Abstract
Antitrypsin deficiency is a primary cause of juvenile liver disease, and it arises from expression of the “Z” variant of the α-1 protease inhibitor (A1Pi). Whereas A1Pi is secreted from the liver, A1PiZ is retrotranslocated from the endoplasmic reticulum (ER) and degraded by the proteasome, an event that may offset liver damage. To better define the mechanism of A1PiZ degradation, a yeast expression system was developed previously, and a gene, ADD66, was identified that facilitates A1PiZ turnover. We report here that ADD66 encodes an ∼30-kDa soluble, cytosolic protein and that the chymotrypsin-like activity of the proteasome is reduced in add66Δ mutants. This reduction in activity may arise from the accumulation of 20S proteasome assembly intermediates or from qualitative differences in assembled proteasomes. Add66p also seems to be a proteasome substrate. Consistent with its role in ER-associated degradation (ERAD), synthetic interactions are observed between the genes encoding Add66p and Ire1p, a transducer of the unfolded protein response, and yeast deleted for both ADD66 and/or IRE1 accumulate polyubiquitinated proteins. These data identify Add66p as a proteasome assembly chaperone (PAC), and they provide the first link between PAC activity and ERAD.
INTRODUCTION
Newly synthesized secreted proteins must pass a stringent quality control checkpoint in the endoplasmic reticulum (ER), which ensures that only properly folded, assembled, and processed proteins transit the secretory pathway (Ellgaard and Helenius, 2003). Polypeptides that fail to pass this checkpoint may be targeted for ER-associated degradation (ERAD), a process in which aberrant proteins are selected and then delivered—or retrotranslocated—to the cytoplasm and degraded by the 26S proteasome (Fewell et al., 2001; Tsai et al., 2002; Kostova and Wolf, 2003; Meusser et al., 2005; Romisch, 2005; Sayeed and Ng, 2005; Nandi et al., 2006). The 26S proteasome is an ∼2.5-MDa multisubunit complex that contains a central 20S proteolytic core particle and two 19S regulatory particles (Voges et al., 1999; Nandi et al., 2006). The 20S core harbors three distinct proteolytic activities—a chymotrypsin-like (CTL), a trypsin-like (TL), and a peptidylglutamyl-peptide hydrolyzing (PGPH) activity—whereas the 19S “cap” (also known as PA700) functions as a gatekeeper at the entrance of the core particle. In addition, the 19S particle contains polyubiquitin-binding subunits, enzymes required for polypeptide deubiquitination, and six AAA ATPases that are thought to unfold and feed polypeptides into the core (Glickman et al., 1998; Voges et al., 1999; Leggett et al., 2002; Verma et al., 2002; Guterman and Glickman, 2004; Soboleva and Baker, 2004).
Some ERAD substrates in humans are mutated versions of wild-type proteins, and not surprisingly, their absence may lead to the onset of specific diseases (Aridor and Hannan, 2000, 2002; Coughlan and Brodsky, 2003). One substrate in which the connection between ERAD and loss-of-function disease is very clear is the Z-variant of α-1-antitrypsin, also known as A1PiZ (or α-1 protease inhibitor, Z). Wild-type A1Pi, originally termed the “M” variant, or A1PiM (Myerowitz et al., 1972), is an ∼53-kDa serine protease inhibitor that is primarily synthesized in and secreted by hepatocytes (Perlmutter, 2002). Serum A1Pi inhibits neutrophil proteases, which are released during inflammatory responses and can mediate proteolysis of the pulmonary connective tissue matrix (Richmond and Zellner, 2005; Rudnick and Perlmutter, 2005). Although secreted A1PiZ retains partial activity, individuals expressing this protein have lower circulating levels of the protein because the E342K mutation compromises its folding in the ER. The resulting decrease in plasma levels of the protease inhibitor lead to antitrypsin deficiency (ATD), which is exemplified by uninhibited neutrophil elastase activity and destruction of the pulmonary extracellular matrix. However, when the A1PiZ variant accumulates, it can form loop-sheet polymers or aggregates that may trigger cirrhosis (Foreman et al., 1984; Perlmutter et al., 1985; Mornex et al., 1986; Verbanac and Heath, 1986; Brantly et al., 1988; McCracken et al., 1989; Lomas et al., 1992; Mast et al., 1992; Kim et al., 1995; Sidhar et al., 1995; Carrell and Lomas, 2002; Parfrey et al., 2003) and hepatocellular carcinoma (Carlson et al., 1989; Rudnick and Perlmutter, 2005). Thus, ATD is also a gain-of-function disease.
The reason only ∼10% of A1PiZ-homozygotes develop liver disease (Sveger, 1988) remains a mystery. However, this phenomenon may arise because a subset of individuals is unable to efficiently clear this aggregation-prone molecule from the ER. Indeed, A1PiZ-expressing fibroblasts from individuals with liver disease degrade the substrate less efficiently than A1PiZ-expressing fibroblasts from healthy homozygotes (Wu et al., 1994). Therefore, factors that alter the efficiency of A1PiZ ERAD may represent genetic modifiers of ATD.
To identify putative ATD modifiers, we developed an A1PiZ yeast expression system (McCracken and Kruse, 1993) because 1) components of the protein quality control machinery are highly conserved, and 2) yeast expression systems for several human disease-causing proteins have led to a better understanding of the pathological consequences of aberrant protein production (Coughlan and Brodsky, 2003). Notably, the A1PiZ yeast expression system helped establish this protein as a bona fide ERAD substrate (Werner et al., 1996) and identified an Hsp70 molecular chaperone in the lumen of the ER, known as BiP, as an important player in A1PiZ turnover (Brodsky et al., 1999). This result was subsequently confirmed in mammalian cells (Cabral et al., 2002; Schmidt and Perlmutter, 2005). We also identified antitrypsin degradation deficient (ADD) mutants by using both a classical genetic (McCracken et al., 1996) and targeted approach (Palmer et al., 2003). One of the genes isolated was ATG6/VPS30, which is required for autophagy (Kametaka et al., 1998), and yeast overexpressing A1PiZ deliver the aggregated protein to the autophagic pathway (Kruse et al., 2006). Similarly, autophagic vesicles are abundant in liver biopsies from individuals with late-stage ATD (Teckman and Perlmutter, 2000), and A1PiZ-expressing autophagy-deficient cell lines degrade A1PiZ less efficiently than wild-type cells (Kamimoto et al., 2006).
Another yeast gene that was isolated is ADD66 (YKL206c). Even though Add66p is required for the degradation of only a subset of ERAD substrates examined (Palmer et al., 2003), the ADD66 transcript is induced by the unfold protein response (UPR) (Travers et al., 2000), which serves as an indicator of ER stress; moreover, the UPR and ERAD provide complementary mechanisms to lessen the effects of aberrant protein accumulation in the ER (Fewell et al., 2001; Patil and Walter, 2001; Schroder and Kaufman, 2005). Because of these observations, and because cells deleted for ADD66 also induce the UPR (Palmer et al., 2003), we suggested that Add66p might play a more general role in ER protein quality control.
In this study, we report that Add66p facilitates the maturation of the 20S proteasome particle. It is vital for maximal proteasome activity and like some other proteasome chaperones (Ramos et al., 1998; Tone and Toh, 2002; Hirano et al., 2005), it can be degraded by the proteasome. Based on these results and on the sequence similarity between ADD66 and the mammalian proteasome assembly chaperone (PAC)2 (Hirano et al., 2005, 2006), we suggest that Add66p is the Saccharomyces cerevisiae PAC2 homologue. Together, these results provide the first direct link between PAC activity and ERAD.
MATERIALS AND METHODS
Strains and Growth Conditions
The Escherichia coli strain used in this study was DH5α (endA1 hsdR17 supE44 thi-1 recA1 gyrA relA1 Δ [lacZYA-argF] U169 deoR [Φ80 dlac Δ lacZ M15]). Bacteria were grown in Luria-Bertani medium supplemented with 50 μg/ml ampicillin for plasmid selection. An add66Δ disruption cassette was obtained by amplifying pRS400 (Brachmann et al., 1998) with the following oligonucleotides: 5′-ACT TCA GGA AAG AAT AGC ACA AAA CCC AAA GGA ACA TAC GCT GTG CGG TAT TTC ACA CCG-3′ and 5′-ATA TAT GCA CTT GTA TAG AAA ACA GAT ATA CTT CTC GGT TAG ATT GTA CTG AGA GTG CAC-3′. ADD66 mutants were obtained as described previously (Brachmann et al., 1998). A pdr5Δ haploid strain was isolated by sporulation and dissection of the yeast pdr5Δ homozygous diploid (Invitrogen, Carlsbad, CA). All other yeast strains used in this study are detailed in Table 1, and they were grown on yeast extract-peptone (YP)-dextrose (YPD) medium or on synthetic complete (SC) medium lacking the indicated nutrient but supplemented with a carbohydrate source to a final concentration of 2%. Yeast were grown at the indicated temperatures, and all genetic and molecular manipulations followed standard published protocols (Adams et al., 1997).
Table 1.
Yeast strains used in this study
| Strain | Relevant genotype | Reference or source |
|---|---|---|
| ADD66 (BY4742) | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Invitrogen |
| add66Δ | MATα add66::kanMX his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Invitrogen |
| ire1Δ | MATα ire1::kanMX his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Invitrogen |
| ire1Δ add66Δ | MATα ire1::kanMX add66::HIS3 his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | This study |
| ADD66(W303) | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1, ura3-1 | |
| add66Δ | MATaadd66::kanMX ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1, ura3-1 | This study |
| CIM5 | MATα ura3-52 lys2-801 ade2-101 his3Δ200 leu2-Δ1 | Ghislain et al. (1993) |
| cim5-1 | MATacim5-1 ura3-52 his3Δ0200 leu2-Δ1 | Ghislain et al. (1993) |
| atg14Δ | MATα atg14::kanMX his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Invitrogen |
| pdr5Δ | MATapdr5::kanMX his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 met15Δ0 | This study |
| JD133 | MATahis3-Δ200 leu2-3,112 lys2-801 trp1-Δ63 ura3-52 UMP1-ha::YIplac128 PRE2-HA::YIplac211 | Ramos et al. (1998) |
| JD134 | MATahis3-Δ200 leu2-3,112 lys2-801 trp1-Δ63 ura3-52 ump1-Δ1::HIS3 PRE2-HA::YIplac211 | Ramos et al. (1998) |
| RJD1144 | MATahis3Δ200 leu2-3,112 lys2-801 trp1Δ63 ura3-52 PRE1-Flag-HIS6::Ylpac211 (URA3) | Verma et al. (2000) |
Detection of Polyubiquitinated Proteins in Yeast
Yeast were transformed with a plasmid engineered for the expression of a ubiquitin-myc fusion protein under the transcriptional control of a copper-inducible promoter (Ecker et al., 1987; subcloned and provided by A. Caplan, Mount Sinai School of Medicine), and transformants were isolated on selective medium (SC-HIS) containing glucose. A culture containing 20 optical density units measured at 600 nm (ODs) of cells was grown to mid-log phase (∼1.0 OD/ml) at 30°C before CuSO4 was added at a final concentration of 100 μM, and the cells were incubated for 1 h. The yeast were harvested and resuspended in 100 μl of sample buffer (1.0% β-mercaptoethanol, 1% SDS, 5% glycerol, 0.05 mg/ml bromphenol blue, and 65 mM Tris, pH 6.8), 0.2 g of glass beads was added, and lysates were prepared by vigorous agitation on a Vortex mixer 10 times for 1 min with a 1-min incubation in an ice bath between each treatment. The total protein in a 5-μl aliquot of each sample was resolved by SDS-polyacrylamide gel electrophoresis (PAGE) and transferred onto nitrocellulose blots. The nitrocellulose was incubated for 30 min, while sandwiched in Whatman filter paper, in boiling double-distilled deionized water. Ubiquitin and Sec61p, a component of the translocon that served as a loading control, were identified by Western blotting by using anti-myc (kindly provided by G. Apodaca and O. Weisz, University of Pittsburgh School of Medicine) and anti-Sec61p (Stirling et al., 1992). Western blots were developed using enhanced chemiluminescence (Pierce Chemical, Rockford, IL) according to the manufacturer's instructions. Images were obtained on a Kodak 440CF Image Station, and the results were quantified using Kodak 1D, version 3.6 imaging software (Eastman Kodak, Rochester, NY).
Expression of Wild Type and the Z Variant of α-1-Antitrypsin and Epitope-tagged Add66p
Yeast were transformed with plasmids containing the genes encoding A1PiM or A1PiZ under the transcriptional control of a galactose-inducible promoter (McCracken et al., 1996), and transformants were isolated on selective medium (SC-URA) containing glucose. As a control, the indicated strains were also transformed with the pYES2 vector (Invitrogen) lacking an insert.
To create an epitope-tagged version of Add66p, a single myc epitope was appended onto the C terminus of Add66p by polymerase chain reaction (PCR) amplification of genomic yeast DNA with the following oligonucleotides: 5′-CGC GGA TCC ATG AGC TGC CTG GTG TTG-3′ (to construct the pGPD vectors; see below) or 5′-CGC GGA TCC TCC TCG ATT TGA CTG GAA AC-3′ (to construct the pRS vector) and 5′-CCC AAG CTT TCA CAG GTC CTC CTC TGA GAT CAG CTT CTG CTC CTC ATT GTA TAA ATC TAC AAA TTT ATC TCT TGC-3′ (the underlined portion encodes the 11-amino acid myc epitope; Evan et al., 1985). The PCR fragment was inserted into the BamH1 and ClaI sites in the pPRS315, pGPD425, and pGPD426 vectors (Sikorski and Hieter, 1989; Mumberg et al., 1995) and the in-frame insertion and integrity of ADD66 were confirmed by DNA sequence analysis. The corresponding vectors or the vectors lacking an insert were introduced into the indicated strains and transformants were isolated on selective media. Next, yeast were grown to mid-log phase, and 2 ODs of cells were harvested and total cell extracts were prepared as published previously (Brodsky et al., 1998). Add66p-myc expression was assessed after SDS-PAGE and by Western blotting, as described above, by using anti-serum against myc.
A Yeast Colony Blot Assay for A1PiZ Accumulation
A colony blot immunoassay was performed to assess A1PiZ levels in wild-type and select mutant yeast as described previously (Palmer et al., 2003). In brief, 3 μl (0.001 OD) of cells from a saturated culture were spotted onto nitrocellulose that had been overlaid onto selective medium containing 2% galactose to induce expression of A1PiZ. After a 36-h incubation at 35°C, the cells were lysed. A1Pi was detected by immunoblotting, and the results were quantified using the Molecular Analyst program (Bio-Rad, Hercules, CA). The signals corresponded to cell and protein levels in the linear range of detection, and previous work established the validity of using the colony blotting protocol as a means to report on AiPiZ turnover in wild-type and the add66Δ mutant (Palmer et al., 2003).
Add66p Localization
The residence of Add66p-myc was assessed by biochemical fractionation using a previously published method, with minor modifications (Kabani et al., 2002). Specifically, detection of Add66p-myc by Western blotting required 1000 ODs of mid-log phase yeast that contained the pGPD425-Add66p-myc expression vector and that had been grown at 30°C.
To assess Add66p-myc localization by indirect immunofluorescence microscopy, a previously described protocol was used (Coughlan et al., 2004) that involved yeast containing either the Add66p-myc expression vector under the transcriptional control of a constitutive promoter (pGPD) or the vector lacking an insert (as a negative control). Images were captured on an Olympus BX60 microscope (Olympus, Tokyo, Japan) fitted with a Hamamatsu C4742-95 digital camera (Hamamatsu, Bridgewater, NJ), and they were analyzed using QED Imaging software (Media Cybernetics, Silver Spring, MD).
Purification of Yeast 26S Proteasomes
FLAG-tagged 26S proteasomes were purified from RJD1144 as described previously (Verma et al., 2000; Saeki et al., 2005; Verma and Deshaies, 2005) with minor modifications. In brief, cells were grown to an OD of ∼3, frozen in liquid nitrogen and ground in a Waring blender. The ground powder (∼20 ml) was thawed with 9 ml of lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 10% glycerol, and 5 mM MgCl2) plus 5 mM ATP, 2× ATP regenerating system (0.02 mg/ml creatine phosphokinase and 20 mM creatine phosphate), 5 mM MgCl2, and 1 mM dithiothreitol (DTT). After centrifugation at 15,000 rpm for 20 min at 4°C in a SS34 rotor (Sorvall, Newton, CT), the supernatant (∼13 ml) was supplemented with 5 mM ATP, 2× ATP regenerating system, and 5 mM MgCl2, and it was incubated with 300 μl of 50% (vol/vol) washed FLAG agarose beads (Sigma-Aldrich, St. Louis, MO) at 4°C for 1.5 h with rocking. The agarose beads were washed twice with 10 ml of lysis buffer plus 2 mM ATP and 1 mM DTT; once with 5 ml of lysis buffer plus 2 mM ATP, 1 mM DTT, and 0.2% Triton X-100; once with 5 ml of lysis buffer plus 2 mM ATP and 1 mM DTT; once with 800 μl of lysis buffer plus 2 mM ATP and 1 mM DTT; and once with 1 ml of 26S elution buffer (25 mM Tris, pH 7.5, 10 mM MgCl2, 150 mM NaCl, and 15% glycerol) plus 2 mM ATP. The bound proteins were eluted with 400 μl of 26S elution buffer plus 2 mM ATP supplemented with 1/50 volume of 5 mg/ml 3xFLAG peptide (Biotechnology Center, University of Pittsburgh) by incubating the solution for 3 h at 4°C with rocking. The eluted proteosomes were enriched to ∼0.9 mg/ml by using a Centricon-30 microconcentrator (Millipore, Billerica, MA), and they were stored at −80°C.
Proteasome Activity Assays and Glycerol Gradient Analysis
Proteasome activity was assessed in clarified yeast cytosols that were obtained from 2 l of the indicated strains grown in selective medium to mid-log phase at 30°C. The cell pellet was washed with water and resuspended in 500 μl of buffer 88 (20 mM HEPES, pH 6.8, 150 mM KOAc, 5 mM MgOAc, and 250 mM sorbitol), and the cell-slurry was frozen in liquid nitrogen. Frozen cells were lysed by grinding with a mortar and pestle in the presence of liquid nitrogen for 12 min, and the cells were thawed in the presence of a minimal amount of buffer 88 containing 1 mM DTT. Unbroken cells and debris were removed by centrifugation in a Sorvall SS34 rotor at 9000 × g for 10 min at 4°C, and the supernatant was clarified by centrifugation at 300,000 × g for 1 h at 4°C. The clarified cytosol was then aliquoted and snap-frozen in liquid nitrogen and stored at −80°C. Total protein was quantified using the Bio-Rad protein assay kit with bovine serum albumin (BSA) as a standard.
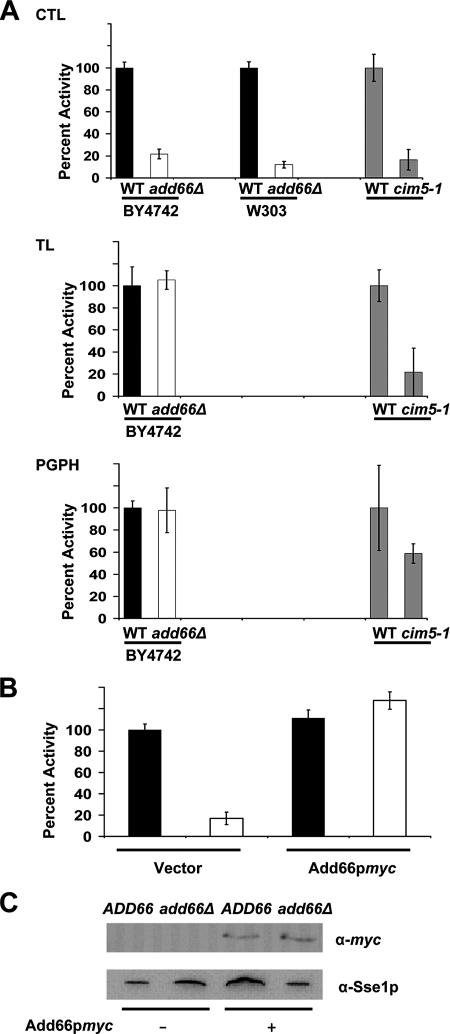
Proteasome activity was determined by modifying a previously described protocol (Glickman et al., 1998). First, a total of 100 μg of clarified cytosol was diluted into 1.8 ml of buffer A (50 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 10% glycerol, 1 mM ATP, and 1 mM DTT) and incubated on ice for 30 min in either the presence or absence of 100 μM MG-132 or leupeptin. Next, a fluorescent substrate to detect each proteasome activity (CTL: Suc-LLVY-AMC [Sigma-Aldrich]; TL: Cbz-AAR-AMC [Calbiochem, San Diego, CA]; PGPH: Cbz-LLE-AMC [Calbiochem]) was added to a final concentration of 100 μM, and the reactions were shifted to 30°C for the indicated times and quenched by the addition of 1% SDS (final concentration). Where noted, a total of 0.5 μg of purified 26S proteasomes was used and treated identically. Fluorescence was determined on an Aminco-Bowman Series 2 luminescence spectrometer (Thermo Electron Corp., Madison, WI) (excitation, 380 nm; emission, 436 nm). The CTL, TL, and PGPH activities were confirmed using the following inhibitors at a final concentration of 100 μM: MG-132 (for CTL and PGPH) and leupeptin (for TL) (Savory and Rivett, 1993; Gaczynska and Osmulski, 2005), and the activity in each reaction and at each time point was obtained after the background fluorescence in the presence of each inhibitor was subtracted from the net fluorescence (Figure 4). When extracts were examined, the data were then normalizing to the activities observed in lysates from the respective isogenic wild-type strains at reaction times of 10 min for the CTL activity, and at 60 min for the TL and PGPH activities (Figure 4). When highly enriched proteasomes were examined, the data were normalized to the activities observed in the absence of inhibitors at 180 min (Supplemental Figure 2, third column), and when lysates were examined in this figure (first and second columns) the CTL, TL, and PGPH activities were normalized to the wild-type activities observed at 180 min in the absence of inhibitors. Maximal, normalized activities are denoted as 100%.
Figure 4.
The chymotrypsin-like activity of the 26S proteasome is reduced in extracts prepared from the add66Δ strain. The CTL, TL, and PGPH activities in clarified extracts from ADD66 (black bar) or add66Δ (white bar) yeast in two different strain backgrounds (BY4742 and W303) were determined. Proteasome activities in CIM5 and cim5-1 (gray bar) strains were used as a positive control. The relative activity was determined by normalizing the fluorescent signals to the levels corresponding to the wild-type strains, as described in Materials and Methods. (B) Constitutive expression of Add66p-myc restores the CTL activity in the add66Δ strain. Wild-type and add66Δ strains were transformed with an empty vector (−) or a vector engineered for the expression of Add66p-myc (+), and the CTL activity was analyzed as described in A. (C) Cytosolic proteins from the strains in B were resolved by SDS-PAGE, and then they were probed for Add66p-myc and Sse1p expression by Western blot analysis. Sse1p served as a loading control.
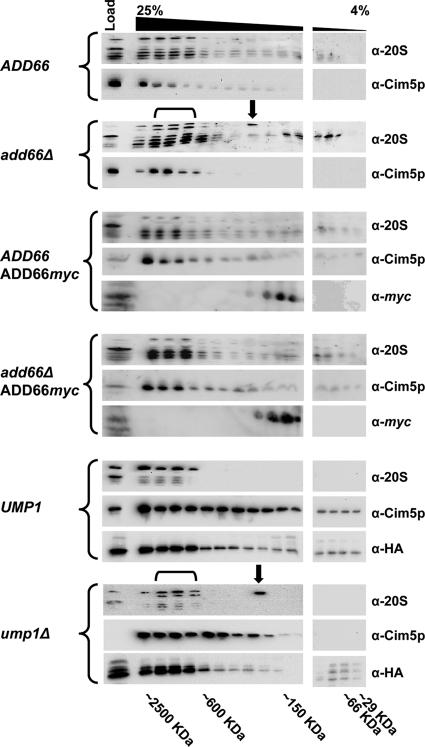
To assess the integrity of the proteasome using glycerol gradient centrifugation, the indicated cells expressing Add66p-myc under its endogenous promoter (Figure 5) or when overexpressed (Supplemental Figure 6) or containing the expression vector lacking an insert were grown in SC-LEU and 2% glucose at 30°C. A total of 100 ODs of mid-log phase cells were harvested and washed once with water and resuspended in 2 ml of buffer A lacking glycerol. (The pdr5Δ strain was incubated with 100 μg of MG-132 for 1 h at 30°C before harvesting.) Cells were lysed as described above, and unbroken cells were removed by centrifugation in a Sorvall SS34 rotor at 9000 × g for 30 min at 4°C. Extracted proteins (5 mg total, as assessed using the Bio-Rad protein assay kit with BSA as the standard) were fractionated on a 30 ml 4–25% linear glycerol gradient in an SW28 rotor (Beckman Coulter, Fullerton, CA) at 83,000 × g for 24 h at 4°C. Molecular mass markers (Sigma-Aldrich) were examined in parallel. One-milliliter fractions were removed, and the refractive index was examined to verify the establishment of a linear gradient. Fractionated proteins were precipitated with trichloroacetic acid (25% final concentration), and they were resolved by SDS-PAGE and analyzed as described above by Western blotting with anti-myc, anti-20S (BIOMOL Research Laboratories, Plymouth Meeting, PA), anti-hemagglutinin (HA) (Roche Molecular Biochemicals, Indianapolis, IN), and anti-Cim5p (Ghislain et al., 1993) antisera.
Figure 5.
Yeast deleted for ADD66 accumulate a 20S intermediate and unprocessed 20S subunits. Cell extracts were prepared from an ADD66 and add66Δ strain containing either a control plasmid or a plasmid engineered for the endogenous expression of Add66p-myc, and from a UMP1 (JD133) and ump1Δ (JD134) strain. In total, 5 mg of protein was then resolved on a linear glycerol gradient (4–25%), and fractions were collected. Proteins in every other fraction were examined for the presence of 20S subunits, a component of the 19S subunit (Cim5p), and Add66p-myc by Western blot analysis. The migrations of molecular mass markers, which were analyzed in parallel, are indicated below the gel, the black downward bracket indicates fractions containing immature 20S subunits (a slower migrating doublet), and the black downward arrow indicates the migration of a 20S assembly intermediate. Note that the later two were observed only in extracts prepared from add66Δ and ump1Δ cells. The immunoreactive HA species in the UMP1 and ump1Δ gradients represents Pre2p (Table 1).
Add66p-myc Degradation Assay
The pdr5Δ yeast strain expressing Add66p-myc was grown in SC-LEU to mid-log phase at 30°C, and protein synthesis was arrested by the addition of cycloheximide to a final concentration of 100 μg/ml. Four ODs of cells were removed at the indicated times. The cells were washed, and total protein was isolated by glass bead lysis as detailed above. Proteins were resolved on either 12.5 or 18% polyacrylamide gels, and they were analyzed and quantified as described above by Western blotting with anti-myc and anti-Sec61p antisera.
Induction of Autophagy
The indicated strains were grown overnight in YPD, and 2.5 ODs of cells were diluted into 10 ml of rich medium (YP with 2% galactose) or nitrogen-depleted media (SC lacking ammonium sulfate but supplemented with 2% galactose) to induce autophagy. After incubation for 5 h at 30°C, equal numbers of cells were harvested, and lysates were prepared by glass bead lysis as described above. Equal amounts of protein (as assessed above) in each sample were resolved by SDS-PAGE and analyzed by Western blotting by using anti-Ape1p (Santa Cruz Biotechnology, Santa Cruz, CA) and anti-Sec61p antisera.
RESULTS
Genetic Interactions between ADD66 and IRE1, a Transducer of the UPR
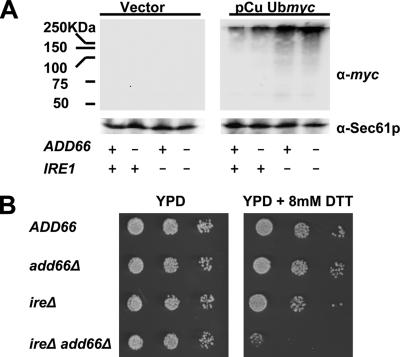
We reported previously on the identification of UPR-target genes in yeast that—when deleted—result in A1PiZ stabilization (Palmer et al., 2003). In turn, the deletion of one gene, ADD66, induced the UPR, suggesting that the corresponding protein might be involved in more general aspects of ER quality control. To further explore the connection between Add66p and the UPR pathway, the following strains were constructed. One strain lacked Ire1p, an ER-resident transmembrane protein that senses a rise in the concentration of misfolded ER lumenal proteins and initiates the transcription of genes whose products lessen ER stress (Cox et al., 1993; Mori et al., 1993; Shamu and Walter, 1996; Credle et al., 2005; Zhou et al., 2006). The ire1Δ strain was examined along with add66Δ yeast and an ire1Δadd66Δ strain. Next, these cells and an isogenic wild-type strain (Table 1) were transformed with either a control plasmid or with a plasmid that expresses a ubiquitin-myc fusion protein under the transcriptional control of a copper inducible promoter. As shown in Figure 1A (right half of figure), we first observed that strains lacking Add66p showed a modest increase in polyubiquitinated proteins compared with the ADD66 strain (∼2-fold in this experiment), and yeast deleted for IRE1—regardless of whether ADD66 was present—accumulated somewhat greater amounts of polyubiquitinated protein. Furthermore, we noted that yeast lacking both ADD66 and IRE1 exhibited a strong, synthetic growth defect when incubated on media containing DTT, a reducing agent that induces the UPR (Figure 1B). These data support the hypothesis that Add66p plays a role in the turnover of polyubiquitinated proteins and ER quality control, and they are consistent with reports indicating that mutations in genes required for both ERAD and the UPR exhibit synthetic phenotypes (Ng et al., 2000; Travers et al., 2000).
Figure 1.
ADD6 and IRE1 synthetically interact. ADD66, add66Δ, ire1Δ, and ire1Δ add66Δ strains were transformed with a control plasmid or a plasmid expressing a ubiquitin-myc fusion protein under the transcriptional control of a copper inducible promoter (pCu Ubmyc). (A) Representative Western blots of extracts from cells containing a vector control or the ubiquitin expression vector are shown. Extracts were prepared after cells had been treated with 100 μM CuSO4 for 1 h at 30°C. Blots were probed with anti-myc and anti-Sec61p (as a loading control). (B) Serial dilutions of the indicated strains (Table 1) were grown on YPD in the presence or absence of 8 mM DTT, as indicated, for 48 h at 30°C. Of note, we chose to examine DTT sensitivity on YPD medium at pH 6.5 to reduce alternate stresses, although previously published work demonstrated a greater sensitivity to DTT in the ire1Δ strain when grown using other conditions (Frand and Kaiser, 1998; Pollard et al., 1998).
In recently published work, autophagy has been shown to play a significant role in mediating A1PiZ degradation in yeast, mammalian cell culture, and mouse models (Teckman and Perlmutter, 2000; Kamimoto et al., 2006; Kruse et al., 2006). Therefore, it was possible that deletion of ADD66 leads to the accumulation of A1PiZ and exhibits synthetic interactions with IRE1 because the autophagic pathway, which is induced by ER stress (Bernales et al., 2006; He et al., 2006; Ogata et al., 2006; Yorimitsu et al., 2006), is compromised. To examine this hypothesis, the maturation of Ape1p, a protease that is targeted to the vacuole during autophagy, was assessed in both ADD66 and add66Δ strains and in a well-characterized autophagy-deficient strain, atg14Δ (Kametaka et al., 1998). As Ape1p enters the vacuole it is proteolytically cleaved, and thus the conversion of immature-Ape1p (“pre-Ape1p”) to mature Ape1p (“m-Ape1p”) can be used to assess induction of autophagy (Suzuki et al., 2002). As anticipated, we found that Ape1p failed to mature in the atg14 mutant, regardless of whether autophagy was induced upon nutrient starvation. In contrast, greater amounts of mature Ape1p were evident in both the wild-type and add66Δ strains upon starvation (Supplemental Figure 1). These data indicate that autophagy is proficient in yeast lacking ADD66.
Add66p Is a Cytosolic, Soluble Protein
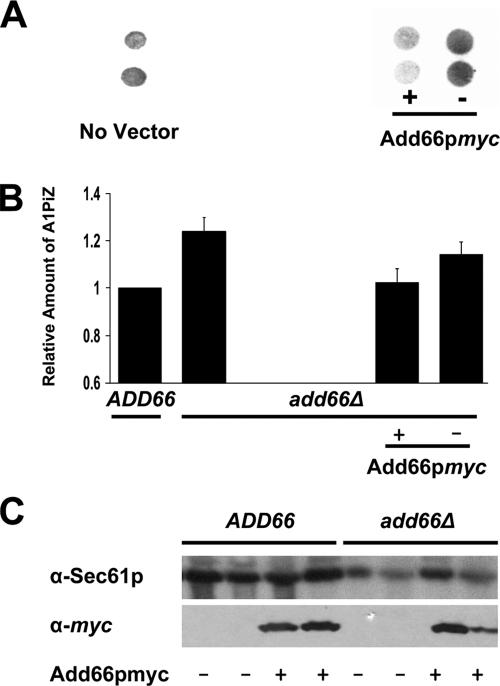
To determine why cells deleted for ADD66 accumulate A1PiZ and polyubiquitinated proteins, we wanted to characterize the gene product. To this end, a sequence encoding the myc epitope was appended onto the C terminus of Add66p, and the construct was cloned into a vector designed for the strong, constitutive expression of the desired protein (pGPD425) (Mumberg et al., 1995) and into a vector in which ADD66 expression was driven by the endogenous promoter (pRS315; see Materials and Methods). To determine whether the tagged protein was active, we assessed whether A1PiZ degradation was restored to wild-type levels in Add66p-myc-expressing add66Δ yeast. To this end, we used a quantitative colony blot assay that reports on A1PiZ expression levels and ERAD, and in fact was used to isolate the ADD mutants (McCracken et al., 1996; Palmer et al., 2003). As shown in Figure 2, A and B, we found that A1PiZ accumulated to wild-type levels when add66Δ cells contained the Add66p-myc expression vector. As expected, Add66p-myc expression was observed only in ADD66 and add66Δ strains that had been transformed with the expression vector (Figure 2C).
Figure 2.
The A1PiZ degradation defect is rescued in add66Δ strains expressing Add66p-myc. (A) A colony-blot immunoassay was performed with anti-antitrypsin antiserum on add66Δ strains expressing A1PiZ and that lacked a vector or that were transformed with an empty vector (−) or with an ADD66-myc expression plasmid (+). (B) The results from three independent colony-blot immunoassays were quantified for wild-type yeast (ADD66) and add66Δ yeast that lacked a vector or that were transformed with the ADD66-myc expression vector (+) or a vector control (−). Data were quantified from signals detected in the linear range of the analysis. (C) Proteins extracts were prepared from ADD66 and add66Δ yeast transformed with a vector control (−) or with the ADD66myc expression plasmid (+) and total proteins were resolved by SDS-PAGE. The blots were probed with anti-myc and anti-Sec61 anti-sera. Duplicate colonies were analyzed and are shown here.
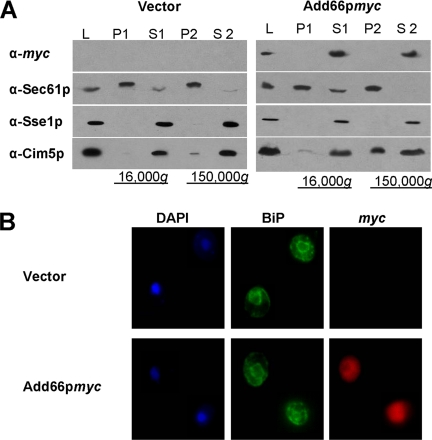
To determine Add66p's residence, lysates were prepared from cells constitutively expressing epitope-tagged Add66p or containing a vector control, and the lysates were analyzed by differential centrifugation. Add66p was found exclusively in a high-speed supernatant (Figure 3A, S2), suggesting cytoplasmic residence. Similar results were observed in strains expressing Add66p-myc expressed under the endogenous promoter (our unpublished data; but see Figure 5). Cytoplasmic residence was further supported through the use of indirect immunofluorescence microscopy: The anti-myc fluorescent signal exhibited a diffuse, cytoplasmic staining, unlike a marker for the ER, BiP, which was evident in perinuclear and peripheral patterns (Figure 3B).
Figure 3.
Add66p is cytosolic. (A) add66Δ strains were transformed with a control plasmid or with a plasmid engineered for the constitutive expression of Add66p-myc. Cell lysates (L) were subjected to 16,000 × g and 150,000 × g centrifugations. Total proteins in the pellets (P1 and P2) and supernatants (S1 and S2) were resolved by SDS-PAGE, and then they were analyzed by Western blot analysis with anti-myc, anti-Sec61p (ER membrane protein), anti-Sse1p (a primarily cytosolic protein; Goeckeler et al., 2002), and anti-Cim5p (a regulatory subunit of the 26S proteasome with cytosolic and ER membrane subcellular localizations) antisera. (B) Indirect immunofluorescence of add66Δ strains transformed with the plasmids described in A were stained with 4,6-diamidino-2-phenylindole (nuclear staining), and then they were probed with anti-BiP (ER perinuclear and peripheral staining) and with anti-myc antisera, and signals were detected as described in Materials and Methods.
Add66p Is Required for Maximal Proteasome Activity
A clue to Add66p's function was provided by a large-scale yeast proteomic analysis in which the gene product was found in a multiprotein complex that included Pre1p (Ho et al., 2002). Pre1p is a subunit in the 20S proteasome core that is one of two subunits that facilitates CTL activity (Heinemeyer et al., 1991; Hilt et al., 1993). More recently, Add66p was identified in a multiprotein complex with Pre5p (Krogan et al., 2006), a nonproteolytic subunit of the 20S proteasome (Heinemeyer et al., 1994), and with Ump1p, a protein required for the maturation of the proteasome core particle (Ramos et al., 1998). These data suggested to us why add66Δ yeast accumulated polyubiquitinated proteins and why synthetic growth defects were observed when mutations in ADD66 and IRE1 were combined and ER stress was induced (Figure 1).
To test directly whether the deletion of ADD66 affected proteasome function, the three proteolytic activities of the proteasome were measured in clarified cytosolic extracts derived from ADD66 and add66Δ strains by modifying an established fluorescence assay (see Materials and Methods). In each set of assays, the background (spontaneous) hydrolysis of the fluorogenic substrate was subtracted, and the resulting values for activity in the mutant lysates were normalized to the activity in the wild-type lysates. As shown in Figure 4A, we observed that the proteasome's CTL activity—which constitutes most of the proteasome's activity—was markedly reduced in extracts from add66Δ yeast (BY4742). To ensure that this effect was not strain specific, the ADD66 gene was ablated in another strain background (W303) and a similar reduction in CTL activity was observed. As a control for these assays, we noted that all three activities were compromised in extracts prepared from a cim5-1 strain (Figure 4A, gray bars), which is known to exhibit delayed protein turnover, an elongated G2/M cell cycle transition, and growth arrest under various stress conditions (Ghislain et al., 1993; Rubin et al., 1998). As another set of controls for this assay, the time dependence of the CTL, TL, and PGPH activity in each extract was measured and compared with the activity in highly enriched 26S proteasomes (Supplemental Figure 2).
We found that the constitutive, strong expression of Add66p-myc in the add66Δ strain complemented the CTL defect and restored activity to wild-type levels (Figure 4, B and C). However, it was unknown whether overexpression of Add66p-myc would present some unforeseen secondary affect on proteasome activity. Therefore, Add66p-myc was also expressed in strains under the control of its endogenous promoter (see Materials and Methods), and lysates were prepared from add66Δ yeast containing a vector control and the expression vector. Here, too, we noted that the CTL defect was complemented (Supplemental Figure 2, ADD66myc).
Combined with the fact that Add66p-myc rescues the A1PiZ degradation defect in add66Δ yeast, these data indicate that Add66p is required for maximal CTL activity, and they strongly suggest that the A1PiZ degradation defect in add66Δ yeast arises from defects in proteasome function.
20S Precursors Accumulate in Yeast Deleted for ADD66
The simplest explanation for the reduced CTL activity in extracts derived from add66Δ yeast is that the number of proteasomes is decreased in this strain. To address this possibility, quantitative immunoblotting was used to measure the levels of six 20S subunits—through the use of a nonspecific polyclonal antiserum—and one 19S subunit (Cim5p) in the add66Δ and wild-type strains (Supplemental Figure 3). However, no significant difference in the relative levels of these subunits was detected.
Pre1p is one of the two essential 20S subunits required for the CTL activity of the proteasome (Heinemeyer et al., 1991; Hilt et al., 1993). Because reduced CTL activity was observed in extracts from ADD66-deleted cells (Figure 4A), it was possible that the absence of the corresponding protein might grossly alter the conformation of Pre1p's substrate binding site. To test this hypothesis, the CTL activity was assayed in extracts from wild-type and add66Δ strains in the presence of increasing concentrations of MG-132 and epoximycin. MG-132 and epoxomicin specifically block the CTL activities in nearly irreversible noncovalent and covalent manners, respectively (Gaczynska and Osmulski, 2005). As shown in Supplemental Figure 4, there was no difference in the apparent KI values for these inhibitors (∼5 × 10−7 M) when titrated into cytosols prepared from wild-type or add66Δ yeast. This result suggests that the conformation of the Pre1p substrate-binding site was not radically altered.
The 26S proteasome is made up of at least 31 different subunits that combine in a spatially and temporally defined manner in various intermediate complexes (Ramos et al., 1998; Voges et al., 1999; Tone et al., 2000; Hirano et al., 2005; Li et al., 2007). Although little is known about the exact mechanism by which the proteins in the cap and the seven distinct α and seven β subunits in the core assemble (but see Discussion), two proteins were identified previously that are required for the proper maturation of the core particle in yeast: Nob1p and Ump1p (Ramos et al., 1998; Tone et al., 2000). Yeast with mutations in NOB1 exhibit defects in the processing of 20S β subunits, which are the central, proteolytic subunits, and in the assembly of the 20S and 26S particles (Tone and Toh, 2002). In addition, as described above, Ump1p coprecipitates with Add66p in a multiprotein complex (Krogan et al., 2006), and mutations in UMP1 affect the function of all three proteolytic activities due to defects in β subunit maturation (Ramos et al., 1998). Thus, it was possible that Add66p participates in proteasome subunit maturation and/or assembly. Moreover, we found that ADD66 is ∼20% identical to PAC2 (Supplemental Figure 5), which facilitates the assembly of the 26S proteasome in mammals (Hirano et al., 2005). PAC2 is also known as CLAST3 and HCCA3, a gene that is up-regulated in hepatic cancers (Wang et al., 2001; Bahar et al., 2002).
Based on these observations and our data presented above, we examined whether 20S proteasome assembly intermediates accumulate in yeast lacking Add66p. To this end, extracts derived from the ADD66 or the add66Δ strains transformed with a control plasmid or with the Add66-myc expression plasmid were fractionated on a glycerol density gradient. A low percentage (4–25%) glycerol gradient was used in this experiment to better resolve early proteasome intermediates, and it was used previously to note assembly intermediates in mammalian cells when PAC2 expression was silenced (Hirano et al., 2005). We first observed that the majority of 20S subunits and a component of the 19S cap resolved at fractions in the gradient that corresponded to particles with a molecular mass of ∼2.5 MDa (Figure 5). This value is in good agreement with the native size of the 26S proteasome. Second, we observed a 20S immunoreactive protein in a distinct, lighter fraction when extracts from the add66Δ strain were examined (see downward arrow, add66Δ, α-20S). This species, which migrated at ∼300 kDa, was absent when extracts from wild-type cells or from add66Δ yeast expressing endogenous levels of Add66p-myc were examined. Third, we found that the same 20S immunoreactive species was present when extracts were resolved from yeast lacking Ump1p (ump1Δ, α-20S), a factor required for 20S maturation (Ramos et al., 1998). Fourth, when extracts were examined from add66Δ or ump1Δ yeast, a 20S, slower-migrating doublet was observed that fractionated at the native size for 26S proteasomes (see downward bracket). Previous work demonstrated that one of the species in the doublet represents an unprocessed β subunit in the 20S proteasome (Ramos et al., 1998). This result suggests similar defects during 20S subunit processing in the add66Δ and ump1Δ strains. And fifth, we found that Add66p resided at a position consistent with a molecular mass of ∼150–300 kDa, suggesting that this ∼30-kDa protein is a component of a multiprotein complex and/or forms higher order oligomers (see Discussion; Li et al., 2007). Overexpression of Add66p-myc was also able to decrease the amount of the immature 20S doublet and of the ∼300-kDa assembly intermediate (Supplemental Figure 6), although the overexpressed Add66p-myc instead resolved in fractions corresponding to a molecular mass of ∼66–150 kDa. Together, these data indicate that yeast deleted for ADD66 accumulate an intermediate in the 20S assembly pathway, as observed in ump1Δ and nob1Δ strains (Ramos et al., 1998; Tone et al., 2000).
Add66p Is Degraded by the 26S Proteasome
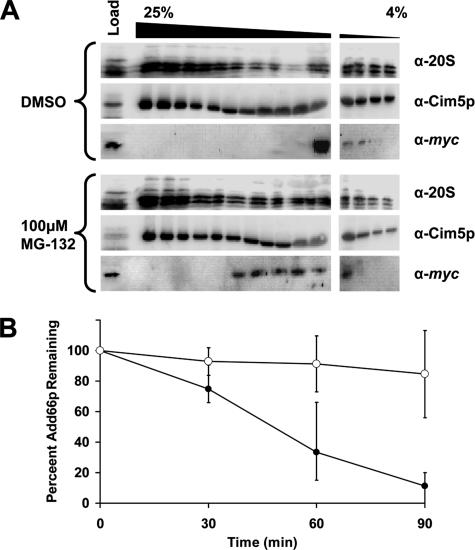
Other proteasome chaperones interact transiently with early proteasome assembly intermediates, and in some cases, they are then degraded by the proteasome (Ramos et al., 1998; Tone et al., 2000; Tone and Toh, 2002; Hirano et al., 2005). We therefore incubated a pdr5Δ Add66p-myc expressing strain with either dimethyl sulfoxide (DMSO) or MG-132. The pdr5Δ strain was used based on the fact that this mutation, like other similarly used mutations, disables a plasma membrane drug efflux pump; therefore, the effect of proteasome inhibitors is magnified (Balzi et al., 1994; Lee and Goldberg, 1996). As shown in Figure 6A, Add66p-myc shifts to fractions containing complexes of greater molecular masses, which contain 20S subunits, when extracts were prepared from cells treated with MG-132. Treatment with MG-132 also results in the accumulation of the slower migrating, 20S “doublets,” as seen when extracts were examined from add66Δ and ump1Δ strains (Figure 5) when the blots were overexposed (our unpublished data). Next, to examine whether Add66p—like several other proteasome chaperones—is degraded by the proteasome, a cycloheximide chase was performed in the presence or absence of MG-132. Once again, the pdr5Δ strain was transformed with a plasmid engineered for the constitutive expression of Add66p-myc, and the cells were incubated with either DMSO or MG-132 for 1 h before the chase (Figure 6B). Although Add66p was rapidly degraded in cells treated with DMSO, we discovered that MG-132 addition resulted in a profound, reproducible stabilization of the protein. To better visualize Add66p-myc in this experiment, it was necessary to overexpress the protein; thus, these data should be interpreted with caution. Nevertheless, the results strongly suggest that Add66p is a proteasome substrate.
Figure 6.
Add66p is degraded by the 26S proteasome. A pdr5Δ strain transformed with a plasmid designed for the constitutive expression of ADD66-myc was incubated either with 100 μM MG-132 or the equivalent volume of DMSO for 1 h at 30°C. (A) Cell extracts from each strain were prepared, and 5 mg of protein was resolved on a linear glycerol gradient (4–25%), and fractions were collected. Proteins in every other fraction were immunoblotted for 20S subunits, a component of the 19S subunit (Cim5p), and Add66p-myc. Molecular mass markers, which were analyzed in parallel, are indicated below the gel. Note that these blots were purposely overexposed (compared with those in Figure 5). (B) The strains described in A were harvested at the indicated time points after the addition of cycloheximide, and cell extracts were prepared and subjected to SDS-PAGE and immunoblotted for Add66p-myc and Sec61p (as a loading control). The amount of Add66p-myc at the start of the chase in each strain, after standardization to the amount of Sec61p at each time point, was set to 100%. ○, MG-132; •, DMSO control.
DISCUSSION
In this report, we present the characterization of Add66p, a protein previously implicated in the ERAD of A1PiZ in yeast (Palmer et al., 2003). We found that strains deleted for ADD66 accumulate polyubiquintinated proteins and grow poorly when they are unable to mount a UPR and challenged with a UPR-inducing agent. These data may be explained by our discovery that the CTL activity of the proteasome is compromised in add66Δ yeast. Yeast deleted for ADD66 also accumulate some of the same proteasome assembly intermediates as those observed when the gene encoding the Ump1p proteasome assembly factor is disabled. Our observations are consistent with the fact that Add66p is found in a multiprotein complex that includes Pre1p and Pre5p—proteins embedded within the 20S core particle—as well as Ump1p itself (Ho et al., 2002; Krogan et al., 2006). Based on these data, we propose that Add66p is a yeast PAC and that it is vital for maximal proteasome function.
The recent characterization of PACs in both yeast and mammals have yielded new insight into the pathway of proteasome biogenesis (Chen and Hochstrasser, 1995, 1996; Heinemeyer et al., 1997; Ramos et al., 1998, 2004; Arendt and Hochstrasser, 1999; Burri et al., 2000; Griffin et al., 2000; Tone et al., 2000; Witt et al., 2000; Tone and Toh, 2002; Hirano et al., 2005, 2006; Li et al., 2007). The symmetrical, 20S barrel-shaped core particle (CP) of the proteasome is made up of two half-proteasome (15S) complexes (Nandi et al., 1997). These half-proteasomes contain a ring of α subunits and a ring of β subunits, three of which are responsible for the CP's proteolytic activity and must be processed. In mammals, after the two 15S complexes combine, the “pro” regions in the β subunits are cleaved and a cohort of PACs—including hUmp1 (also known as POMP or Proteassemblin), PAC1, PAC2, and PAC3—participate in complex formation. PAC1 and PAC2 have been proposed to maintain the assembly competence of the α ring, and they are degraded after assembly of the two half-proteasomes; in contrast, PAC3 assists in the assembly of the β ring before dissociating. PAC3 also recruits hUmp1, which later catalyzes the dimerization of the two half-proteasomes. In yeast, Nob1p facilitates the association of the 19S particle to the CP (Tone and Toh, 2002; Hirano et al., 2005, 2006). Based on our results and data derived from studies of PAC2 function in mammalian cells (Hirano et al., 2005, 2006), we propose that Add66p is the S. cerevisiae homologue of PAC2: 1) the proteins are ∼20% identical throughout their sequence (Supplemental Figure 5), and several conserved residues are present in PAC2 homologues in all species (our unpublished data); 2) both factors are proteasome substrates; 3) lowering protein levels (by RNA interference in mammalian cells) or completely ablating the gene (in yeast) leads to the accumulation of CP precursors, increases the concentration of polyubiquitinated proteins, and decreases the proteasome's CTL activity; and 4) MG-132 treatment in each cell type results in the accumulation of the protein in denser fractions after glycerol gradient centrifugation.
While this article was in revision, Hochstrasser and colleagues reported that Add66p (which they named Pba2, for proteasome biogenesis-associated polypeptide 2) associates with another protein (Pba1) to form a stable complex (Li et al., 2007); the Add66p–Pba1 complex resolves with distinct proteasome assembly intermediates, and it was found that deletion of ADD66 restores the growth of an ump1Δ mutant, which is consistent with antagonistic action between Ump1p and the PACs. They also reported that the concentration of a pro-β5 assembly intermediate increased in the add66Δ mutant. Combined with the results reported here and with studies in mammalian cells (Hirano et al., 2005, 2006), these data provide support that Pba1 and Add66p are the yeast homologues of PAC1 and PAC2, respectively.
In contrast to prior studies of PAC function in mammalian cells and yeast, we have determined the importance of Add66p on the degradation of a distinct class of proteins. More specifically, we have discovered a link between PAC activity and ERAD. In previous work, we noted that Add66p facilitates the degradation of some ERAD substrates (A1PiZ and the cystic fibrosis transmembrane conductance regulator) but not others (carboxypeptidase Y* and pro-α factor) (Palmer et al., 2003). At first glance, these data and the results presented in this article may seem at-odds, i.e., if the proteasome is partially disabled in add66Δ mutants, why is the turnover of all ERAD substrates not affected similarly, especially because any one of the proteasome's three activities may be sufficient to remove an ERAD substrate (Oberdorf et al., 2001)? Consistent with data from other laboratories, we suggest two possible answers to this question.
First, we propose that the rate-limiting step during ERAD differs for unique classes of substrates. Notably, ERAD can be envisaged as a five-step temporal and—for some substrates—spatial processes, consisting of substrate identification, retrotranslocation, polyubiquintination, deubquintination, and degradation. In reality, each of these steps is likely further broken down into multiple, discrete kinetic events. Regardless, the overall rate of substrate degradation, as for any multistep process, is established by the rate-limiting step. During the degradation of some proteasome substrates, it has been shown that deubiquitination is rate limiting (Yao and Cohen, 2002; Guterman and Glickman, 2004; Hanna et al., 2006). During ERAD, the removal of a specific substrate class (proteins in the “ERAD-L” family) takes significantly longer than the removal of other substrates (proteins in the “ERAD-C” family), possibly because only ERAD-L substrates can transit to the Golgi apparatus before ER retrieval and degradation (Vashist et al., 2001). Thus, effects arising from impaired proteasome function (Figure 4) might be masked by the robust activity of factors acting at an alternative, rate-limiting step.
Second, it has been proposed that proteasome isoforms exist within cells: Those that are stable and those that recycle (Tone and Toh, 2002). In fact, it was reported later that the proteasome disassembles after ATP hydrolysis and substrate degradation (Babbitt et al., 2005). Combined with the concept that specific proteasome subpopulations may recognize distinct classes of substrates (Fujimuro et al., 1998), it is possible that A1PiZ is degraded by only one of these “classes.” Formally, then, deletion of ADD66 might compromise the assembly of a unique proteasome subset that is required for the degradation of some but not all substrates. Alternatively, the absence of Add66p might alter the proteasome's interaction with the ER membrane. Different ERAD substrates might be delivered from the ER in different conformations, only some of which may require close apposition of the proteasome with the ER and possibly Sec61p (Kalies et al., 2005).
A surprising aspect of this study was that deletion of ADD66 seemed to affect the CTL activity of the 26S proteasome, whereas there were no obvious defects in the TL and PGPH activities. This is in contrast to the fact that all three activities decrease in an ump1Δ strain (Ramos et al., 1998). We propose two explanations for this phenomenon. First, Ump1p may have a global effect on proteasome activity due to its role as an assembly checkpoint during the dimerization of half proteasomes (Li et al., 2007). Thus, deletion of UMP1 may result in a significant accumulation of defective dimers or other intermediates, thereby reducing the level of functional 26S proteasomes. In contrast, the Add66p dependence during proteasome maturation may give rise to subtle changes in proteasome assembly or even in the conformations of individual, catalytic subunits, or associated subunits. Thus, there might be qualitative, perhaps minor differences in the architecture of CTL-requiring subunits in proteasomes between wild-type and add66Δ cells. Consistent with this model, more severe proteasome assembly defects were noted in ump1Δ versus add66Δ (pba2Δ) mutants when intermediates were resolved by gel filtration chromatography (Ramos et al., 1998; Li et al., 2007). Second, more trivially, we note that neither the TL nor PGPH activity was measured when specific proteasome assembly factors were ablated (Tone and Toh, 2002; Hirano et al., 2005, 2006). Therefore, it remains possible that unique effects on proteolytic activities do exist when related PACs are disabled, at least in some cell types or under specific conditions.
One goal of our long-term studies is to identify conserved yeast genes that impact the ERAD of distinct substrates. This undertaking is particularly relevant for A1PiZ, given that only a small percentage of A1PiZ homozygotes develop severe liver disease (Sveger, 1988). It is thought that both environmental and genetic modifiers play a role in determining the onset and severity of liver disease (Wu et al., 1994; Perlmutter, 2002). Therefore, it is critical that genetic polymorphisms or secondary mutations that alter A1PiZ quality control are identified. Intriguingly, we found that ∼15% of add66Δ cells expressing A1PiZ are inviable (data not shown; but see Supplemental Figure 7 for an example). In principle, one might be able to co-opt this phenotype to screen for second-site or suppressor mutations. It is also possible that the observed genetic penetrance arises from stochastic variability in some cells relative to others: Recent insight into phenotypic variation have led to a greater understanding of the “noise” that accounts for the stochastic nature of protein production and phenotypic variation, especially in single-cell organisms (Raser and O'Shea, 2004; Newman et al., 2006; Samoilov et al., 2006).
Another explanation for why a subpopulation of cells exhibits growth defects (and why a subpopulation of individuals with ATD develops liver disease) is that alternate mechanisms exist to clear the ER of A1PiZ. Indeed, it is now established that the autophagic pathway can degrade A1PiZ in mammalian cells (Teckman and Perlmutter, 2000; Kamimoto et al., 2006) and in yeast (Kruse et al., 2006) when the protein is overexpressed. However, we found that autophagy is active in add66Δ cells (Supplemental Figure 1), suggesting that variations in this alternate mode of protein quality control do not contribute to the toxic effects of A1PiZ, at least in yeast. In the future, it will be important to measure how variations in the efficiency of autophagy—as well as variations in the relative steady-state levels of A1PiZ—impact A1PiZ clearance and liver disease in individuals afflicted with ATD.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ray Deshaies, Jürgen Dohmen, Jiyang O-Wang, Mark Hochstrasser, and William Saunders for various strains and plasmids used in this study. We also thank Gerry Apodaca, Avrom Caplan, Jeffrey Hildebrand, Jeffrey Lawrence, Carl Mann, Kunio Nakatsukasa, and Ora Weisz for reagents and equipment that were critical for the success of this project. This work was supported by grant GM-75061 (to J.L.B.) and grants HL-037784 and DK-052526 (to D.H.P.) from the National Institutes of Health; by grant MCB-0110331 (to J.L.B. and A.A.M.) from the National Science Foundation; and by a grant from the Alpha-1 Foundation (to J.L.B. and D.H.P.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-01-0034) on July 18, 2007.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Adams A., Gottschling D. E., Kaiser C. A., Stearns T. Plainview, NY: Cold Spring Harbor Laboratory Press; 1997. Methods in Yeast Genetics. [Google Scholar]
- Arendt C. S., Hochstrasser M. Eukaryotic 20S proteasome catalytic subunit propeptides prevent active site inactivation by N-terminal acetylation and promote particle assembly. EMBO J. 1999;18:3575–3585. doi: 10.1093/emboj/18.13.3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aridor M., Hannan L. A. Traffic jam: a compendium of human diseases that affect intracellular transport processes. Traffic. 2000;1:836–851. doi: 10.1034/j.1600-0854.2000.011104.x. [DOI] [PubMed] [Google Scholar]
- Aridor M., Hannan L. A. Traffic jams II: an update of diseases of intracellular transport. Traffic. 2002;3:781–790. doi: 10.1034/j.1600-0854.2002.31103.x. [DOI] [PubMed] [Google Scholar]
- Babbitt S. E., et al. ATP hydrolysis-dependent disassembly of the 26S proteasome is part of the catalytic cycle. Cell. 2005;121:553–565. doi: 10.1016/j.cell.2005.03.028. [DOI] [PubMed] [Google Scholar]
- Bahar R., O-Wang J., Kawamura K., Seimiya M., Wang Y., Hatano M., Okada S., Tokuhisa T., Watanabe T., Tagawa M. Growth retardation, polyploidy, and multinucleation induced by Clast3, a novel cell cycle-regulated protein. J. Biol. Chem. 2002;277:40012–40019. doi: 10.1074/jbc.M205345200. [DOI] [PubMed] [Google Scholar]
- Balzi E., Wang M., Leterme S., Van Dyck L., Goffeau A. PDR5, a novel yeast multidrug resistance conferring transporter controlled by the transcription regulator PDR1. J. Biol. Chem. 1994;269:2206–2214. [PubMed] [Google Scholar]
- Bernales S., McDonald K. L., Walter P. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 2006;4:e423. doi: 10.1371/journal.pbio.0040423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Hieter P., Boeke J. D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Brantly M., Courtney M., Crystal R. G. Repair of the secretion defect in the Z form of alpha 1-antitrypsin by addition of a second mutation. Science. 1988;242:1700–1702. doi: 10.1126/science.2904702. [DOI] [PubMed] [Google Scholar]
- Brodsky J. L., Lawrence J. G., Caplan A. J. Mutations in the cytosolic DnaJ homologue, YDJ1, delay and compromise the efficient translation of heterologous proteins in yeast. Biochemistry. 1998;37:18045–18055. doi: 10.1021/bi980900g. [DOI] [PubMed] [Google Scholar]
- Brodsky J. L., Werner E. D., Dubas M. E., Goeckeler J. L., Kruse K. B., McCracken A. A. The requirement for molecular chaperones during endoplasmic reticulum-associated protein degradation demonstrates that protein export and import are mechanistically distinct. J. Biol. Chem. 1999;274:3453–3460. doi: 10.1074/jbc.274.6.3453. [DOI] [PubMed] [Google Scholar]
- Burri L., Hockendorff J., Boehm U., Klamp T., Dohmen R. J., Levy F. Identification and characterization of a mammalian protein interacting with 20S proteasome precursors. Proc. Natl. Acad. Sci. USA. 2000;97:10348–10353. doi: 10.1073/pnas.190268597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabral C. M., Liu Y., Moremen K. W., Sifers R. N. Organizational diversity among distinct glycoprotein endoplasmic reticulum-associated degradation programs. Mol. Biol. Cell. 2002;13:2639–2650. doi: 10.1091/mbc.E02-02-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson J. A., Rogers B. B., Sifers R. N., Finegold M. J., Clift S. M., DeMayo F. J., Bullock D. W., Woo S. L. Accumulation of PiZ alpha 1-antitrypsin causes liver damage in transgenic mice. J. Clin. Invest. 1989;83:1183–1190. doi: 10.1172/JCI113999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrell R. W., Lomas D. A. Alpha1-antitrypsin deficiency–a model for conformational diseases. N. Engl. J. Med. 2002;346:45–53. doi: 10.1056/NEJMra010772. [DOI] [PubMed] [Google Scholar]
- Chen P., Hochstrasser M. Biogenesis, structure and function of the yeast 20S proteasome. EMBO J. 1995;14:2620–2630. doi: 10.1002/j.1460-2075.1995.tb07260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P., Hochstrasser M. Autocatalytic subunit processing couples active site formation in the 20S proteasome to completion of assembly. Cell. 1996;86:961–972. doi: 10.1016/s0092-8674(00)80171-3. [DOI] [PubMed] [Google Scholar]
- Coughlan C. M., Brodsky J. L. Yeast as a model system to investigate protein conformational diseases. Methods Mol. Biol. 2003;232:77–90. doi: 10.1385/1-59259-394-1:77. [DOI] [PubMed] [Google Scholar]
- Coughlan C. M., Walker J. L., Cochran J. C., Wittrup K. D., Brodsky J. L. Degradation of mutated bovine pancreatic trypsin inhibitor in the yeast vacuole suggests post-endoplasmic reticulum protein quality control. J. Biol. Chem. 2004;279:15289–15297. doi: 10.1074/jbc.M309673200. [DOI] [PubMed] [Google Scholar]
- Cox J. S., Shamu C. E., Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- Credle J. J., Finer-Moore J. S., Papa F. R., Stroud R. M., Walter P. On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 2005;102:18773–18784. doi: 10.1073/pnas.0509487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker D. J., Khan M. I., Marsh J., Butt T. R., Crooke S. T. Chemical synthesis and expression of a cassette adapted ubiquitin gene. J. Biol. Chem. 1987;262:3524–3527. [PubMed] [Google Scholar]
- Ellgaard L., Helenius A. Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 2003;4:181–191. doi: 10.1038/nrm1052. [DOI] [PubMed] [Google Scholar]
- Evan G. I., Lewis G. K., Ramsay G., Bishop J. M. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol. Cell. Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fewell S. W., Travers K. J., Weissman J. S., Brodsky J. L. The action of molecular chaperones in the early secretory pathway. Annu. Rev. Genet. 2001;35:149–191. doi: 10.1146/annurev.genet.35.102401.090313. [DOI] [PubMed] [Google Scholar]
- Foreman R. C., Judah J. D., Colman A. Xenopus oocytes can synthesize but do not secrete the Z variant of human alpha 1-antitrypsin. FEBS Lett. 1984;168:84–88. doi: 10.1016/0014-5793(84)80211-2. [DOI] [PubMed] [Google Scholar]
- Frand A. R., Kaiser C. A. The ERO1 gene of yeast is required for oxidation of protein dithiols in the endoplasmic reticulum. Mol. Cell. 1998;1:161–170. doi: 10.1016/s1097-2765(00)80017-9. [DOI] [PubMed] [Google Scholar]
- Fujimuro M., Tanaka K., Yokosawa H., Toh-e A. Son1p is a component of the 26S proteasome of the yeast Saccharomyces cerevisiae. FEBS Lett. 1998;423:149–154. doi: 10.1016/s0014-5793(98)00084-2. [DOI] [PubMed] [Google Scholar]
- Gaczynska M., Osmulski P. A. Small-molecule inhibitors of proteasome activity. Methods Mol. Biol. 2005;301:3–22. doi: 10.1385/1-59259-895-1:003. [DOI] [PubMed] [Google Scholar]
- Ghislain M., Udvardy A., Mann C. S. cerevisiae 26S protease mutants arrest cell division in G2/metaphase. Nature. 1993;366:358–362. doi: 10.1038/366358a0. [DOI] [PubMed] [Google Scholar]
- Glickman M. H., Rubin D. M., Fried V. A., Finley D. The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol. Cell. Biol. 1998;18:3149–3162. doi: 10.1128/mcb.18.6.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeckeler J. L., Stephens A., Lee P., Caplan A. J., Brodsky J. L. Overexpression of yeast Hsp110 homolog Sse1p suppresses ydj1-151 thermosensitivity and restores Hsp90-dependent activity. Mol. Biol. Cell. 2002;13:2760–2770. doi: 10.1091/mbc.02-04-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin T. A., Slack J. P., McCluskey T. S., Monaco J. J., Colbert R. A. Identification of proteassemblin, a mammalian homologue of the yeast protein, Ump1p, that is required for normal proteasome assembly. Mol. Cell. Biol. Res. Commun. 2000;3:212–217. doi: 10.1006/mcbr.2000.0213. [DOI] [PubMed] [Google Scholar]
- Guterman A., Glickman M. H. Complementary roles for Rpn11 and Ubp6 in deubiquitination and proteolysis by the proteasome. J. Biol. Chem. 2004;279:1729–1738. doi: 10.1074/jbc.M307050200. [DOI] [PubMed] [Google Scholar]
- Hanna J., Hathaway N. A., Tone Y., Crosas B., Elsasser S., Kirkpatrick D. S., Leggett D. S., Gygi S. P., King R. W., Finley D. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell. 2006;127:99–111. doi: 10.1016/j.cell.2006.07.038. [DOI] [PubMed] [Google Scholar]
- He C., Song H., Yorimitsu T., Monastyrska I., Yen W. L., Legakis J. E., Klionsky D. J. Recruitment of Atg9 to the preautophagosomal structure by Atg11 is essential for selective autophagy in budding yeast. J. Cell Biol. 2006;175:925–935. doi: 10.1083/jcb.200606084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemeyer W., Fischer M., Krimmer T., Stachon U., Wolf D. H. The active sites of the eukaryotic 20 S proteasome and their involvement in subunit precursor processing. J. Biol. Chem. 1997;272:25200–25209. doi: 10.1074/jbc.272.40.25200. [DOI] [PubMed] [Google Scholar]
- Heinemeyer W., Kleinschmidt J. A., Saidowsky J., Escher C., Wolf D. H. Proteinase yscE, the yeast proteasome/multicatalytic-multifunctional proteinase: mutants unravel its function in stress induced proteolysis and uncover its necessity for cell survival. EMBO J. 1991;10:555–562. doi: 10.1002/j.1460-2075.1991.tb07982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemeyer W., Trondle N., Albrecht G., Wolf D. H. PRE5 and PRE6, the last missing genes encoding 20S proteasome subunits from yeast? Indication for a set of 14 different subunits in the eukaryotic proteasome core. Biochemistry. 1994;33:12229–12237. doi: 10.1021/bi00206a028. [DOI] [PubMed] [Google Scholar]
- Hilt W., Enenkel C., Gruhler A., Singer T., Wolf D. H. The PRE4 gene codes for a subunit of the yeast proteasome necessary for peptidylglutamyl-peptide-hydrolyzing activity. Mutations link the proteasome to stress- and ubiquitin-dependent proteolysis. J. Biol. Chem. 1993;268:3479–3486. [PubMed] [Google Scholar]
- Hirano Y., Hayashi H., Iemura S., Hendil K. B., Niwa S., Kishimoto T., Kasahara M., Natsume T., Tanaka K., Murata S. Cooperation of multiple chaperones required for the assembly of mammalian 20S proteasomes. Mol. Cell. 2006;24:977–984. doi: 10.1016/j.molcel.2006.11.015. [DOI] [PubMed] [Google Scholar]
- Hirano Y., Hendil K. B., Yashiroda H., Iemura S., Nagane R., Hioki Y., Natsume T., Tanaka K., Murata S. A heterodimeric complex that promotes the assembly of mammalian 20S proteasomes. Nature. 2005;437:1381–1385. doi: 10.1038/nature04106. [DOI] [PubMed] [Google Scholar]
- Ho Y., et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- Kabani M., Beckerich J. M., Brodsky J. L. Nucleotide exchange factor for the yeast Hsp70 molecular chaperone Ssa1p. Mol. Cell. Biol. 2002;22:4677–4689. doi: 10.1128/MCB.22.13.4677-4689.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalies K. U., Allan S., Sergeyenko T., Kroger H., Romisch K. The protein translocation channel binds proteasomes to the endoplasmic reticulum membrane. EMBO J. 2005;24:2284–2293. doi: 10.1038/sj.emboj.7600731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kametaka S., Okano T., Ohsumi M., Ohsumi Y. Apg14p and Apg6/Vps30p form a protein complex essential for autophagy in the yeast, Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:22284–22291. doi: 10.1074/jbc.273.35.22284. [DOI] [PubMed] [Google Scholar]
- Kamimoto T., Shoji S., Hidvegi T., Mizushima N., Umebayashi K., Perlmutter D. H., Yoshimori T. Intracellular inclusions containing mutant alpha1-antitrypsin Z are propagated in the absence of autophagic activity. J. Biol. Chem. 2006;281:4467–4476. doi: 10.1074/jbc.M509409200. [DOI] [PubMed] [Google Scholar]
- Kim J., Lee K. N., Yi G. S., Yu M. H. A thermostable mutation located at the hydrophobic core of α1-antitrypsin suppresses the folding defect of the Z-type variant. J. Biol. Chem. 1995;270:8597–8601. doi: 10.1074/jbc.270.15.8597. [DOI] [PubMed] [Google Scholar]
- Kostova Z., Wolf D. H. For whom the bell tolls: protein quality control of the endoplasmic reticulum and the ubiquitin-proteasome connection. EMBO J. 2003;22:2309–2317. doi: 10.1093/emboj/cdg227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan N. J., et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Kruse K. B., Brodsky J. L., McCracken A. A. Characterization of an ERAD gene as VPS30/ATG6 reveals two alternative and functionally distinct protein quality control pathways: one for soluble Z variant of human α-1 proteinase inhibitor (A1PiZ) and another for aggregates of A1PiZ. Mol. Biol. Cell. 2006;17:203–212. doi: 10.1091/mbc.E04-09-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. H., Goldberg A. L. Selective inhibitors of the proteasome-dependent and vacuolar pathways of protein degradation in Saccharomyces cerevisiae. J. Biol. Chem. 1996;271:27280–27284. doi: 10.1074/jbc.271.44.27280. [DOI] [PubMed] [Google Scholar]
- Leggett D. S., Hanna J., Borodovsky A., Crosas B., Schmidt M., Baker R. T., Walz T., Ploegh H., Finley D. Multiple associated proteins regulate proteasome structure and function. Mol. Cell. 2002;10:495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- Li X., Kusmierczyk A. R., Wong P., Emili A., Hochstrasser M. beta-Subunit appendages promote 20S proteasome assembly by overcoming an Ump1-dependent checkpoint. EMBO J. 2007;26:2339–2349. doi: 10.1038/sj.emboj.7601681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomas D. A., Evans D. L., Finch J. T., Carrell R. W. The mechanism of Z alpha 1-antitrypsin accumulation in the liver. Nature. 1992;357:605–607. doi: 10.1038/357605a0. [DOI] [PubMed] [Google Scholar]
- Mast A. E., Enghild J. J., Salvesen G. Conformation of the reactive site loop of alpha 1-proteinase inhibitor probed by limited proteolysis. Biochemistry. 1992;31:2720–2728. doi: 10.1021/bi00125a012. [DOI] [PubMed] [Google Scholar]
- McCracken A. A., Karpichev I. V., Ernaga J. E., Werner E. D., Dillin A. G., Courchesne W. E. Yeast mutants deficient in ER-associated degradation of the Z variant of alpha-1-protease inhibitor. Genetics. 1996;144:1355–1362. doi: 10.1093/genetics/144.4.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken A. A., Kruse K. B. Selective protein degradation in the yeast exocytic pathway. Mol. Biol. Cell. 1993;4:729–736. doi: 10.1091/mbc.4.7.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken A. A., Kruse K. B., Brown J. L. Molecular basis for defective secretion of the Z variant of human alpha-1-proteinase inhibitor: secretion of variants having altered potential for salt bridge formation between amino acids 290 and 342. Mol. Cell. Biol. 1989;9:1406–1414. doi: 10.1128/mcb.9.4.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meusser B., Hirsch C., Jarosch E., Sommer T. ERAD: the long road to destruction. Nat. Cell Biol. 2005;7:766–772. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- Mori K., Ma W., Gething M. J., Sambrook J. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell. 1993;74:743–756. doi: 10.1016/0092-8674(93)90521-q. [DOI] [PubMed] [Google Scholar]
- Mornex J. F., Chytil-Weir A., Martinet Y., Courtney M., LeCocq J. P., Crystal R. G. Expression of the alpha-1-antitrypsin gene in mononuclear phagocytes of normal and alpha-1-antitrypsin-deficient individuals. J. Clin. Invest. 1986;77:1952–1961. doi: 10.1172/JCI112524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D., Muller R., Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- Myerowitz R. L., Handzel Z. T., Robbins J. B. Human serum 1-antitrypsin: isolation and demonstration of electrophoretic and immunologic heterogeneity. Clin. Chim. Acta. 1972;39:307–317. doi: 10.1016/0009-8981(72)90049-6. [DOI] [PubMed] [Google Scholar]
- Nandi D., Tahiliani P., Kumar A., Chandu D. The ubiquitin-proteasome system. J. Biosci. 2006;31:137–155. doi: 10.1007/BF02705243. [DOI] [PubMed] [Google Scholar]
- Nandi D., Woodward E., Ginsburg D. B., Monaco J. J. Intermediates in the formation of mouse 20S proteasomes: implications for the assembly of precursor beta subunits. EMBO J. 1997;16:5363–5375. doi: 10.1093/emboj/16.17.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman J. R., Ghaemmaghami S., Ihmels J., Breslow D. K., Noble M., DeRisi J. L., Weissman J. S. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 2006;441:840–846. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- Ng D. T., Spear E. D., Walter P. The unfolded protein response regulates multiple aspects of secretory and membrane protein biogenesis and endoplasmic reticulum quality control. J. Cell Biol. 2000;150:77–88. doi: 10.1083/jcb.150.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberdorf J., Carlson E. J., Skach W. R. Redundancy of mammalian proteasome beta subunit function during endoplasmic reticulum associated degradation. Biochemistry. 2001;40:13397–13405. doi: 10.1021/bi011322y. [DOI] [PubMed] [Google Scholar]
- Ogata M., et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol. Cell. Biol. 2006;26:9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer E. A., Kruse K. B., Fewell S. W., Buchanan S. M., Brodsky J. L., McCracken A. A. Differential requirements of novel A1PiZ degradation deficient (ADD) genes in ER-associated protein degradation. J. Cell Sci. 2003;116:2361–2373. doi: 10.1242/jcs.00439. [DOI] [PubMed] [Google Scholar]
- Parfrey H., Mahadeva R., Lomas D. A. alpha(1)-Antitrypsin deficiency, liver disease and emphysema. Int. J. Biochem. Cell Biol. 2003;35:1009–1014. doi: 10.1016/s1357-2725(02)00250-9. [DOI] [PubMed] [Google Scholar]
- Patil C., Walter P. Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr. Opin. Cell Biol. 2001;13:349–355. doi: 10.1016/s0955-0674(00)00219-2. [DOI] [PubMed] [Google Scholar]
- Perlmutter D. H. Liver injury in alpha1-antitrypsin deficiency: an aggregated protein induces mitochondrial injury. J. Clin. Invest. 2002;110:1579–1583. doi: 10.1172/JCI16787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlmutter D. H., Kay R. M., Cole F. S., Rossing T. H., Van Thiel D., Colten H. R. The cellular defect in alpha 1-proteinase inhibitor (alpha 1-PI) deficiency is expressed in human monocytes and in Xenopus oocytes injected with human liver mRNA. Proc. Natl. Acad. Sci. USA. 1985;82:6918–6921. doi: 10.1073/pnas.82.20.6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard M. G., Travers K. J., Weissman J. S. Ero1p: a novel and ubiquitous protein with an essential role in oxidative protein folding in the endoplasmic reticulum. Mol. Cell. 1998;1:171–182. doi: 10.1016/s1097-2765(00)80018-0. [DOI] [PubMed] [Google Scholar]
- Ramos P. C., Hockendorff J., Johnson E. S., Varshavsky A., Dohmen R. J. Ump1p is required for proper maturation of the 20S proteasome and becomes its substrate upon completion of the assembly. Cell. 1998;92:489–499. doi: 10.1016/s0092-8674(00)80942-3. [DOI] [PubMed] [Google Scholar]
- Ramos P. C., Marques A. J., London M. K., Dohmen R. J. Role of C-terminal extensions of subunits beta2 and beta7 in assembly and activity of eukaryotic proteasomes. J. Biol. Chem. 2004;279:14323–14330. doi: 10.1074/jbc.M308757200. [DOI] [PubMed] [Google Scholar]
- Raser J. M., O'Shea E. K. Control of stochasticity in eukaryotic gene expression. Science. 2004;304:1811–1814. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond R. J., Zellner K. M. Alpha1-Antitrypsin deficiency: incidence and implications. Dimens. Crit. Care Nurs. 2005;24:255–260. doi: 10.1097/00003465-200511000-00001. quiz 261–252. [DOI] [PubMed] [Google Scholar]
- Romisch K. Endoplasmic reticulum-associated degradation. Annu. Rev. Cell Dev. Biol. 2005;21:435–456. doi: 10.1146/annurev.cellbio.21.012704.133250. [DOI] [PubMed] [Google Scholar]
- Rubin D. M., Glickman M. H., Larsen C. N., Dhruvakumar S., Finley D. Active site mutants in the six regulatory particle ATPases reveal multiple roles for ATP in the proteasome. EMBO J. 1998;17:4909–4919. doi: 10.1093/emboj/17.17.4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudnick D. A., Perlmutter D. H. Alpha-1-antitrypsin deficiency: a new paradigm for hepatocellular carcinoma in genetic liver disease. Hepatology. 2005;42:514–521. doi: 10.1002/hep.20815. [DOI] [PubMed] [Google Scholar]
- Saeki Y., Isono E., Toh E. A. Preparation of ubiquitinated substrates by the PY motif-insertion method for monitoring 26S proteasome activity. Methods Enzymol. 2005;399:215–227. doi: 10.1016/S0076-6879(05)99014-9. [DOI] [PubMed] [Google Scholar]
- Samoilov M. S., Price G., Arkin A. P. From fluctuations to phenotypes: the physiology of noise. Sci. STKE. 2006;2006:re17. doi: 10.1126/stke.3662006re17. [DOI] [PubMed] [Google Scholar]
- Savory P. J., Rivett A. J. Leupeptin-binding site (s) in the mammalian multicatalytic proteinase complex. Biochem. J. 1993;289:45–48. doi: 10.1042/bj2890045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayeed A., Ng D. T. Search and destroy: ER quality control and ER-associated protein degradation. Crit. Rev. Biochem. Mol. Biol. 2005;40:75–91. doi: 10.1080/10409230590918685. [DOI] [PubMed] [Google Scholar]
- Schmidt B. Z., Perlmutter D. H. Grp78, Grp94, and Grp170 interact with alpha1-antitrypsin mutants that are retained in the endoplasmic reticulum. Am. J. Physiol. 2005;289:G444–G455. doi: 10.1152/ajpgi.00237.2004. [DOI] [PubMed] [Google Scholar]
- Schroder M., Kaufman R. J. The mammalian unfolded protein response. Annu. Rev. Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- Shamu C. E., Walter P. Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J. 1996;15:3028–3039. [PMC free article] [PubMed] [Google Scholar]
- Sidhar S. K., Lomas D. A., Carrell R. W., Foreman R. C. Mutations which impede loop/sheet polymerization enhance the secretion of human alpha 1-antitrypsin deficiency variants. J. Biol. Chem. 1995;270:8393–8396. doi: 10.1074/jbc.270.15.8393. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soboleva T. A., Baker R. T. Deubiquitinating enzymes: their functions and substrate specificity. Curr. Protein Pept. Sci. 2004;5:191–200. doi: 10.2174/1389203043379765. [DOI] [PubMed] [Google Scholar]
- Stirling C. J., Rothblatt J., Hosobuchi M., Deshaies R., Schekman R. Protein translocation mutants defective in the insertion of integral membrane proteins into the endoplasmic reticulum. Mol. Biol. Cell. 1992;3:129–142. doi: 10.1091/mbc.3.2.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Kamada Y., Ohsumi Y. Studies of cargo delivery to the vacuole mediated by autophagosomes in Saccharomyces cerevisiae. Dev. Cell. 2002;3:815–824. doi: 10.1016/s1534-5807(02)00359-3. [DOI] [PubMed] [Google Scholar]
- Sveger T. The natural history of liver disease in alpha 1-antitrypsin deficient children. Acta Paediatr. Scand. 1988;77:847–851. doi: 10.1111/j.1651-2227.1988.tb10767.x. [DOI] [PubMed] [Google Scholar]
- Teckman J. H., Perlmutter D. H. Retention of mutant alpha(1)-antitrypsin Z in endoplasmic reticulum is associated with an autophagic response. Am. J. Physiol. 2000;279:G961–G974. doi: 10.1152/ajpgi.2000.279.5.G961. [DOI] [PubMed] [Google Scholar]
- Tone Y., Tanahashi N., Tanaka K., Fujimuro M., Yokosawa H., Toh-e A. Nob1p, a new essential protein, associates with the 26S proteasome of growing saccharomyces cerevisiae cells. Gene. 2000;243:37–45. doi: 10.1016/s0378-1119(99)00566-1. [DOI] [PubMed] [Google Scholar]
- Tone Y., Toh E. A. Nob1p is required for biogenesis of the 26S proteasome and degraded upon its maturation in Saccharomyces cerevisiae. Genes Dev. 2002;16:3142–3157. doi: 10.1101/gad.1025602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers K. J., Patil C. K., Wodicka L., Lockhart D. J., Weissman J. S., Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- Tsai B., Ye Y., Rapoport T. A. Retro-translocation of proteins from the endoplasmic reticulum into the cytosol. Nat. Rev. Mol. Cell Biol. 2002;3:246–255. doi: 10.1038/nrm780. [DOI] [PubMed] [Google Scholar]
- Vashist S., Kim W., Belden W. J., Spear E. D., Barlowe C., Ng D. T. Distinct retrieval and retention mechanisms are required for the quality control of endoplasmic reticulum protein folding. J. Cell Biol. 2001;155:355–368. doi: 10.1083/jcb.200106123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbanac K. M., Heath E. C. Biosynthesis, processing, and secretion of M and Z variant human alpha 1-antitrypsin. J. Biol. Chem. 1986;261:9979–9989. [PubMed] [Google Scholar]
- Verma R., Aravind L., Oania R., McDonald W. H., Yates J. R., 3rd, Koonin E. V., Deshaies R. J. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- Verma R., Chen S., Feldman R., Schieltz D., Yates J., Dohmen J., Deshaies R. J. Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol. Biol. Cell. 2000;11:3425–3439. doi: 10.1091/mbc.11.10.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R., Deshaies R. J. Assaying degradation and deubiquitination of a ubiquitinated substrate by purified 26S proteasomes. Methods Enzymol. 2005;398:391–399. doi: 10.1016/S0076-6879(05)98032-4. [DOI] [PubMed] [Google Scholar]
- Voges D., Zwickl P., Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- Wang Z. X., Hu G. F., Wang H. Y., Wu M. C. Expression of liver cancer associated gene HCCA3. World J. Gastroenterol. 2001;7:821–825. doi: 10.3748/wjg.v7.i6.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner E. D., Brodsky J. L., McCracken A. A. Proteasome-dependent endoplasmic reticulum-associated protein degradation: an unconventional route to a familiar fate. Proc. Natl. Acad. Sci. USA. 1996;93:13797–13801. doi: 10.1073/pnas.93.24.13797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt E., Zantopf D., Schmidt M., Kraft R., Kloetzel P. M., Kruger E. Characterisation of the newly identified human Ump1 homologue POMP and analysis of LMP7(beta 5i) incorporation into 20 S proteasomes. J. Mol. Biol. 2000;301:1–9. doi: 10.1006/jmbi.2000.3959. [DOI] [PubMed] [Google Scholar]
- Wu Y., Whitman I., Molmenti E., Moore K., Hippenmeyer P., Perlmutter D. H. A lag in intracellular degradation of mutant alpha 1-antitrypsin correlates with the liver disease phenotype in homozygous PiZZ alpha 1-antitrypsin deficiency. Proc. Natl. Acad. Sci. USA. 1994;91:9014–9018. doi: 10.1073/pnas.91.19.9014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao T., Cohen R. E. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- Yorimitsu T., Nair U., Yang Z., Klionsky D. J. Endoplasmic reticulum stress triggers autophagy. J. Biol. Chem. 2006;281:30299–30304. doi: 10.1074/jbc.M607007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Liu C. Y., Back S. H., Clark R. L., Peisach D., Xu Z., Kaufman R. J. The crystal structure of human IRE1 luminal domain reveals a conserved dimerization interface required for activation of the unfolded protein response. Proc. Natl. Acad. Sci. USA. 2006;103:14343–14348. doi: 10.1073/pnas.0606480103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.