including drug-resistance genes and positive transcriptional regulators using annotations from the Saccharomyces Genome Database (http://www.yeastgenome.org). From the remaining data, 857 interactions that occurred exclusively between nuclear and nonnuclear proteins as annotated by the Gene Ontology project (GO; http://www.geneontology.org/) were not analyzed further (Supplementary Material 1). The possibility exists that some of these represent bona fide in vivo interactions by proteins that shuttle between the nucleus and cytoplasm or whose localizations have not been thoroughly characterized.
Table 1.
List of bait proteins used in this study
| DNA architecture |
| Chromatin assembly factor I: Rlf2 (Cac1), Cac2, Msi1(Cac3) |
| Cohesin: Irr1 (Scc3), Med1 (Scc1), Rec8, Smc1, Smc3 |
| Cse4 |
| Scm3 |
| Sgo1 |
| Sir1 |
| Kinetochore |
| CBF3 complex: Cbf2 (Ndc10), Cep3, Ctf13, Skp1 |
| MTW1 complex: Dsn1, Mtw1, Nnf1, Nsl1 |
| SPC105 complex: Spc105, Ydr532c |
| CTF19 complex: Ame1, Chl4, Ctf3, Ctf19, Iml3 (Mcm19), Mcm16, Mcm21, Mcm22, Nkp1, Nkp2, Okp1 |
| NDC80 complex: Nuf2, Spc24, Spc25, Tid3 (Ndc80) |
| DAM1 complex: Ask1, Dad1, Dad2, Dad3, Dad4, Dam1, Duo1, Hsk3, Spc19, Spc34 |
| Monopolin complex: Mam1, Lrs4, Csm1 |
| Cbf1 |
| Cnn1 |
| Mif2 |
| Regulators |
| Anaphase-promoting complex: Ama1, Apc1, Apc2, Apc4, Apc5, Apc9, Apc11, Cdc16, Cdc20, Cdc23, Cdc26, Cdc27, Cdh1, Doc1, Mnd2, Swm1 |
| Ipl1 complex: Bir1, Ipl1, Sli15 |
| Protein phosphatase type 1: Glc7, Glc8 |
| Spindle assembly checkpoint: Mad1, Mad2, Mad3, Bub1, Bub3, Mps1 |
| Cdc28 |
| Esp1 |
| Kin3 |
| Pds1 |
| Cdc14 early anaphase release |
| FEAR: Cdc5, Cdc14, Net1, Slk19, Spo12 |
| Microtubule-associated proteins |
| Kinesins and associated proteins: Cin8, Kar3, Kip1, Kip2, Kip3, Vik1 |
| MAPs: Ase1, Bik1, Bim1, Mhp1, Stu1, Stu2 |
| Tubulin: Tub1, Tub2, Tub3, Tub4 |
| Spindle pole body |
| SPB: Cdc31, Cmd1, Mps2, Mps3, Spc42, Spc97, Spc98, Spc110 |
The common names of proteins are listed in parentheses after the standard names where necessary. Nonfunctional bait constructs are shown in italics.
Protein interactions, n = 1526, covering 730 distinct prey proteins were tallied after filtering. Of these pair-wise combinations, 604 (39.6%) were detected by multiple screens. These multiple-hit interactions comprise the core data set of this study (Supplementary Materials 2 and 3). Three hundred three (41.5%) of the prey proteins interacted with multiple bait proteins. Although they may represent weak or indirect, but otherwise meaningful interactions, the single hit data were segregated from the main data set and were not analyzed further in this investigation (Supplementary Material 1). The number of multiple hit interactions per bait construct ranged from 1 to 52, with a mean of 7.4.
Intersection with Published Databases
To evaluate the novelty of the data obtained in this study, we compared our 604 multiple-hit interactions with those in a database of physical protein interactions available in the Saccharomyces Genome Database. A direct comparison with previously published yeast two-hybrid data including two comprehensive genomic studies (Uetz et al., 2000; Ito et al., 2001) revealed only 58 interactions in common (Supplementary Material 4). Thus, >90% of the interactions detected were novel. The greater number of interactions for the mitotic proteins used in this study, compared with those reported in previous studies, could be partially attributed to differences in experimental design. In the genome-wide studies, multiple baits were pooled and tested against a complementary pool of preys, whereas each bait in this study was tested individually against every protein in the prey library. We also compared our results with interactions reported in the database of physical interactions detected by tandem affinity capture and mass spectrometry (MS; Rigaut et al., 1999; Puig et al., 2001). Although the methodologies of protein–protein interaction detection are different and the affinity capture technique is predicted to preferentially detect stable complexes (Gavin et al., 2002), the 65 protein–protein interactions shared by our study and the affinity capture-MS datasets was slightly higher than the overlap with the two-hybrid database. The higher level of intersection of our data set with data derived from an independent and orthogonal method argues for the validity of our data set. Importantly, the vast majority of interactions presented here represent novel findings.
Evaluation of Dam1 Complex Interactions
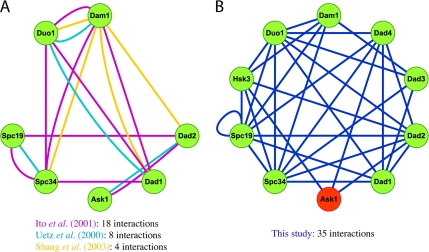
To further evaluate the comprehensiveness of this study, we closely analyzed the protein network generated for subunits of the well-defined Dam1 kinetochore complex. Purified from yeast, the Dam1 complex consists of 10 essential proteins (Dam1p, Duo1p, Dad1p, Spc19p, Spc34p, Ask1p, Dad2p, Dad3p, Dad4p, and Hsk3p) that localize to the kinetochore and spindle microtubules (Cheeseman et al., 2001a,b; Li et al., 2002). Each subunit purifies stoichiometrically in the complex, and multiple Dam1 complexes can oligomerize into stable ring structures that may be required to form stable attachments between chromosomes and spindle microtubules (Miranda et al., 2005; Westermann et al., 2005). Additional biochemical and yeast two-hybrid studies had also previously identified many of the subunits of the complex (Figure 1A; Uetz et al., 2000; Ito et al., 2001; Ikeuchi et al., 2003; Shang et al., 2003). Of the two comprehensive yeast two-hybrid projects, one study (Ito et al., 2001) reported 18 interactions between seven subunits, compared with the eight interactions between the same seven subunits identified by the other study (Uetz et al., 2000). The Spc19p–Spc34p reciprocal interactions reported in the latter study were isolated from those of the other subunits, such that they could not be inferred to be part of the larger Dam1 complex. Some proteins, such as Ask1p, had only a single interaction with another member of the complex. Whether Ask1p was an integral component of the Dam1 complex or a protein with a separate function was obscured.
Figure 1.
Comparison of intra-Dam1 complex interactions detected from genome-wide and focused two-hybrid screens. Protein interaction networks of subunits within the Dam1 complex derived from yeast two-hybrid studies and an in vitro expression experiment were generated with Cytoscape network visualization software. The bait construct for Ask1p (shown in red) was lethal to yeast and could not be screened. (A) The interactions reported from previous comprehensive two-hybrid screens identified seven of the 10 Dam1 complex subunits. (B) The network of interactions found by this study identified all 10 subunits of the Dam1 complex. The very high number of interactions detected between subunits is consistent with their association as a protein complex.
In contrast, our study identified 35 interactions between all 10 subunits of the Dam1 complex (Figure 1B). The pattern of interconnectivity was apparent even though several two-hybrid constructs were missing from the analysis. The Ask1-bait construct was nonfunctional. Also, because interactions of the Dam1-bait construct were published previously, this bait was excluded from our study (Shang et al., 2003). Finally, Hsk3p, Dad3p, and Dad4p were only screened as baits because the construction of the prey library predated their identification as ORFs. Despite these limitations, every subunit had at least three two-hybrid interactions with other Dam1 complex components. The extensiveness of the interaction network connecting these 10 proteins strongly supports the conclusion that they form a stable structure in vivo.
Interactions among Subunits of Defined Protein Complexes
As with the Dam1 complex, multiple interactions were detected among subunits of other biochemically well-defined kinetochore complexes. Fifteen protein–protein interactions were detected within the 11-subunit Ctf19 complex. There was no enrichment of connections among proteins of the proposed COMA (Ctf19p-Okp1p-Mcm21p-Ame1p) subcomplex (De Wulf et al., 2003), although it should be noted that the Ame1p and Mcm21p bait constructs were nonfunctional. Of the six interactions within the Ctf19 complex involving COMA subunits, three were with non-COMA subunits. Within the four-member Mtw1 and Ndc80 kinetochore complexes, there were four and three internal interactions, respectively. The ratio of internal two-hybrid interactions to the number of subunits for these kinetochore complexes was significantly lower than was obtained for the Dam1 complex. This is partially attributed to complications with nonfunctional Dsn1p and Nsl1p bait constructs and self-activation of the Ndc80p-AD fusion protein. In addition, the elaborate network of interactions within the Dam1 complex might include indirect linkages. Nevertheless, the two-hybrid data appeared consistent with available biochemical data regarding the composition of previously annotated kinetochore complexes and provides further biological validation for the specificity of our screen.
Patterns of Interactions between Kinetochore and Chromatin-associated Proteins
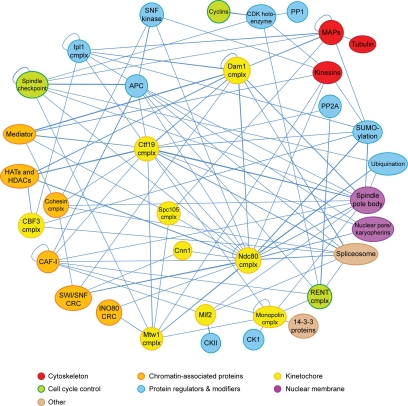
Considered in its entirety, the wealth of yeast two-hybrid data forms a dauntingly complex network (Supplementary Material 2). Whether this reflects the occurrence of indirect interactions or the reality of a multitude of protein–protein interactions possible under a variety of environmental and temporal conditions within a dividing cell is unclear. To simplify the overall interaction network, an interaction map was generated in which well-characterized processes and multisubunit complexes were represented as single nodes (Figure 2). Multiple hits between a protein and different subunits within a particular complex increase confidence that the interactions are relevant biologically. The simplified interaction network reveals high connectivity among proteins implicated in spindle function, especially at the level of chromosome attachment to the spindle.
Figure 2.
A simplified spindle protein interaction network. This simplified network includes proteins with demonstrated spindle or chromosome functions as well as uncharacterized proteins that interact with multiple spindle proteins. Proteins that belong to the same complex or functional process are grouped into single nodes. cmplx, complex; APC, anaphase-promoting complex; CK1, casein kinase I; CKII, casein kinase II; CAF-I, chromatin assembly factor I; CRC, chromatin remodeling complex; CDK, cyclin-dependent kinase; HATs, histone acetyltransferases; HDACs, histone deacetylases; MAPs, microtubule-associated proteins; PP1, protein phosphatase I; PP2A, protein phosphatase 2A.
A high level of connectivity exists between kinetochore components and other chromatin-associated proteins (Table 2). One example of this is Mif2p, a homolog of the mammalian CENP-C inner kinetochore protein (Meluh and Koshland, 1995, 1997). It copurifies with the Mtw1 kinetochore complex and histone proteins including the centromeric histone H3 variant, Cse4p/CENP-A (Westermann et al., 2003). Our data registered single-hit interactions of Mif2p with Hta2p and Htb1p, possibly indicating an indirect or transient connection between this inner kinetochore protein and these nucleosome subunits. Mif2p also interacted with Rlf2p, a subunit of chromatin assembly factor I (CAF-I), whose redundant function with the histone regulatory genes (HIR) pathway is required in the deposition of Cse4p at centromeres (Sharp et al., 2002). Interactions of Mif2p with the Mtw1 complex and the CBF3 inner kinetochore complex (Ndc10p, Ctf13p, Cep3p, and Skp1p), with which it shows synthetic phenotypes, were not detected (Meluh and Koshland, 1995). The Gal4p fusion domains might interfere with Mif2p's binding site for other kinetochore proteins. Finally, Mif2p specifically associated with two subunits of the ubiquitous and highly conserved casein kinase 2, Cka2p and Ckb2p. Intriguingly, Mif2p is a phosphoprotein in vivo whose phosphorylation is essential for its function (Westermann et al., 2003). Altogether, these data reinforce a model in which a Mif2p-Cse4p–containing nucleosome interaction bridges the chromosome and peripheral kinetochore elements.
Table 2.
Notable interactions between spindle and chromatin-associated proteins
| Prey name | Bait name | Bait description |
|---|---|---|
| SWI/SNF Chromatin remodeling complex interactions | ||
| SNF6 | SPC25 | Ndc80 kinetochore complex |
| SNF6 | ESP1 | Separase |
| SWI1 | SMC1 | Cohesin complex |
| SWI1 | DAD4 | Dam1 kinetochore complex |
| SWI1 | CNN1 | Kinetochore |
| SWI1 | SPC24 | Ndc80 kinetochore complex |
| SWI1 | SPC25 | Ndc80 kinetochore complex |
| SWI1 | TID3 | Ndc80 kinetochore complex |
| SWI3 | NUF2 | Ndc80 kinetochore complex |
| Ino80 chromatin remodeling complex interactions | ||
| IES3 | MTW1 | Mtw1 kinetochore complex |
| NHP10 | NNF1 | Mtw1 kinetochore complex |
| NHP10 | NUF2 | Ndc80 kinetochore complex |
| SAGA histone acetylation interactions | ||
| ADA2 | SPC25 | Ndc80 kinetochore complex |
| AHC2 | NKP2 | Ctf19 kinetochore complex |
| AHC2 | DAD1 | Dam1 kinetochore complex |
| AHC2 | HSK3 | Dam1 kinetochore complex |
| SGF73 | CEP3 | CBF3 complex |
| SGF73 | SPC25 | Ndc80 kinetochore complex |
| TAF9 | APC1 | Anaphase-promoting complex |
| RPD3 deacetylase interactions | ||
| DEP1 | SPC25 | Ndc80 kinetochore complex |
| PHO23 | NKP2 | Ctf19 kinetochore complex |
| PHO23 | DAD4 | Dam1 kinetochore complex |
| PHO23 | SPC25 | Ndc80 kinetochore complex |
| RXT3 | CEP3 | CBF3 complex |
| SDS3 | SPC25 | Ndc80 kinetochore complex |
| Interactions with other deacetylases | ||
| CPR1 | MCM22 | Ctf19 kinetochore complex |
| HDA2 | HSK3 | Dam1 kinetochore complex |
| HDA2 | NUF2 | Ndc80 kinetochore complex |
| HDA2 | SPC24 | Ndc80 kinetochore complex |
| Mediator complex interactions | ||
| CSE2 | CIN8 | Kinesin |
| MED11 | DAD2 | Dam1 kinetochore complex |
| MED4 | CIN8 | Kinesin |
| MED6 | SPC25 | Ndc80 kinetochore complex |
| MED7 | SPC105 | Kinetochore |
| MED8 | HSK3 | Dam1 kinetochore complex |
| PGD1 | NKP2 | Ctf19 kinetochore complex |
| PGD1 | DAD1 | Dam1 kinetochore complex |
| PGD1 | DAD2 | Dam1 kinetochore complex |
| PGD1 | HSK3 | Dam1 kinetochore complex |
| SRB7 | MCD1 | Cohesin complex |
| SRB7 | NKP2 | Ctf19 kinetochore complex |
| SRB7 | DAD4 | Dam1 kinetochore complex |
Strikingly, a significant number of DNA-associated proteins exhibited multiple interactions with kinetochore components. One highly represented class of interactors was chromatin-remodeling factors. These ATP-dependent complexes generally serve to modulate nucleosome positioning, integration, and removal from chromatin for processes such as gene transcription and repairing DNA damage (Shen et al., 2000; Mohrmann and Verrijzer, 2005). Both the Ino80 and SWI/SNF chromatin remodeling complexes exhibited multiple interactions with kinetochore proteins, but not with Cse4p or Mif2p inner kinetochore proteins. Subunits of the Ino80 complex had two interactions with the Mtw1 complex, a central kinetochore element, and the SWI/SNF chromatin remodeling complex had five interactions with the Ndc80/Hec1 complex, an outer kinetochore element. A third class of chromatin remodeler, the abundant RSC complex, was not appreciably represented in our interaction network. Its absence is consistent with the locus and operational specificity exhibited by chromatin remodeling complexes despite their shared function and is suggestive of specific protein interactions rather than a general connection to a cellular process (Chai et al., 2005). Although the anchorage of central and outer kinetochore complexes to centromeres is believed to be mediated by inner kinetochore proteins, it is possible that their deposition is facilitated by the repositioning of centromeric and neighboring nucleosomes. If the Ino80 and SWI/SNF complexes function redundantly in this process, this role may have so far gone undetected.
Another class of enzymatic chromatin structure modifiers that was highly represented in our screen was the histone acetyltransferases/deacetylases. These histone-modifying proteins help to regulate gene transcription, gene silencing, DNA replication, and DNA repair via modification of lysine residues on the amino-terminal tails of histones (Kurdistani and Grunstein, 2003). Our protein interaction network exhibited connectivity between this class of histone modifiers and central/outer kinetochore elements. The SAGA acetyltransferase complex had two interactions with the Spc25p subunit of the Ndc80/Hec1 kinetochore complex. Ahc2p, a proposed subunit of SAGA, interacted with two subunits of the Dam1 complex and with Nkp2p, a protein in the Ctf19 complex. These interactions with acetyltransferases were complemented by a similar pattern of interactivity with histone deacetylases. Both Pho23p and Sds3p of the Rpd3 deacetylase complex interacted with Spc25p. In addition, Pho23p also interacted with Dad4p of the Dam1 complex and Nkp2p. Another histone deacetylase, Hda2p, interacted with two subunits of the Ndc80/Hec1 complex and Hsk3p of the Dam1 complex. These interactions are indicative of a specific relationship between histone acetyltransferases/deacetylases and particular components of the kinetochore.
The detection of interactions between kinetochore proteins and the transcriptional Mediator complex was particularly intriguing. Mediator is a 20+ subunit coactivator that can be biochemically divided into head, middle, and tail domains (Biddick and Young, 2005). It has been shown to recruit RNA polymerase II (RNAPII) to promoters and can physically bridge transcriptional activators with RNAPII via its tail and head/middle domains respectively (Kim et al., 1994; Bhoite et al., 2001). Pgd1p/Med3p, a subunit of the tail domain, had three interactions with subunits of the Dam1 kinetochore complex and one interaction with Nkp2p of the Ctf19 kinetochore complex. In addition, Srb7p, Med8p, and Med11p associated with Dam1 complex subunits. Nkp2p also interacted with Srb7p of the middle domain. The ability of Gal4p, the transcriptional activator used in the two-hybrid fusion constructs, to bind to the tail domain of Mediator was considered as a possible source of false-positive results (Park et al., 2000). However, the specific affinity of multiple bait constructs of the Dam1 complex for Pgd1p argues against the possibility of the nonspecific recruitment of Mediator and the RNAPII holoenzyme to Gal4p-binding sites. Additionally, the Pgd1p-prey fusion construct is not a common false positive in other screens using the same prey library.
She1p and Other MAPs Interact with the Dam1 and Aurora Kinase Complexes
One protein that warranted further investigation was She1p, a mostly uncharacterized protein that exhibited interactions with Duo1p and Spc34p of the Dam1 outer kinetochore complex and that was previously demonstrated to interact with Dam1p (Shang et al., 2003). It also interacted with the yeast INCENP homolog, Sli15p, which, in conjunction with the Aurora B kinase, Ipl1p, phosphorylates Dam1p, Spc34p, and Ask1p of the Dam1 complex (Cheeseman et al., 2002). The specificity of She1p interactions with the Dam1 complex and its effector, taken together with their common localization to nuclear microtubules, strongly suggests a previously unrecognized function for this protein in mitosis (Hofmann et al., 1998; Huh et al., 2003).
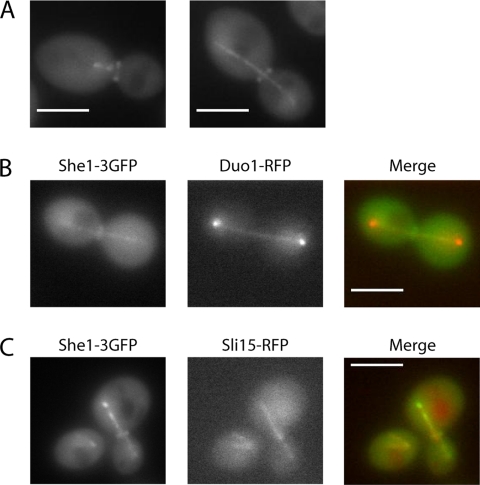
To further validate novel protein–protein interactions detected by the yeast two-hybrid system and to obtain clues to possible functions, we used fluorescent microscopy to localize fluorescently tagged proteins within live cells. She1p was C-terminally tagged with three tandem GFPs for increased fluorescence. She1-3GFP localized to the mitotic spindle at all stages of spindle assembly and to nuclear microtubules during G1 (Figure 3A). She1-3GFP colocalized with RFP-tagged versions of both Duo1p and Sli15p along the spindle, as predicted by the two-hybrid data (Figure 3, B and C). She1-3GFP staining was present along the length of the spindle, consistent with the localization pattern of the Dam1 complex and the localization pattern of the Ipl1 complex before late anaphase (Hofmann et al., 1998; Biggins et al., 1999). However, She1p did not localize exclusively to the spindle midzone with Ipl1p-Sli15p during late anaphase (Buvelot et al., 2003; Pereira and Schiebel, 2003). We also found that She1p localizes to the bud neck throughout mitosis (Figure 3A). She1p is recruited to the bud site early during bud formation and persists through the large-budded stage. Cross-sectional images show brighter staining at the edges of the bud neck compared with the middle, indicative of a ring-shaped structure. Thus, fluorescent microscopy revealed that She1p localizes to the same mitotic structure as its two-hybrid interacting partners and additionally localizes to the bud neck, which is shown here for the first time.
Figure 3.
She1-3GFP localizes to the mitotic spindle and the bud neck. (A) Localization of She1-3GFP during metaphase and anaphase. (B) She1-3GFP (green) colocalizes with Duo1-RFP (red) on the mitotic spindle. (C) She1-3GFP (green) colocalizes with Sli15-RFP (red) on microtubules. Bar, 4 μm.
In addition, subunits of the Dam1 complex exhibited multiple interactions with other MAPs. Bim1p and Stu2p are plus-end tracking proteins implicated in microtubule stability and elongation. Both of these proteins interacted with the Spc34p and Duo1p subunits. We also detected two-hybrid interactions between Bim1p and the Aurora kinase proteins, Ipl1p and Sli15p, and with She1p. Finally, Bim1p and Stu2p interacted with each other in our screen. This intricate web of interconnectivity between these microtubule-associated and kinetochore-localized proteins strongly implies a shared mechanistic function, presumably at the microtubule plus ends.
Phosphorylation State Dependency of Ndc80p Interactions
The mitotic machinery is tightly regulated to ensure faithful chromosome segregation. Protein modifications such as peptide cleavage, phosphorylation, ubiquitination, and sumoylation have a central role in signaling for progression through the cell cycle. Modification-dependent protein–protein interactions are one way in which these signals might be recognized. Although the yeast two-hybrid method has widely been used to detect interactions between proteins, its capacity to screen for modification-dependent interactions has not been well utilized. We tested two phospho-mutant forms of Ndc80p/Hec1, part of the KMN network of kinetochore proteins that is required to form a stable attachment with microtubules, for altered two-hybrid interactions (Kotwaliwale and Biggins, 2006). The N-terminus of Ndc80p/Hec1, including Ser100 in budding yeast Ndc80p, is phosphorylated in vitro by the Aurora B kinase (Cheeseman et al., 2002, 2006; DeLuca et al., 2006) and may electrostatically modulate the protein's affinity for microtubules (Wei et al., 2007).
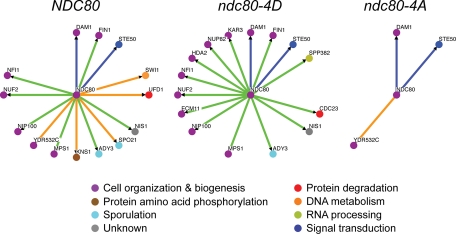
We performed a comparative two-hybrid screen of wild-type Ndc80p and alleles harboring mutations of the four N-terminal Ipl1/Aurora B consensus sites (T54, T74, S95, and S100) to mimic their phosphorylated and dephosphorylated states, against the entire yeast genome (Figure 4). The majority of Ndc80p-interacting proteins exhibited specificity for one or two of the Ndc80p forms. The ndc80-4A mutant had significantly fewer protein–protein interactions than the other forms, but these included interactions with two other kinetochore proteins, Dam1p and YDR532c, suggesting that phosphorylation by the Ipl1/AuroraB kinase is not required for the association of these kinetochore components. Although wild-type Ndc80p and Ndc80-4D had a comparable number of interactions with other proteins, there were notable differences in their yeast two-hybrid interaction maps, including their ability to bind to YDR532c and Kar3p, a kinesin. Using the yeast two-hybrid method, we were able to efficiently create an interaction profile for mutants mimicking different modifications of Ndc80p and to categorize the interactors based on their preferential affinity for a particular form of the bait protein.
Figure 4.
Comparison of protein interaction maps of Ndc80p phospho-mutants. The protein interaction maps shown here summarize the results of yeast two-hybrid screens performed using wild-type Ndc80p and phospho-mutants that mimic the phosphorylation and dephosphorylation of the four N-terminal Ipl1/Aurora B consensus sites, as baits. The color of the nodes corresponds with their GO Process classification. The interactions are classified as being common to all three alleles (blue lines), exclusive of the ndc80-4A allele (green lines), or exclusive of the ndc80-4D allele (orange lines). The protein interaction maps shown here were generated with OSPREY (http://biodata.mshri.on.ca/osprey; Breitkreutz et al., 2003).
DISCUSSION
A Two-Hybrid Screen of Proteins Implicated in Spindle Function Uncovered 604 Protein–Protein Interactions
Understanding how the mitotic spindle functions depends on the identification of the proteins involved in its composition and regulation and on determining how each protein is positioned within a basic organizational framework. The budding yeast S. cerevisiae is amenable to such an undertaking because of the availability of an annotated genome and procedures for systematic studies. Although genome-wide screens for protein–protein interactions have been conducted previously and in principle should have uncovered most of the interactions involving the mitotic spindle, the large scale of such studies required compromises in their execution and scoring that likely lead to a significant number of false-negative results.
To investigate spindle-mediated chromosome segregation in depth, we have conducted a two-hybrid screen that focuses on that cellular process. Each bait construct averaged almost twice as many interactions as were found in previous large-scale screens (Uetz et al., 2000; Ito et al., 2001), supporting the idea that these screens were not saturating. By individually testing each pair-wise protein–protein combination in a focused study, we could detect a greater number of spindle-related physical interactions.
The improved coverage achieved here can be attributed to the more focused scope of our study, which allowed for pairwise testing of all possible bait interactions. By focusing on a single cellular process, we could expend more effort to optimize and troubleshoot individual screens. Some baits that exhibited no interactions were recloned with the DBD fused to the opposite end of the protein. Other baits required scaling of selection conditions to balance the suppression of false positives with the avoidance of false negatives. These factors allowed our two-hybrid investigation to uncover new protein–protein interactions within the budding yeast spindle.
The Spindle Two-Hybrid Screen Successfully Identified Known Complexes
To gauge the effectiveness of this investigation, it is useful to compare the physical interaction network generated here for spindle protein complexes to those generated in other studies. A number of investigations have identified interactions between subunits of the Dam1 kinetochore complex, making these proteins useful as a comparative template for this study.
The spindle two-hybrid screen presented here detected over twice as many pair-wise interactions between subunits of the Dam1 complex as other previously published two-hybrid screens (Uetz et al., 2000; Ito et al., 2001). All 10 subunits identified by biochemical purification (Cheeseman et al., 2002; Miranda et al., 2005; Westermann et al., 2005) were also detected by this study, attesting to the sensitivity of this genomic survey. The integrity of this protein complex is made apparent by the multiple interactions made by each subunit with other subunits of the Dam1 complex. This analysis was facilitated in part by the recent identification of many new short ORFs in the yeast genome, including DAD3 and DAD4 of the Dam1 complex, that were not included in older genomic libraries. In the case of the Dam1 kinetochore complex, this two-hybrid study proved to be more sensitive than previous efforts, which raises the expectation for the detection of novel spindle interactions. The spindle two-hybrid screen also identified She1p as a novel binding partner of the Dam1 complex. Other than being localized to the mitotic spindle (Huh et al., 2003), this nonessential protein is largely uncharacterized. We demonstrated by fluorescence microscopy that She1p colocalizes with Duo1p and Sli15p on the mitotic spindle, but does not share with these proteins the enriched localization at the spindle poles where kinetochores cluster during anaphase. This suggests that the role of She1p may be related to the spindle integrity function of the Dam1 complex rather than to its kinetochore function. We further found that She1p localizes to the yeast bud neck in a ring-shaped structure, but that it does not appear to interact with the Dam1 complex or Ipl1 complex in that area. Its localization to two structures essential for cell division suggests a novel function, the nature of which awaits further investigation.
In addition, a local network of interactions connected She1p, the Dam1 complex, and the Ipl1 complex with the microtubule plus-end tracking proteins (+TIPs) Stu2p and Bim1p. It has been proposed that a combination of Stu2p, Bim1p, and a third +TIP, Bik1p, act together and, partially redundantly, to modulate kinetochore-microtubule dynamics (Wolyniak et al., 2006). This control is important because a newly captured chromosome bound to the lateral surface of a microtubule can become detached if the microtubule shrinks beyond the attachment point (Tanaka et al., 2005). The formation of an end-on attachment between a kinetochore and microtubule plus end is postulated to be mediated by the Dam1 complex. The ability of these various proteins to physically interact may be indicative of a cooperative function for the establishment and/or maintenance of end-on attachments.
Discovery of Novel Interactions between Chromatin-associated Proteins and Spindle Proteins
The 604 pair-wise interactions mapped by the spindle two-hybrid screen presented an opportunity to reveal heretofore-undiscovered mechanisms important for spindle function. By congregating the interaction network nodes based on biochemically characterized physical associations and on participation in specific, narrowly defined cellular processes, patterns signifying the novel convergence of nuclear processes were observed. One of the most striking convergences was that of kinetochore proteins with chromatin-associated proteins, which has implications for the formation of kinetochores on newly replicated chromosomes. Although the current data support a model wherein the yeast kinetochore is organized around the association of Mif2p to centromere-specific nucleosomes containing Cse4p in a CBF3-dependent manner (Westermann et al., 2007), our results suggest that the establishment of kinetochores on centromeric DNA might be more complex.
That remodeling of DNA is important for kinetochore loading has previously been shown by the dependence of chromosome segregation fidelity on the function of either of two redundant chromatin remodeling pathways: the CAF-I and HIR pathways (Sharp et al., 2002). It is possible that regulation of the underlying centromeric chromatin structure is an integral part of kinetochore function before and/or after its establishment. In support of such a possibility, null mutants of subunits of the NuA4 acetyltransferase have synthetic genetic interactions with kinetochore alleles, exhibit sensitivity to the microtubule-destabilizing drug benomyl, and display elevated levels of mini-chromosome mis-segregation (Krogan et al., 2004). Also, the Schizosaccharomyces pombe histone deacetylase Mis16 and its human homologues, RbAp46/48, are required for loading of Cnp1/CENP-A/Cse4p onto centromeres and for prevention of hyper-acetylation of centromeric histones (Hayashi et al., 2004). The two-hybrid data reported here raise the possibility that histone acetyltransferases/deacetylases may have an even more extensive role in kinetochore function than just remodeling the centromere for protein deposition.
Subunits of the Mediator complex, a transcriptional activator, were also implicated in spindle function by their interactions with spindle components, especially Dam1 complex subunits. The lack of similarly dense interaction networks with other transcriptional complexes suggests that the interactions between spindle proteins and the Mediator complex are specific. The possibility of a functional connection between Mediator and the kinetochore is bolstered by previous studies of Cse2p, a Mediator subunit originally identified by its requirement for chromosome segregation fidelity. cse2 mutants exhibited chromosome nondisjunction and mitotic arrest, which were synergistically exacerbated in combination with point mutations in centromeric DNA (Xiao et al., 1993; Xiao and Fitzgerald-Hayes, 1995). In its role as a transcriptional activator, Mediator has been shown to have histone-acetyltransferase activity, leading to chromatin remodeling (Lorch et al., 2000). Whether the role of Mediator subunits in chromosome segregation is linked to their transcriptional function or results from an independent function is not known.
The observation that chromatin remodeling proteins, histone acetyltransferases/deacetylases, and the Mediator transcriptional activator had specific physical interactions with the spindle machinery suggests a previously unrecognized functional relationship. The observations that only a small subset of the known classes of transcriptional helpers exhibited interactions in this screen, that their physical associations were specific to particular kinetochore complexes, and that they were not common false positives all indicate that these proteins were not activating the two-hybrid assay with their transcription-regulating properties. Whether their chromatin-modifying abilities are essential for their function with spindle proteins and whether these proteins form the same complexes used in transcriptional regulation when interacting with spindle proteins is not known. Although it is possible that these interactions have revealed a novel function for these chromatin-associated proteins, this network of interactions is also consistent with the hypothesis that specific chromatin effectors play a role in the establishment and/or maintenance of kinetochores. By possessing the abilities to associate with both DNA and kinetochore proteins, these chromatin effectors are well positioned to execute a variety of possible tasks including the recruitment, establishment, and/or maintenance of kinetochores. If functional redundancy exists among these proteins, it would explain how their mitotic functions have escaped characterization thus far.
Using a Yeast Two-Hybrid Screen to Investigate the Role of Protein Modification
In addition to screening for interactions between wild-type proteins in vegetative cells, we wanted to test how protein modifications may be studied by detecting associated alterations to their two-hybrid interaction profiles. To this end, we screened wild-type Ndc80p and two mutants of Ndc80p that mimicked the phosphorylated and unphosphorylated states of four Ipl1/Aurora B consensus sites. Interestingly, most of the proteins found in this screen interacted preferentially with one of the two mutants (ndc80-4D). A notable exception was that Dam1p strongly interacted with all forms of Ndc80p tested. It was previously shown that Ndc80p binds to an S-to-A phospho-mutant of Dam1p, but not to the corresponding S-to-D phospho-mutant (Shang et al., 2003). These results suggest that the interaction between Dam1p and Ndc80p is regulated by the phosphorylation state of the former protein and that Ndc80p's phosphorylation state controls other interactions.
YDR532c, which forms a complex with Spc105p (KNL-1) of the KMN network (Nekrasov et al., 2003), stands out as being the only protein that exhibits a strong interaction with Ndc80-4A, but not with Ndc80-4D. This result is consistent with the model of phosphorylation by Aurora B/Ipl1p weakening the integrity of the kinetochore-microtubule interface. In contrast, Kar3p, a member of the kinesin-14 family implicated in the transport of newly captured chromosomes along microtubules (Tanaka et al., 2005), interacts only with the 4D variant of Ndc80p. This is significant because, in nocodazole-treated cells, Kar3p colocalizes with Ndc80p, specifically on chromosomes detached from the mitotic spindle (Tytell and Sorger, 2006). Although it is unclear whether Ipl1p becomes activated in nocodazole-treated cells, this modification-specific interaction might be a mechanism for the localization of Kar3p to kinetochores inactivated by Ipl1p.
Although the genomes of several model organisms have been systematically screened using the yeast two-hybrid method, there are compelling reasons to use this technique for smaller, focused screens. Besides detecting potential binding partners for a protein, yeast two-hybrid screens can also be used to investigate the roles of protein modifications. Using this technique, we report the discovery of novel protein–protein interactions and effects of protein phosphorylation that provide insights into the mechanistic workings of the mitotic spindle.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Hector Aldaz, Jeffrey Woodruff, and Ann Marie Faust for discussions; Randall Tyers for critical reading of the manuscript; Yidi Sun and Roger Tsien for plasmids; and Peter Yang for constructive contributions. This work was supported by grants from the National Institute of General Medical Sciences to G.B. (GM-47842), a grant to the Yeast Research Center from the National Center for Research Resources of the National Institutes of Health (Comprehensive Biology, Exploiting the Yeast Genome; PHS P41 RR11823), and a Research Starter Grant from the PhRMA Foundation (T.H.). S.F. is an investigator of the Howard Hughes Medical Institute.
Abbreviations used:
- AD
activation domain
- DBD
DNA-binding domain
- MAP
microtubule-associated protein.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-06-0536) on July 18, 2007.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Ayscough K. R., Drubin D. G. Cell Biology: A Laboratory Handbook. Vol. 2. San Diego: Academic Press; 1998. Immunofluorescence microscopy of yeast cells; pp. 477–485. [Google Scholar]
- Bhoite L. T., Yu Y., Stillman D. J. The Swi5 activator recruits the Mediator complex to the HO promoter without RNA polymerase II. Genes Dev. 2001;15:2457–2469. doi: 10.1101/gad.921601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddick R., Young E. T. Yeast mediator and its role in transcriptional regulation. CR Biol. 2005;328:773–782. doi: 10.1016/j.crvi.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Biggins S., Severin F. F., Bhalla N., Sassoon I., Hyman A. A., Murray A. W. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 1999;13:532–544. doi: 10.1101/gad.13.5.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitkreutz B. J., Stark C., Tyers M. Osprey: a network visualization system. Genome Biol. 2003;4:R22. doi: 10.1186/gb-2003-4-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buvelot S., Tatsutani S. Y., Vermaak D., Biggins S. The budding yeast Ipl1/Aurora protein kinase regulates mitotic spindle disassembly. J. Cell Biol. 2003;160:329–339. doi: 10.1083/jcb.200209018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai B., Huang J., Cairns B. R., Laurent B. C. Distinct roles for the RSC and Swi/Snf ATP-dependent chromatin remodelers in DNA double-strand break repair. Genes Dev. 2005;19:1656–1661. doi: 10.1101/gad.1273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I. M., Anderson S., Jwa M., Green E. M., Kang J., Yates J. R., 3rd, Chan C. S., Drubin D. G., Barnes G. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 2002;111:163–172. doi: 10.1016/s0092-8674(02)00973-x. [DOI] [PubMed] [Google Scholar]
- Cheeseman I. M., Brew C., Wolyniak M., Desai A., Anderson S., Muster N., Yates J. R., Huffaker T. C., Drubin D. G., Barnes G. Implication of a novel multiprotein Dam1p complex in outer kinetochore function. J. Cell Biol. 2001a;155:1137–1145. doi: 10.1083/jcb.200109063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I. M., Chappie J. S., Wilson-Kubalek E. M., Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- Cheeseman I. M., Enquist-Newman M., Muller-Reichert T., Drubin D. G., Barnes G. Mitotic spindle integrity and kinetochore function linked by the Duo1p/Dam1p complex. J. Cell Biol. 2001b;152:197–212. doi: 10.1083/jcb.152.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wulf P., McAinsh A. D., Sorger P. K. Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev. 2003;17:2902–2921. doi: 10.1101/gad.1144403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca J. G., Gall W. E., Ciferri C., Cimini D., Musacchio A., Salmon E. D. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- Gavin A. C., et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- Gavin A. C., et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- Hayashi T., Fujita Y., Iwasaki O., Adachi Y., Takahashi K., Yanagida M. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell. 2004;118:715–729. doi: 10.1016/j.cell.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Hazbun T. R., et al. Assigning function to yeast proteins by integration of technologies. Mol. Cell. 2003;12:1353–1365. doi: 10.1016/s1097-2765(03)00476-3. [DOI] [PubMed] [Google Scholar]
- Hofmann C., Cheeseman I. M., Goode B. L., McDonald K. L., Barnes G., Drubin D. G. Saccharomyces cerevisiae Duo1p and Dam1p, novel proteins involved in mitotic spindle function. J. Cell Biol. 1998;143:1029–1040. doi: 10.1083/jcb.143.4.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Ikeuchi A., Sasaki Y., Kawarasaki Y., Yamane T. Exhaustive identification of interaction domains using a high-throughput method based on two-hybrid screening and PCR-convergence: molecular dissection of a kinetochore subunit Spc34p. Nucleic Acids Res. 2003;31:6953–6962. doi: 10.1093/nar/gkg888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Chiba T., Ozawa R., Yoshida M., Hattori M., Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. J., Bjorklund S., Li Y., Sayre M. H., Kornberg R. D. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- Kotwaliwale C., Biggins S. Microtubule capture: a concerted effort. Cell. 2006;127:1105–1108. doi: 10.1016/j.cell.2006.11.032. [DOI] [PubMed] [Google Scholar]
- Krogan N. J., et al. Regulation of chromosome stability by the histone H2A variant Htz1, the Swr1 chromatin remodeling complex, and the histone acetyltransferase NuA4. Proc. Natl. Acad. Sci. USA. 2004;101:13513–13518. doi: 10.1073/pnas.0405753101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan N. J., et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Kurdistani S. K., Grunstein M. Histone acetylation and deacetylation in yeast. Nat. Rev. Mol. Cell Biol. 2003;4:276–284. doi: 10.1038/nrm1075. [DOI] [PubMed] [Google Scholar]
- Li Y., Bachant J., Alcasabas A. A., Wang Y., Qin J., Elledge S. J. The mitotic spindle is required for loading of the DASH complex onto the kinetochore. Genes Dev. 2002;16:183–197. doi: 10.1101/gad.959402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lorch Y., Beve J., Gustafsson C. M., Myers L. C., Kornberg R. D. Mediator-nucleosome interaction. Mol. Cell. 2000;6:197–201. doi: 10.1016/s1097-2765(00)00021-6. [DOI] [PubMed] [Google Scholar]
- Meluh P. B., Koshland D. Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol. Biol. Cell. 1995;6:793–807. doi: 10.1091/mbc.6.7.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh P. B., Koshland D. Budding yeast centromere composition and assembly as revealed by in vivo cross-linking. Genes Dev. 1997;11:3401–3412. doi: 10.1101/gad.11.24.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millson S. H., Truman A. W., Piper P. W. Vectors for N- or C-terminal positioning of the yeast Gal4p DNA binding or activator domains. BioTechniques. 2003;35:60–64. doi: 10.2144/03351bm06. [DOI] [PubMed] [Google Scholar]
- Miranda J. J., De Wulf P., Sorger P. K., Harrison S. C. The yeast DASH complex forms closed rings on microtubules. Nat. Struct. Mol. Biol. 2005;12:138–143. doi: 10.1038/nsmb896. [DOI] [PubMed] [Google Scholar]
- Mogilner A., Wollman R., Civelekoglu-Scholey G., Scholey J. Modeling mitosis. Trends Cell Biol. 2006;16:88–96. doi: 10.1016/j.tcb.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Mohrmann L., Verrijzer C. P. Composition and functional specificity of SWI2/SNF2 class chromatin remodeling complexes. Biochim. Biophys. Acta. 2005;1681:59–73. doi: 10.1016/j.bbaexp.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Nekrasov V. S., Smith M. A., Peak-Chew S., Kilmartin J. V. Interactions between centromere complexes in Saccharomyces cerevisiae. Mol. Biol. Cell. 2003;14:4931–4946. doi: 10.1091/mbc.E03-06-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. M., Kim H. S., Han S. J., Hwang M. S., Lee Y. C., Kim Y. J. In vivo requirement of activator–specific binding targets of mediator. Mol. Cell. Biol. 2000;20:8709–8719. doi: 10.1128/mcb.20.23.8709-8719.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G., Schiebel E. Separase regulates INCENP-Aurora B anaphase spindle function through Cdc14. Science. 2003;302:2120–2124. doi: 10.1126/science.1091936. [DOI] [PubMed] [Google Scholar]
- Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E., Wilm M., Seraphin B. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- Shang C., Hazbun T. R., Cheeseman I. M., Aranda J., Fields S., Drubin D. G., Barnes G. Kinetochore protein interactions and their regulation by the Aurora kinase Ipl1p. Mol. Biol. Cell. 2003;14:3342–3355. doi: 10.1091/mbc.E02-11-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp J. A., Franco A. A., Osley M. A., Kaufman P. D. Chromatin assembly factor I and Hir proteins contribute to building functional kinetochores in S. cerevisiae. Genes Dev. 2002;16:85–100. doi: 10.1101/gad.925302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., Mizuguchi G., Hamiche A., Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000;406:541–544. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- Sun Y., Carroll S., Kaksonen M., Toshima J. Y., Drubin D. G. PtdIns(4,5)P2 turnover is required for multiple stages during clathrin- and actin-dependent endocytic internalization. J. Cell Biol. 2007;177:355–367. doi: 10.1083/jcb.200611011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Mukae N., Dewar H., van Breugel M., James E. K., Prescott A. R., Antony C., Tanaka T. U. Molecular mechanisms of kinetochore capture by spindle microtubules. Nature. 2005;434:987–994. doi: 10.1038/nature03483. [DOI] [PubMed] [Google Scholar]
- Tong A. H., et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- Tytell J. D., Sorger P. K. Analysis of kinesin motor function at budding yeast kinetochores. J. Cell Biol. 2006;172:861–874. doi: 10.1083/jcb.200509101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P., et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Wei R. R., Al-Bassam J., Harrison S. C. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat. Struct. Mol. Biol. 2007;14:54–59. doi: 10.1038/nsmb1186. [DOI] [PubMed] [Google Scholar]
- Westermann S., Avila-Sakar A., Wang H. W., Niederstrasser H., Wong J., Drubin D. G., Nogales E., Barnes G. Formation of a dynamic kinetochore-microtubule interface through assembly of the Dam1 ring complex. Mol. Cell. 2005;17:277–290. doi: 10.1016/j.molcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- Westermann S., Cheeseman I. M., Anderson S., Yates J. R., 3rd, Drubin D. G., Barnes G. Architecture of the budding yeast kinetochore reveals a conserved molecular core. J. Cell Biol. 2003;163:215–222. doi: 10.1083/jcb.200305100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S., Drubin D. G., Barnes G. Structures and functions of yeast kinetochore complexes. Annu. Rev. Biochem. 2007;76:563–591. doi: 10.1146/annurev.biochem.76.052705.160607. [DOI] [PubMed] [Google Scholar]
- Wolyniak M. J., Blake-Hodek K., Kosco K., Hwang E., You L., Huffaker T. C. The regulation of microtubule dynamics in Saccharomyces cerevisiae by three interacting plus-end tracking proteins. Mol. Biol. Cell. 2006;17:2789–2798. doi: 10.1091/mbc.E05-09-0892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z., McGrew J. T., Schroeder A. J., Fitzgerald-Hayes M. CSE1 and CSE2, two new genes required for accurate mitotic chromosome segregation in Saccharomyces cerevisiae. Mol. Cell. Biol. 1993;13:4691–4702. doi: 10.1128/mcb.13.8.4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z. X., Fitzgerald-Hayes M. Functional interaction between the CSE2 gene product and centromeres in Saccharomyces cerevisiae. J. Mol. Biol. 1995;248:255–263. doi: 10.1016/s0022-2836(95)80048-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.