Abstract
Spermatogenesis in Marsilea vestita is a rapid process that is activated by placing dry microspores into water. Nine division cycles produce seven somatic cells and 32 spermatids, where size and position define identity. Spermatids undergo de novo formation of basal bodies in a particle known as a blepharoplast. We are interested in mechanisms responsible for spermatogenous initial formation. Mago nashi (Mv-mago) is a highly conserved gene present as stored mRNA and stored protein in the microspore. Mv-mago protein increases in abundance during development and it localizes at discrete cytoplasmic foci (Mago-dots). RNA interference experiments show that new Mv-mago protein is required for development. With Mv-mago silenced, asymmetric divisions become symmetric, cell fate is disrupted, and development stops. The α-tubulin protein distribution, centrin translation, and Mv-PRP19 mRNA distribution are no longer restricted to the spermatogenous cells. Centrin aggregations, resembling blepharoplasts, occur in jacket cells. Mago-dots are undetectable after the silencing of Mv-mago, Mv-Y14, or Mv-eIF4AIII, three core components of the exon junction complex (EJC), suggesting that Mago-dots are either EJCs in the cytoplasm, or Mv-mago protein aggregations dependent on EJCs. Mv-mago protein and other EJC components apparently function in cell fate determination in developing male gametophytes of M. vestita.
INTRODUCTION
For the past several years, we have been interested in the process of de novo basal body formation in spermatids of Marsilea vestita, a water fern. Spermatogenesis is a rapid, synchronous, and precise process in Marsilea, in which cell sizes, locations, and fates are constant among gametophytes (Sharp, 1914; Mizukami and Gall, 1966; Hepler, 1976; Myles and Hepler, 1977, 1982; Hyams et al., 1983; Pennell et al., 1988; Hart and Wolniak, 1998, 1999; Wolniak et al., 2000; Klink and Wolniak, 2000, 2001). The microspore starts out as a one-cell gametophyte, which is dry when it is obtained from the plant. When it is placed in water, the gametophyte undergoes a series of nine rapid cell division cycles to produce seven sterile cells and 32 spermatids. Four of the first five divisions are unequal, and they produce the smaller somatic cells and two large spermatogenous initials that form the germline. The two cell types formed are distinctly different, because the somatic cells lose the ability to divide, but the spermatogenous initials keep dividing. Later, each of the spermatids undergoes an elaborate differentiation process that includes the de novo synthesis of basal bodies in a particle known as a blepharoplast (Sharp, 1914; Hepler, 1976), the assembly of a complex cytoskeleton, nuclear and cell reshaping, and the production of ∼140 cilia. The entire process reaches completion in ∼11 h at 20°C (Hepler, 1976; Myles and Hepler, 1977).
Spermatogenesis in M. vestita relies almost entirely on mRNA and proteins that are present in the microspore before its desiccation. In vivo radiolabeling experiments (Hart and Wolniak, 1998), mRNA isolations, in vitro translation experiments (Hart and Wolniak, 1999), and morphological analyses of spores treated with α-amanitin and cycloheximide (Klink and Wolniak, 2001) show that there is little, if any, new transcription required for spermiogenesis to reach completion; like many spermatogenous systems, the male gametophyte is transcriptionally quiescent during its brief period of development. In a transcriptionally quiescent system with no cell movements, division asymmetries underlie different paths of differentiation of spermatogenous and sterile cells of the gametophyte. Obviously, precise control over the plane of cell division will affect patterns of development in the gametophyte. At a more subtle level, division asymmetries result from nonrandom distributions of cytoplasmic components between daughter cells, which could produce differences in translational capacities that lead to altered fates. Thus, the rearrangement of the cytoplasm, including segregation of transcripts and proteins, apparently plays an important role in establishing cell fate before cytokinesis in these unequal cell divisions.
We recently used in situ hybridization and immunolocalization assays to compare the distributions of transcripts and proteins in specific cells of the gametophyte during development (Tsai et al., 2004). Among more than 20 mRNAs analyzed, the majority of these transcripts are equally abundant in all cells of the gametophyte. Only two of these transcripts, one encoding a prespliceosome like molecule, Mv-Prp19, and the other encoding an RNA helicase, Mv-eIF4AIII, are present in spermatogenous cells, but absent from jacket cells. In contrast to most transcripts, the relative abundance of proteins in spermatogenous and sterile cells were strikingly different. Tubulin proteins are abundant in the dry spore, and they become heavily concentrated in the spermatogenous initials. This is not surprising, because the spermatogenous cells assemble a complex cytoskeleton and a ciliary apparatus. Centrin protein, which is essential for the formation of blepharoplasts (Klink and Wolniak, 2001), is translated from stored mRNA only in the spermatogenous cells, although its transcript is abundant in all cells of the gametophyte. The appearance of blepharoplast-like particles is a clear manifestation of distributional asymmetries that underlie cell fate determination in this simple gametophyte. We are interested in understanding how, in the absence of cell movement, the gametophyte rapidly establishes an axis and partitions sterile jacket cells from its spermatogenous initials.
As a first step in identifying mechanisms that control various aspects of cellular differentiation and cell fate in this gametophyte, we have focused on proteins and genes that have been linked to the control of cell fate in other organisms. Mago nashi is a gene that was originally found as a maternal effect mutation in Drosophila melanogaster that disrupts the anterior–posterior axis formation and formation of germ cells in the embryo (Boswell et al., 1991). The mago nashi gene was later characterized as a posterior group gene that functions in the localization of oskar mRNA and Staufen protein. It is involved in the polarization of the microtubule cytoskeleton and the migration of the nucleus (Newmark and Boswell, 1994; Micklem et al., 1997; Newmark et al., 1997). Mago nashi is a highly conserved protein (Newmark et al., 1997) and in humans its homologue was found to be part of the exon–exon junction complex (EJC), which is deposited onto the pre-mRNA during splicing, 20–24 nucleotides upstream of exon–exon junctions, in a sequence independent manner (Le Hir et al., 2000, 2001a,b; Kataoka et al., 2001; Kim et al., 2001; Shibuya et al., 2004). The EJC is attached to the mRNA by an RNA helicase, eIF4AIII. Mago, together with its binding partner Y14, inhibit the ATPase activity of this RNA helicase, which keeps it locked onto the mRNA (Ballut et al., 2005). Mago, Y14 and eIF4AIII are three proteins of the heterotetreamer that make up the core of the EJC, whereas the other components of the EJC are transiently associated with it (Palacios et al., 2004; Shibuya et al., 2004; Tange et al., 2005). As part of the EJC, mago protein is found in the nucleus (Micklem et al., 1997; Newmark et al., 1997), where it can be localized in nuclear speckles (Degot et al., 2004), which are interchromatin regions enriched in pre-mRNA and proteins of the splicing machinery (Lamond and Spector; 2003). Depending on the proteins that associate with the core complex, the EJC can play a role in export of mRNA from the nucleus (Luo and Reed, 1999; Zhou et al., 2000; Le Hir et al., 2001b), transcript quality control via the nonsense-mediated degradation pathway (NMD) (for review, see Tange et al., 2004), and the enhancing of translation (Nott et al., 2003; Wiegand et al., 2003; Nott et al., 2004).
In this article, we characterize a mago nashi homologue in M. vestita that we call Mv-mago. We use RNA interference (RNAi) (Fire et al., 1998; Klink and Wolniak, 2000, 2001) to show that newly translated Mv-mago protein functions in an essential role during spermiogenesis in the male gametophyte. We present evidence that Mv-mago protein is required for the proper control of the plane of cell division in the gametophyte, especially during the normally asymmetric divisions that give rise to the sterile jacket cells. Moreover, we provide evidence that during or before the asymmetric divisions, Mv-mago is involved in the redistribution of mRNAs of proteins involved in RNA processing. We show that centrin, which is normally only translated in the spermatogenous cells, is translated in jacket cells and spermatogenous cells after knockdowns of Mv-mago. Remarkably, these jacket cells exhibit centrin staining patterns that resemble blepharoplasts and basal bodies, organelles that normally are never made anywhere in the organism except in spermatids. In normal gametophytes, we show that Mv-mago becomes aggregated in dots in the cytoplasm at the end of the division cycles in all cells in the gametophyte. RNAi treatments with double-stranded RNA (dsRNA) from Mv-mago, Mv-Y14, or Mv-eIF4AIII all eliminate this punctate staining pattern, and they suggest that Mv-mago protein is functioning with these other proteins in EJCs. We suspect Mv-mago protein is involved in cytoplasmic rearrangements that affect transcript distributions and localized translations in cell fate determination and spermiogenesis in the male gametophyte of M. vestita.
MATERIALS AND METHODS
Growth and Fixation of Microspores
Dry sporocarps were collected from M. vestita sporophytes that had been raised in seven ponds at the greenhouse facilities at the University of Maryland (College Park, MD). Sporocarps were ground in a commercial coffee grinder, and microspores were separated from the debris with sieves (Hepler, 1976; Klink and Wolniak, 2001; Tsai and Wolniak, 2001). Gametophytes were cultured in commercial spring water in a ratio of 8 mg of dry microspores/10 ml of H2O in 50-ml flasks with continuous, gentle rotational shaking at 20°C (Hepler, 1976; Klink and Wolniak, 2001; Tsai and Wolniak, 2001). Fixation protocols were modified from Klink and Wolniak (2001) and Tsai and Wolniak (2001). In short, at the time of fixation, gametophytes were filtered out of the solution onto a polyester filter (BioDesign Cell MicroSieves; BioDesign, Carmel, NY) with a Buchner funnel and vacuum pump. The filter with the microspores was transferred to a metal mortar, 100 μl of fixative [4% paraformaldehyde in 50 mM piperazine-N,N′-bis(2-ethanesulfonic acid), pH 7.0] was added and the spores cracked by the mechanical force from a hammer blow to a metal pestle by using two stacked 50-μm brass spacers to prevent complete squashing of the microspores (Hepler, 1976; Klink and Wolniak, 2003). Microspores were then washed from the filter with 15 ml of fixative into a 50-ml conical centrifuge tube, and they were left for 2 h at 4°C. After this, the microspores were rinsed three times in phosphate-buffered saline buffer, pH 7.4, by spinning down the spores for a short time in a tabletop centrifuge at low speed. Spores were transferred to 2-ml centrifuge tubes and dehydrated in an ethanol series of 10, 25, 50, 75, and 3 × 100% for 2 h each except for the second 100% rinse, which ran overnight. Spores were infiltrated with methacrylate embedding media and polymerized in essence as described by Baskin et al. (1992) by using BEEM capsules with a pyramidal tip. Modifications included 2 h of infiltration time and five replacements of the100% methacrylate mixture of which the fourth replacement ran overnight. After polymerization of the plastic in UV light at 4°C, semithin sections were made using a glass knife on an ultramicrotome, and the sections were placed on a drop of water on a microscope slide. Sections were relaxed by holding a chloroform-saturated swab in proximity to the section. Then, the water drop was allowed to dry on a heating block at 40°C, so the sections would adhere to the glass.
Identification of cDNAs from M. vestita
An M. vestita cDNA library was constructed from mRNAs isolated at all stages of gametophyte development (Hart and Wolniak, 1998). In vivo excisions and mass excisions were performed using the ExAssist/SOLR system and the pBluescript plasmid as described by the manufacturer (Stratagene, La Jolla, CA). Clones were sequenced at the DNA sequencing facility in the Center for Biosystems Research, part of the Maryland Biotechnology Institute on campus, or with an Applied Biosystem 3100 genetic analyzer housed in a core instrumentation lab in the Cell Biology and Molecular Genetics (University of Maryland) according to manufacturer's instructions (Applied Biosystems, Foster City, CA). More than 1500 clones were sequenced and identified by BLAST searches. The cDNA clones were entered into GenBank with their initial designation or name: Mv-mago (AF329672), Mv-Y14 (EU009956), Mv-eIF4AIII (CF867680), and Mv-Prp19 (AF484839).
RNAi
dsRNA was made as described by Tsai and Wolniak (2001) except that the polymerase chain reaction (PCR) product was sometimes cleaned by ethanol precipitation before resuspension in RNase-free water. Products were analyzed by spectrophotometry and gel electrophoresis. dsRNA was generated by first heating each single-stranded (ssRNA) to 80°C for 5 min, followed by placing it on ice for 10 min and incubating equal amounts of the sense and antisense RNA together at 37°C for 2 h. The quality of dsRNA was checked by gel electrophoresis before each experiment. RNAi experiments were performed on an orbiting shaker in a 2-ml Microfuge tube by adding the dsRNA to the microspores at the start of imbibition in a concentration of 200 μg/ml (Klink and Wolniak, 2001; Tsai and Wolniak, 2001). After 1 h, the spores were transferred to 50-ml flasks containing 10 ml of commercial spring water in a rotating water bath as described above. At various times, spores were fixed and processed as described above. For each dsRNA, at least three RNAi experiments were performed, and for each insert, we made at least three batches of dsRNA.
In Situ Hybridizations
In situ hybridizations were performed according to protocols described by Tsai and Wolniak (2001). RNA probes were labeled by substituting half of the dUTP with digoxigenin-11-dUTP (Roche Diagnostics, Indianapolis, IN). Modifications to our earlier protocol include the use of silanated slides (KD Medical, Columbia, MD), relaxation of the sections by holding a chloroform saturated cotton swab in proximity to the sections and drying of the slides on a heat block at 40°C. Slides were treated with acetone, proteinase K, paraformaldehyde, and triethanolamine according to procedures by Steel et al. (1998). Hybridization procedures and visualization of the probe with nitro-blue tetrazolium and 5-bromo-4-chloro-3 indolyl-phosphate were performed as described by Tsai and Wolniak (2001).
Generation of a Polyclonal Anti-Mv-Mago Antibody
A glutathione S-transferase (GST)-Mv-mago expression plasmid was constructed; the mago EcoRI–Xho fragment was digested from Mvu34 and inserted into the expression vector pGEX-4T-1 (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom). Products of the ligation reaction were transformed into Escherichia coli DH5α and screened by restriction mapping and protein expression (see below). Log phase cultures of E. coli BL21(DE3) containing pGEX-mago were induced with isopropyl-α-d-thiogalactopyranoside (Smith and Johnson, 1988). Bacterial cultures were pelleted, resuspended, and lysed by sonication. The fusion protein was isolated using glutathione agarose as described by Hardin and Wolniak (1998). The eluted fusion protein was then digested with 1 U of thrombin (Promega, Madison, WI) at room temperature for 16 h.
Digested protein samples were subjected to preparative electrophoresis on 12.5% polyacrylamide-SDS gels. The Mv-mago protein fraction was gel-purified and used for polyclonal antiserum production (Rockland, Gilbertsville, PA). The antiserum was batch affinity purified using purified Mv-mago protein that had been bound to the glutathione agarose beads (Hardin and Wolniak, 1998). The antiserum was tested on immunoblots with protein isolates from microspores 4 h into development by using ECF chemiluminescence (GE Healthcare), with detection on a STORM 860 PhosphorImager (GE Healthcare) at a dilution of 1:50 (Hardin and Wolniak, 1998; Klink and Wolniak, 2001). For immunoblot analyses of gametophyte polypeptides, soluble proteins were isolated from identical populations of growing gametophytes at regular time intervals by using a Dounce homogenizer to fracture the spore walls. We pooled polypeptide fractions from each time point, and we dissolved them in SDS buffer at 100°C as described previously (Hart and Wolniak, 1998). One hundred micrograms of protein from each sample was loaded into separate wells of a polyacrylamide gel, and proteins were separated electrophoretically as described previously (Hart and Wolniak, 1998; Klink and Wolniak, 2001). Immunoblots were prepared as described by Klink and Wolniak (2003) and analyzed as described above. Every blot assay was repeated with newly isolated protein samples.
Cytology and Immunocytochemistry
Sections where either observed unstained with differential interference contrast (DIC) microscopy, or they were stained with Toluidine Blue O and viewed with bright-field microscopy (O'Brien and McCully, 1981) Immunocytochemistry was performed as described by Baskin and Wilson (1997), except that the etching time was 20 min, incubation time with the antibodies was 1 h, and after the first set of rinses after etching, blocking solution (Hardin and Wolniak, 2001) was added to the slides for 1 h. Excess blocking solution was rinsed off by quickly dipping the slides in phosphate-buffered saline containing 0.05% Tween 20 (PBST). Primary antibodies used were the above-described anti-Mv-mago (1:50), anti-centrin monoclonal 20H5 directed against Chlamydomonas reinhardtii (1:100) (kind gift from Jeff Salisbury, Mayo Clinic, Rochester, MN), and monoclonal anti-α-tubulin (1:20) (ab-1; Calbiochem, San Diego, CA). Our previous immunolocalization studies (Klink and Wolniak, 2001; Tsai et al., 2004) showed that the distribution of α-tubulin matches that of β-tubulin in our gametophytes; the loss of a specific commercially available anti-β-tubulin antibody prompted us to switch to the anti-α-tubulin probe. The secondary antibody used was an Alexa Fluor 594 (Invitrogen, Carlsbad, CA). All antibodies were diluted in PBST 0.05%. Fluorescence microscopy was performed with a Zeiss Axioskop (Carl Zeiss, Jena, Germany) with a standard Texas Red filter set. Paired fluorescence and phase contrast images were made of at least 100 gametophytes from each sample.
RESULTS
A. marsilea Homologue of Mago Nashi
We have isolated an apparent homologue of Drosophila mago nashi, which we initially identified as MvU34 in a random screen of a cDNA library made from mRNA originally isolated from sequential growth stages of the male gametophytes of M. vestita (Hart and Wolniak, 1999). On the basis of sequence homologies, we have named the cDNA Mv-mago (GenBank accession no. AF329672). The nucleic acid sequence comparisons reveal striking similarities among Mago nashi genes in widely divergent organisms (Figure 1). The alignment of Mv-mago with other species shows that the Mv-mago cDNA probably lacks a short nucleotide stretch that encodes a few amino acids at the amino terminal end. Our attempts to isolate a longer Mv-mago cDNA from our library have been unsuccessful thus far. The Mv-mago cDNA encodes a predicted sequence for a 152 amino-acid protein (Figure 1). The predicted amino acid sequence for Mv-mago matches the extraordinarily high level of conservation found in Mago nashi nucleotide sequences observed among diverse species; BLAST analyses reveal that Mv-mago is 76% identical with the predicted protein of D. melanogaster (Figure 1).
Figure 1.
Alignment of the deduced amino acid sequence from Mv-mago a cDNA insert from a male gametophyte library of M. vestita. Residues identical to Mv-mago sequences are indicated as dashes. The amino acid sequences are aligned with the homologs from the following organisms: Arabidopsis thaliana (No. U89959), Xenopus leavis (No. AF007860), Homo sapiens (No. AF035940), Drosophila melanogaster (No. U03559), Caenorhabditis elegans (No. AF007861), and Schizosaccharomyces pombe (No. A1022070). Within the coding region there is a 76% identity between predicted amino acid sequences of Drosophila and M. vestita. Judging by alignments with mago nashi proteins from other organisms, Mv-mago is a truncated clone that appears to lack only a few amino acids at the amino terminal end. It lacks the start sequence at 5′ end, but is predicted to encode a 152-amino acid protein. This sequence has been deposited in the GenBank database under the accession number AF329672.
Localization of Mv-mago mRNA and Protein
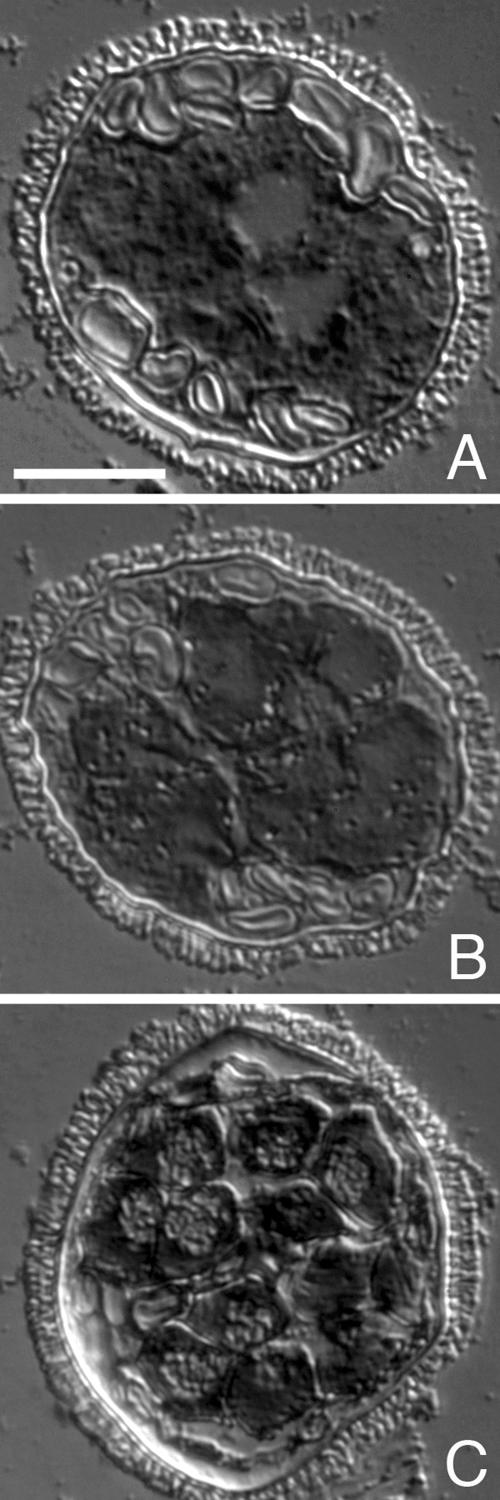
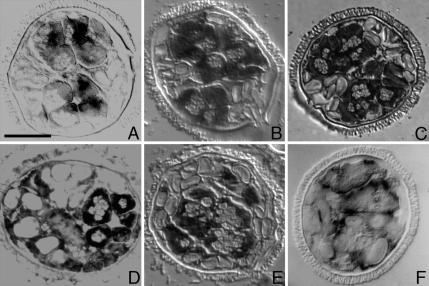
We have performed a series of assays that enable us to localize Mv-mago mRNA in gametophytes at different stages in the development. Digoxigenin-labeled Mv-mago antisense riboprobes were hybridized in situ with sectioned, untreated gametophytes that had been fixed at different stages of development, up to 8 h. Mv-mago mRNA is present in dry spores and throughout development (Figure 2, A–C). The transcript exhibits an evenly distributed staining pattern that was apparent when jacket cells and spermatogenous cells were formed (Figure 2A). The pattern persists later in development toward the end of the cell division cycles as gamete differentiation commences (Figure 2B) and even later as the gametes become coiled cells (Figure 2C).
Figure 2.
Localization of Mv-mago mRNA during spermiogenesis in gametophytes of M. vestita. Gametophytes were fixed at 2 h (A), 4 h (B), and 8 h (C) after the dry microspores were placed in water. Mv-mago mRNA distribution was observed using a digoxygenin labeled Mv-mago RNA probe and hybridized in situ to sectioned gametophytes. Mv-mago mRNAs appeared to be present at all time points, and are uniformly distributed in all the cells of the gametophyte. Bar, 25 μm.
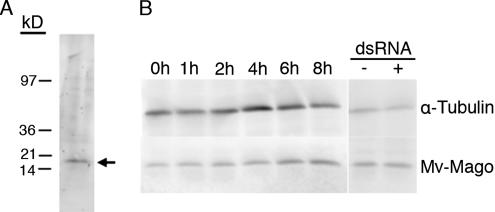
To assess the abundance and distribution of Mv-mago protein during spermiogenesis, we expressed a GST–Mv-mago fusion protein in bacteria for the generation of a polyclonal antiserum in rabbits. The serum was then column affinity purified. On immunoblots, the antibody bound to isolated fern polypeptides with an apparent molecular mass of 20 kDa (Figure 3A), approximately the predicted mass of 17 kDa for Mago protein in Drosophila (Newmark and Boswell, 1994). Fern gametophyte proteins were isolated at various times during development, and they were probed with the anti-Mv-mago antibody on immunoblots (Figure 3B). We were surprised to find a low level of detectable binding in newly hydrated microspores, indicating that some Mv-mago protein is present in the dry spore; this level of staining increased slightly after 6 h (Figure 3B). In contrast to rapid and dramatic centrin protein accumulation in gametophytes (Hart and Wolniak, 1998; Klink and Wolniak, 2001), the intensity of anti-Mv-mago antibody binding on the immunoblots is weak and increases only modestly over time (Figure 3B), suggesting that Mv-mago protein is neither abundant nor rapidly accumulating in the developing gametophyte. This binding pattern shows that some Mv-mago protein is present in gametophytes at the onset of development and that it remains present even after RNAi-induced silencing treatments (below).
Figure 3.
Anti-Mv-mago antibody labeling on an immunoblots. Anti-Mv-mago antibody labeling on an immunoblot of fern gametophyte polypeptides isolated 4 h into development (A). The protein isolate was loaded onto a polyacrylamide gel, separated electrophoretically and transferred to a blot. The isolates were probed with the polyclonal antibody raised in rabbits against Mv-mago and affinity purified. ECF fluorescence was detected with a STORM 860 Phosphorescence-Fluorescence Imaging Scanner (Molecular Dynamics). The antibody binds to one band of approximately 20 kDa, which is the predicted size of mago nashi protein in Drosophila. Immunoblot analyses of proteins isolated from fern gametophytes at various time points during development (B). This blot was labeled with anti-α-tubulin antibody (upper panel), then stripped, and labeled with anti-Mv-mago antibody (lower panel). This binding is uniform from the onset of development with anti-α-tubulin antibody. Binding with anti-Mv-mago antibody reveals the presence of Mv-mago protein in the gametophyte from the onset of development, with a slight increase in abundance over time. Right panel: protein isolates were obtained at 4 h of development after Mv-mago silencing. The binding of anti-α-tubulin antibody and anti-Mv-mago antibody both appear to be similar to binding levels in untreated controls. The uniformity of binding by the anti-tubulin antibody reveals that Mv-mago silencing has no effect on the abundance of tubulin in the gametophyte. After Mv-mago silencing, we can still detect Mv-mago protein in our isolates, a result consistent with the finding that some Mv-mago protein was already present when the dsRNA was added to the spores.
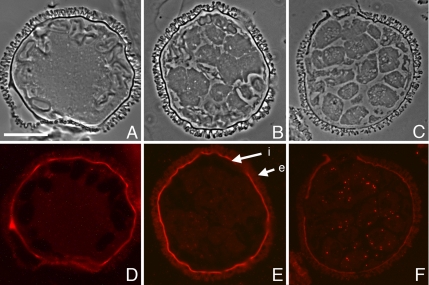
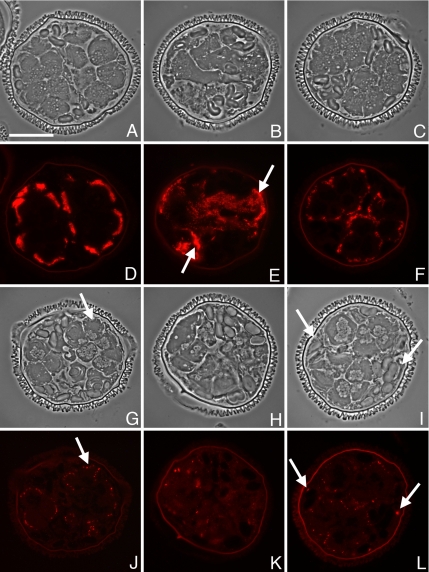
Indirect immunofluorescence was performed on fixed, sectioned gametophytes with our purified anti-Mv-mago antibody. Paired phase contrast and fluorescence images are presented as Figure 4. The cytoplasm exhibits a low level of autofluorescence that decreases during development of the gametophyte, and unstained portions of the gametophyte exhibit autofluorescence from the spore wall, especially the intine layer (Figure 4E, arrows). During the first 4 h of development, the staining with the anti-Mv-mago antibody was low, but it was diffuse and detectable above background autofluorescence (Figure 4, A and D). More than 4 h into development, at the point when all divisions are finished and spermatids start to differentiate, the anti Mv-mago antibody staining becomes more distinctly apparent as small, punctae (Mago-dots) in the cytoplasm (Figure 4, B and E). These Mago-dots become brighter later during gamete maturation (Figure 4, C and F), and they persist to at least 11 h after the start of development. Although the Mago-dots are present in both jacket cells and spermatids, they were most prominent in the spermatogenous cells. There was some variation in the numbers of Mago-dots that were visible in each cell, but on average, there were ∼10 of these punctae in each spermatid in a semithin section of the spore. The punctae were not associated with any specific or recognizable cytoplasmic component of the cell observed with phase contrast or DIC microscopy (e.g., a cell plate, plastids, or the multilayered structure). When gametophytes were costained with 4,6-diamidino-2-phenylindole (DAPI), we found no Mago-dots in the nuclei of spermatogenous or sterile cells.
Figure 4.
Mv-mago protein distributions in normal, developing gametophytes. Fixed, sectioned gametophytes were immunolabeled using an affinity purified polyclonal antibody raised in rabbits against expressed Mv-mago protein. Alexa Fluor 594-conjugated goat anti-rabbit antibody was used for detection with fluorescence microscopy. Paired phase contrast (A–C) and fluorescence (D–F) micrographs were taken of each gametophyte. Presented data are for gametophytes fixed at 2 h (A and D), 4 h (B and E), and 8 h (C and F) after the dry microspores were placed in water. Minor levels of autofluoresence are present in the cytoplasm and the spore wall often autofluoresces brightly (E; i = intine, e = exine) with the excitation/emission bandwidths for Alexa Fluor 594, though the autofluorescence signals from the gametophyte are lower with this combination than with others currently available. Early in development, antibody staining for Mv-mago protein is weak but detectable in the gametophyte (D), and after the cell division cycles have reached completion, staining with the antibody becomes more prominent and is localized in dots in the cytoplasm of both jacket and spermatogenous cells (E). This staining pattern persists through development and the punctae (Mago-dots) become more prominent as the spermatids mature (F). Bar, 25 μm.
RNAi-induced Silencing of Mv-mago Results in Altered Gametophyte Development
We used RNAi to reduce the abundance of newly synthesized Mv-mago protein in the gametophyte. The RNAi phenocopy of a null mutant could demonstrate whether and when new Mv-mago protein is involved in gametophyte development. We used the same RNAi protocol developed earlier for male gametophytes of M. vestita (Klink and Wolniak, 2001; Tsai and Wolniak, 2001; Tsai et al., 2004). dsRNA was generated by in vitro transcription by using the Mv-mago cDNA as a template, and it was present in the aqueous culture medium at the time the dry spores were added (Klink and Wolniak, 2001). The effectiveness of the dsRNA to destroy the stored mRNA of Mv-mago was verified by in situ hybridization assays on RNAi-Mv-mago cell sections by using digoxigenin-labeled riboprobes, because we lack sufficient numbers of cells in an RNAi experiment for the production of mRNA isolates required with Northern blots. The shortest interval between dsRNA addition and fixation for in situ hybridization was 30 min, and even in these cells, we could not detect Mv-mago mRNA, a result suggesting that there is rapid depletion of the stored mRNA in these gametophytes (data not presented). In control experiments, in situ hybridization assays showed that the abundance of β-tubulin was unaltered after the addition of dsRNA made from Mv-mago cDNA, indicating that this RNAi treatment did not destroy mRNA in general (Klink and Wolniak, 2001, 2003). Because the microspores contain large quantities of stored mRNA (Hart and Wolniak 1998, 1999), and because the gametophytes develop in the presence of α-amanitin (Klink and Wolniak, 2001), it is reasonable to suspect that the Mv-mago mRNA is present in the dry spore as a stored transcript. It seems that the Mv-mago transcript is susceptible to degradation after the introduction of Mv-mago dsRNA into the cytoplasm of the spore at the time of its hydration.
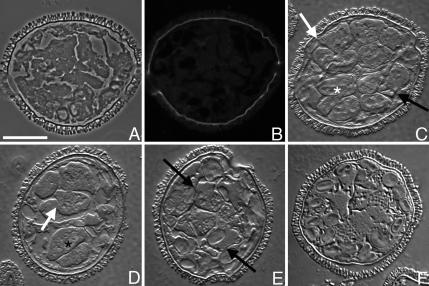
Changes in the abundance of Mv-mago protein are difficult to detect after Mv-mago silencing (Figure 3B), because there is Mv-mago protein present in the dry spore and because gametophytes do not translate large amounts of this protein during development. We treated gametophytes at the onset of development with dsRNA derived from Mv-mago cDNA, and we fixed them 8 h later for in situ immunolabeling (Figure 5, A and B) to determine whether the RNAi treatment affected the apparent distribution of Mv-mago protein in the gametophytes. As with untreated gametophytes, we observed only low levels of diffuse anti-Mv-mago antibody labeling in the cytosol of gametophytes after the RNAi treatments, irrespective of the time of fixation (Figure 5B). Importantly, no Mago-dots (Figure 4F) were seen in gametophytes treated with Mv-mago dsRNA (Figure 5B).
Figure 5.
Mv-mago RNAi treatments abolish anti-Mv-mago antibody staining and alter cell fate determination in the gametophyte. Dry microspores were placed into water containing dsRNA derived from Mv-mago cDNA, and the gametophytes were fixed after 8 h of development, embedded in methacrylate and sectioned. Paired phase contrast (A) and fluorescence (B) of fixed gametophytes stained with anti Mv-mago antibody was used to detect Mv-mago protein distributions after silencing. Weak fluorescence was sometimes visible, but no Mago-dots were detectable after Mv-mago silencing. DIC (C–F) microscopy was used to reveal cell structure. Observations of at least 100 spores showed four different phenocopies with a range of severity (C–F), but with similar defects: incomplete cell divisions (C and D, white arrows), altered plane of cell division (C, black arrow) causing atypical cell arrangement (white star), incorrect localization of starch-bearing plastids (black star), and unusual distributions of growing plastids. The altered planes of cell division are most apparent in the formation of the jacket cells, which are larger than normal because the divisions have become more symmetric (E, black arrows). The majority of the gametophytes in all phenocopy classes showed a loss of cell identity (E); the distinctions between jacket cells and spermatogenous cells are lost or reduced after treatment with Mv-mago dsRNA. The most severe phenocopy (F) appears to result from dsRNA toxicity. Bar, 25 μm.
Observations were made of multiple sections through each of at least 100 spores that had been treated with Mv-mago dsRNA to understand the altered patterns of development in three dimensions. Four overarching patterns of development were observed. Approximately 10% of the gametophytes were similar to untreated gametophytes; at first glance, the jacket cells formed correctly and the correct number of spermatogenous cells were present (Figure 5C). However, closer observations revealed subtle anomalies in the gametophytes: divisions may have occurred in unusual places in the spermatogenous cells (black arrows), altering the relative sizes of spermatogenous and sterile cells (white star). In addition, some incomplete cell plates were present, so some of the cells are not completely separated from each other (white arrow). Approximately 30% of the gametophytes underwent seemingly correct divisions early in development when jacket cells were formed, but they clearly showed anomalous planes of cytokinesis in the divisions of the spermatogenous cells (Figure 5D), incomplete separation (white arrow), and a decreased number of spermatogenous cells (Figure 5D). Moreover, some of the large starch-bearing plastids, normally localized in the jacket cells, occasionally occur in spermatogenous-like cells (Figure 5D, black star). For the majority of gametophytes (>50%), cells of undetermined fate reside within the spore walls (Figure 5E). These gametophytes exhibit incorrectly placed planes of cell division. Incomplete cell plates from the third, fourth, and fifth division cycles were also common. (These divisions normally produce the jacket cells.) The net result is that presumptive jacket cells situated on the periphery of the spermatogenous cell mass are much larger than normal jacket cells would be in untreated controls. The loss of asymmetry that produces large, presumptive jacket cells also produces smaller spermatogenous cells in the central portion of the gametophyte. The extent to which cell fate is affected by changes in cell volume were accompanied by anomalous distributions of large starch bearing plastids, which normally occur only in the jacket cells, and of the growing plastids, which normally occur only in the spermatids late in development. A small segment of the population, making up <10% of the spores, underwent a few oddly placed divisions, often without complete partitioning by cell plates (Figure 5F). These gametophytes were not included in our analyses of effects.
Although each of these phenocopies seems to represent a unique response to the dsRNA addition, the underlying defects are the same: the gametophytes exhibit incorrect placement of one or more cell plates, incomplete cell plates, and incorrect localization of starch-bearing plastids, and unusual distributions of the growing plastids. The cells that exhibit early arrest (Figure 5F) are either showing signs of dsRNA toxicity (Klink and Wolniak, 2001), or they are revealing that early translation of Mv-mago mRNA is necessary for the gametophytes to progress through the early division cycles. We attribute the anomalies observed later in development either to differences in the effective concentration of dsRNA by variations in uptake at the time of spore hydration or to differences in the amount of mago protein present in the gametophyte at the onset of development.
Mago RNAi Affects the Distributions of Specific Transcripts and Proteins in the Gametophyte
Because the identities of the cells in the majority of the gametophytes after RNAi treatments with Mv-mago dsRNA were equivocal by morphological analysis alone, we analyzed distributions of specific mRNAs and proteins necessary for the development of spermatogenous cells. Previously, we (Tsai et al., 2004) found only two transcripts among >20 mRNAs screened from our cDNA library by in situ hybridization that were present in spermatogenous cells but essentially absent from jacket cells. The majority of transcripts were equally abundant in all cells of the gametophyte. Both messages localized in spermatogenous cells encode proteins that are involved in pre-mRNA splicing. One mRNA encodes a pre-spliceosome protein we named Mv-Prp19, on the basis of BLAST search analysis. The second mRNA encodes an RNA helicase that we named Mv-eIF4AIII, again on the basis of BLAST search analyses. After 8 h of development, in situ hybridization assays show that both Mv-PRP-19 and Mv-eIF4AIII reside predominantly in the spermatogenous cells of untreated gametophytes (Figure 6, A and B; Tsai et al., 2004). However, after Mv-mago RNAi treatment, these mRNAs were visible in all the cells of the gametophyte (Figure 6, D and E). Although there was more staining in the smaller, presumptive spermatogenous cells, no distinct pattern of the transcript localization was seen between the two cell types in these treated gametophytes.
Figure 6.
Mv-mago RNAi treatments alter transcript distributions in gametophytes. Mv-mago silencing results in the dispersion of mRNAs encoding both Mv-Prp19 and Mv-eIF4AIII through all cells of the gametophyte and alters the localization pattern of Mv-β-tubulin mRNA. Gametophytes were treated with dsRNA derived from Mv-mago at the time the spores were placed in water and fixed 8 h later. Distributions of mRNA were observed using a digoxygenin labeled Mv-mago RNA probe and hybridized in situ to sectioned gametophytes. In normally developing gametophytes, mRNA of Mv-Prp19 (A) and Mv-eIF4AIII (B) is only localized in the spermatogenous cells but Mv-β-tubulin mRNA (C) is distributed equally in all cells of the gametophyte. In Mv-mago RNAi treated spores Mv-Prp19 (D) and Mv-eIF4AIII (E) mRNA is distributed throughout the whole gametophyte and Mv-β-tubulin mRNA (F) is nonuniformly distributed in the gametophyte. Images were obtained with DIC microscopy. Bar, 25 μm.
Multiple tubulins function in spermatogenous cell development as components in an elaborate microtubule cytoskeleton that underlies a extensive ciliary apparatus making up ∼140 axonemes (Myles and Hepler, 1977). Tubulin proteins are abundant in the microspore (Hart and Wolniak, 1998; Klink and Wolniak, 2003), and these proteins become localized in the spermatogenous initials as the division phase progresses in the gametophyte (Klink and Wolniak, 2001; Tsai et al., 2004). In contrast, mRNAs encoding α- and β-tubulin are present in all cells of the developing gametophyte (Tsai et al., 2004; Figure 6C), with new tubulin translation preceding ciliogenesis in the developing spermatids (Hart and Wolniak, 1998; Klink and Wolniak, 2001; Tsai et al., 2004). The distribution of β-tubulin mRNA is also uniform in all cells of the gametophyte after RNAi Mv-mago treatments (Figure 6F); however, unlike the untreated gametophytes, the distribution pattern of β-tubulin mRNA was not uniform throughout the cytoplasm (Figure 6F). The regions of the treated gametophytes presenting the strongest signals seem to be the more centrally positioned (albeit anomalously shaped) spermatogenous cells (Figure 6F).
Eight hours into development, α-tubulin protein is exclusively present in the spermatogenous cells, aggregated mostly on the outer edges where the microtubule ribbon is being assembled (Figure 7, A and D). After Mv-mago RNAi treatments, α-tubulin is present in some cells and not in others without any clear distinction to size or position (Figure 7, B and E). Some aggregations of anti-α-tubulin antibody staining are present in some of the cells of the gametophyte (Figure 7E, white arrows). In spores treated with Mv-mago dsRNA and exhibiting only subtle anomalies, α-tubulin protein was concentrated in the spermatogenous cells, and it was aggregated on the outer edges of the cell, although the overall level of labeling was lower than that of control cells (Figure 7, C and F).
Figure 7.
Immunolocalizations of α-tubulin and centrin proteins in Mv-mago RNAi treated microspores show a loss of cell identity. Gametophytes were treated with dsRNA derived from Mv-mago at the time the spores were placed in water and fixed 8 h later. The distribution of α-tubulin and centrin proteins were observed in sections of the gametophytes using Alexa Fluor 594 conjugated to the secondary antibody. Paired phase contrast (A–C and G–I) and fluorescence (D–F and J–L) micrographs are presented. In normally developing gametophytes, α-tubulin is localized at the outer edges of the spermatogenous cells where the microtubule ribbon is localized (A and D) 8 h into development. In Mv-mago RNAi-treated gametophytes, α-tubulin protein is present in some cells with no discernable localization pattern and with occasional randomly-distributed aggregations visible of antibody staining (B and E, white arrows). In Mv-mago RNAi-treated gametophytes with minor defects, there is weak α-tubulin staining at the outer edges of the spermatogenous cells (C and F). In normal gametophytes, centrin is only translated in spermatogenous cells and becomes localized near the basal bodies of the motile apparatus (G and J, white arrow). In treated gametophytes where cell identity is lost, centrin is translated and localized in all cells (H and K). In treated gametophytes with minor defects, centrin is also translated and localized in jacket cells as well as the spermatogenous cells (I and L, white arrows). Bar, 25 μm.
Another essential component for sperm development is centrin protein. In untreated gametophytes, centrin mRNA is present in all cells of the gametophyte (Tsai et al., 2004), but centrin protein is only translated in spermatogenous cells, ∼4 h into development (Klink and Wolniak, 2001; Tsai and Wolniak, 2001), when it becomes localized in the blepharoplast as a large and distinctive spot in the cytoplasm (Klink and Wolniak, 2001). In untreated cells fixed later in development (Figure 7, G and J), anti-centrin antibody also labels the basal bodies of the developing spermatid, which are visible as bright and discrete points at the anterior end of each spermatid (Figure 7J, white arrow). In RNAi Mv-mago-treated cells, the distribution of centrin mRNA (data not presented) was similar that of β-tubulin mRNA (Figure 6, C and F); in untreated gametophytes, both transcripts were uniformly distributed in all cells of the gametophyte, and after Mv-mago silencing, transcripts were detectable in all cells, but they were not spread out in a uniform pattern.
In contrast to normal gametophytes, centrin protein in gametophytes treated with Mv-mago dsRNA is translated everywhere (Figure 7, H, I, K, and L). In these spores with ambiguous cell fates, centrin protein becomes aggregated at apparently random spots in all cells throughout the spore (Figure 7, H and K). Remarkably, these spots resemble blepharoplasts and (clustered) basal bodies. Even in gametophytes showing only mild morphological anomalies late in development, centrin is translated in all cells of the gametophyte, with detectable centrin aggregates both in presumptive spermatogenous cells and presumptive jacket cells (Figure 7, I and L, white arrows) that resemble single or clustered basal bodies in untreated gametophytes (Figure 7, I and L).
RNAi Treatments Using cDNAs from Mv-Y14, Mv-Eif4aIII, and Mv-Prp19
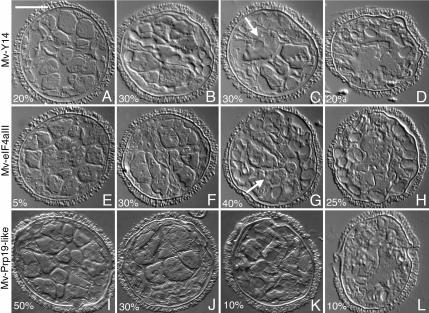
In addition to Mv-mago and Mv-eIF4AIII, BLAST analyses of isolated cDNAs have also enabled us to identify Mv-Y14, a third core component of the EJC (Ballut et al., 2005). RNAi experiments were performed, using dsRNAs derived from Mv-Y14 and Mv-eIF4AIII cDNAs to determine whether the disruption of various EJC components resulted in developmental anomalies that are similar to those observed with Mv-mago RNAi treatments. For RNAi treatments using dsRNAs made from Mv-Y14 or Mv-eIF4AIII, we observed four patterns of anomalous development that resembled gametophytes treated with Mv-mago dsRNA, and with similar frequencies of occurrence (compare Figure 5 with Figure 8, A–H). The most prominent developmental anomaly observed with Mv-Y14 and with Mv-eIF4AIII dsRNAs was that the jacket cells are larger than normal jacket cells in untreated gametophytes. This effect is similar to changes in jacket cells observed with Mv-mago dsRNA. The underlying cause for this change is the mislocalization of cell division planes, which were more symmetric than in normal gametophytes during jacket cell formation (Figure 8, B, C, F, and G). Like the Mv-mago dsRNA treatments, we saw the variations in the responses among gametophytes; dsRNA from Mv-Y14 and Mv-eIF4AIII exerted minimal effects on some of the gametophytes, and the division patterns seemed essentially normal (Figure 8, A and E). In these gametophytes, incomplete divisions were present with a much lower frequency than in Mv-mago RNAi-treated spores (Figure 8, A and E). In the majority of the gametophytes (Figure 8, B, C, F, and G), spermatogenous cells were oddly shaped, too few in number, and they formed atypical clusters within the spore. Incomplete divisions were readily observed in the more severely affected gametophytes (Figure 8, C and G, arrows). In contrast to Mv-mago dsRNA-treated spores, the large starch-bearing plastids were always present in oversized jacket cells near the periphery of the spore. The small plastids were always present in the smaller, more centrally located presumptive spermatogenous cells (Figure 8, B, C, F, and G). Therefore, it was easier to identify a cell based on its appearance and location after RNAi of Mv-Y14 and Mv-eIF4AII than after Mv-mago. As in Mv-mago RNAi treatments, some gametophytes were severely altered (Figure 8, D and H), presumably because of dsRNA toxicity.
Figure 8.
RNAi treatments of microspores with EJC and splicesome dsRNAs alter development in different ways. Gametophytes were treated with dsRNA derived from Mv-Y14 (A–D), Mv-eIF4aIII (E–H), and Mv-Prp19 (I–L) at time the spores were placed in water and were fixed after 8 h. Four different phenocopies with a range from mild (A, E, and I) to severe defects (D, H, and L) were observed for the RNAi treatments using each insert. We found that specific responses were exhibited by portions of each microspore population, and we include these percentages with each image presented. dsRNAs made from Mv-Y14 and Mv-eIF4aIII both exhibited incomplete cell divisions (C and G, white arrows) and altered planes of cell division, causing an atypical cell arrangement within the spore walls. Thus, the silencing of Mv-Y14 and Mv-eIF4aIII results in defects similar to those observed with silencing of Mv-mago. dsRNAs made from Mv-Prp19 caused altered planes of cell division in the spermatogenous cells (I) and more severe phenocopies showed a loss of cell divisions (J and K). The defects caused by RNAi treatments employing dsRNA made from the pre-spliceosome component Mv-Prp19 are discernably different from those observed after silencing of any EJC core component. The most severe phenocopies (D, H, and L) are thought to be caused by dsRNA toxicity. The images were obtained with DIC. Bar, 25 μm.
The similarities in anomalous patterns of development leads us to suspect that Mv-mago works in conjunction with other components of the EJC. For all of the Mv-Y14 and Mv-eIF4AIII RNAi treatments, we were unable to detect punctate staining with anti-Mv-mago antibody, a result that suggests Mv-Y14 and Mv-eIF4AIII proteins are associated with or are essential for the appearance of the Mago-dots seen in untreated gametophytes (Figure 4, E and F). These results will be explored in greater depth elsewhere.
We wanted to know whether the developmental anomalies are caused by a general splicing malfunction or whether they are specific to the EJC. The addition of dsRNA made from Mv-Prp19 to spores resulted in developmental anomalies that were different from those observed with Mv-mago, Mv-Y14, or Mv-eIF4AIII. The majority of the spores had atypically arranged spermatogenous cells, which resulted from incorrect division planes, but the jacket cells were normal in size, numbers, and positions within the spore (Figure 8I). Approximately 30% of the spores exhibited oversized jacket cells, and the spermatogenous cells were grossly abnormal, as if particular division cycles had never occurred (Figure 8J). Only a small percentage of the gametophytes (<10%) resembled Mv-mago RNAi-treated spores in that they had incomplete cell formation, incomplete cell placement, abnormally large jacket cells, and dislocations of starch-bearing plastids (Figure 8K). We attribute extreme abnormalities to be caused by dsRNA toxicity (Figure 8L).
DISCUSSION
Our isolated mago nashi homologue from a Marsilea microspore cDNA library exhibits a predicted amino acid sequence that is >70% identical to Mago nashi in flies, worms, fungi, and mammals (Figure 1). RNAi experiments, in situ hybridizations, and immunolocalizations show that Mv-mago is important for the normal development of the male gametophyte. From the onset of gametophyte development, some Mv-mago protein is present in the cytoplasm of the microspore, and it remains uniformly distributed in the cytoplasm as the spermatogenous and jacket cells are formed. Mv-mago protein increases are apparent after the division phase has ended (Figure 3B) when it becomes localized in cytoplasmic punctae, Mago-dots, in both jacket and spermatogenous cells (Figure 4).
RNA silencing reveals that new Mv-mago protein synthesis occurs during the cell division phase in the gametophyte, because the treatment causes the normally asymmetric divisions that give rise to jacket cells to become more symmetric. With Mv-mago silencing the presumptive jacket cells are larger than normal and ultimately do not senesce as in normal gametophytes (Sharp, 1914; Mizukami and Gall, 1966; Hepler, 1976). Simultaneously, presumptive spermatogenous cells fail to mature, and punctate cytoplasmic staining with anti-Mv-mago antibody is undetectable.
Because basal body formation occurs exclusively in spermatogenous cells (Sharp, 1914; Mizukami and Gall, 1966; Hepler, 1976), it is clear that centrin translation, blepharoplast formation, and basal body assembly are tangibly unique aspects of fate specification for spermatogenous cells in the gametophyte. These components are not found in the immediately adjacent jacket cells of the normal gametophyte, although both spermatogenous and jacket cells arise from the same progenitor. The normal pattern of centrin translation (Klink and Wolniak, 2001) is altered after Mv-mago dsRNA treatment of microspores; centrin protein is ubiquitous in all cells of the gametophyte (Figure 7K). Later in development, centrin aggregates into particles in sterile jacket cells that resemble blepharoplasts and presumptive basal bodies (Figure 7, K and L). The appearance of these blepharoplast-like particles in jacket cells is in striking contrast to knockdowns of housekeeping genes or of genes that affect cell division directly (Tsai and Wolniak, 2001; Klink and Wolniak, 2003), where blepharoplast and basal body formation is suppressed in all cells of the gametophyte in spite of the fact that centrin translation occurs at its normal time and accumulates to essentially normal levels (Tsai and Wolniak, 2001).
Mago-Dots in the Cytoplasm of the Spermatids
In several organisms, Mago nashi has been localized within the nucleus (Micklem et al., 1997; Newmark et al., 1997), often in nuclear speckles (Degot et al., 2004), where little or no DNA is present, and where splicing factors are thought to be stored (Lamond and Spector, 2003). In Marsilea male gametophytes, Mv-mago proteins aggregate into Mago-dots (to distinguish them from nuclear speckles) at the end of the cell division phase, alongside increases in the abundance of numerous proteins (Klink and Wolniak, 2003).
We suspect that Mv-mago protein is a temporal and spatial translation regulator during spermatid differentiation. Dry spores contain large quantities of stored mRNAs, whose translation drives development in the absence of new transcription (Hart and Wolniak, 1998, 1999; Klink and Wolniak, 2001, 2003). Like many rapidly developing systems (Leatherman and Jongens, 2003), spermatogenesis is transcriptionally quiescent in Marsilea. So, although the absence of detectable anti-Mv-mago antibody staining in the nuclei of the spermatids was surprising, it may be the result of unusual gamete nucleus remodeling as Mago-dots occur in the cytosol, or the result of unusual pre-mRNA processing that is linked to translational control for a large number of stored transcripts. In vitro translation experiments demonstrate that stored transcripts are not immediately available for translation when the spores are hydrated (Hart and Wolniak, 1998), so some level of mRNA processing could be a necessary prerequisite for bursts of translation that occur at specific times during development (Klink and Wolniak, 2001, 2003). Late in development, most if not all of the mRNAs become undetectable by in situ hybridization (Tsai et al., 2004), so considerable mRNA sequestration or degradation occurs as spermatids mature. Mago-dots remain prominent during this phase of development. Mv-mago protein might be recruited for the silencing or sequestering of transcripts.
RNAi treatments specific to Mv-Y14 and Mv-eIF4AIII also cause Mago-dots to disappear from the gametophytes. Because mago, Y14, and eIF4AIII are three of the core components of the EJC (Ballut et al., 2005), Mago-dots could be cytoplasmic aggregations of EJCs. We suspect they control patterns of transcript distribution and translation in the cytoplasm that underlie spermiogenetic cell fate differentiation, as EJC are known to function elsewhere (Hachet and Ephrussi, 2004). EJC components also interact with the nonsense-mediated degradation (NMD) pathway (Kim et al., 2001; Lykke-Andersen et al., 2001; Gehring et al., 2003) where the dots could be localized foci for NMD in the spermatid cytoplasm. Recent results link the EJC with the targeting of premature termination coding-containing mRNAs to p-bodies in yeast (Sheth and Parker, 2006). We think a role for Mago-dots in NMD is less likely, because the dots are undetectable during early phases of gametophyte development.
Patterns of Localized Translation in the Gametophyte
The EJC core associates with factors that play distinct roles in mRNA nuclear export, cytoplasmic localization, translation, and quality control (for review, see Tange et al., 2004). Although we cannot tie EJC nuclear functions with changes observed in gametophytes, the normal localizations of Mv-Prp19 and Mv-eIF4AIII mRNAs in spermatogenous cells are lost in Mv-mago knockdowns, showing that Mv-mago and perhaps the EJCs affect the cytoplasmic transport of these transcripts. Thus, Mv-mago protein already present in the spore at the onset of development could control the timing of appearance and abundance of new EJC components and thereby alter the abundance and distribution of many proteins necessary for spermatid maturation.
The aggregation of newly translated centrin protein in jacket cells after Mv-mago knockdowns shows that Mv-mago protein is involved in restricting the translation of centrin to spermatogenous cells of normal gametophytes. Because centrin protein normally accumulates in spermatogenous cells after the asymmetric divisions produce the jacket cells (Klink and Wolniak, 2001), we can exclude the possibility of protein segregation occurring during the cell division phase. Centrin mRNA is abundant in both spermatogenous and jacket cells through 8 h of development (Tsai et al., 2004), so perhaps the control of translation for centrin and other proteins underlies developmental differences between spermatogenous and sterile cells. Like centrin, β-tubulin mRNA is found throughout the gametophyte (Tsai et al., 2004), but late in development tubulin translation is restricted to spermatids (Klink and Wolniak, 2001). Mv-Mago knockdowns disrupt tubulin protein distributions, revealing effects on proteins already present, and on locations where new translation of tubulin will occur. Translational differences between adjacent spermatogenous and sterile cells could result from differences in the stabilization of transcripts. We demonstrated striking differences in polyadenylated transcript abundance from spermatogenous and sterile cells of the gametophyte (Tsai et al., 2004); although levels of specific transcripts were comparable in both cell types, polyadenylated transcripts were abundant in the spermatogenous cells and barely detectable in the adjacent sterile cells.
The loss of Mago nashi proteins in the cytoplasm affects microtubule organization, which in turn alters the localization patterns of certain transcripts (Micklem et al., 1997; Newmark et al., 1997). Kinesin heavy chain has been found to contribute to the translocation of oskar mRNA and other components to the posterior end of the oocyte (Palacios and St. Johnston, 2002). Because the positioning and apportionment of plastids and mitochondria within cells of the male gametophyte have long been linked to changes in cytoskeletal organization (Wolniak, 1976), the unusual redistribution of large starch-containing plastids in spermatogenous cells of Mv-mago knockdowns (Figure 5) suggests that Mv-mago contributes to the control of large-scale polarity via the microtubule cytoskeleton and its motor proteins. Ultrastructural studies of Marsilea male gametophytes show that ribosome density is much higher in normal spermatogenous cells than jacket cells (Hepler, 1976); thus, a disrupted cytoskeleton could affect gametophytic ribosomal distribution.
Mv-mago nashi functions at multiple levels to control gametophyte development in M. vestita. Knockdowns of Mv-mago, Mv-Y14, and Mv-eIF4AIII link EJC functions in mRNA processing with translational patterns that control cell fate in the gametophyte. In the absence of Mv-mago protein, the normal patterns of stored β-tubulin accumulation and localized centrin translation in the spermatogenous cells are disrupted. Insofar as precise division planes define cell size, position, and identities, and compositional differences underlie the distinct fates of spermatogenous and jacket cells, Mv-mago protein seems to be essential for cell fate determination in M. vestita gametophytes.
ACKNOWLEDGMENTS
We are grateful to Dr. Jeff Salisbury for providing anti-centrin antibody, to Dr. Vincent Klink for help with anti-Mv-mago antibody purification, and to Dr. Peter Hepler for the original supply of sporocarps. Matthew Thompson, Vincent Klink, Faten Deeb, and Jeffrey Molk provided input, comments, suggestions, and encouragement at various stages of this project. We acknowledge support for this work from National Science Foundation grant MCB 0234423 and from the Maryland Agricultural Experiment Station.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-11-0979) on July 18, 2007.
REFERENCES
- Baskin T., Busby C. H., Fowke L. C., Sammut M., Gubler F. Improvements in immunostaining samples embedded in methacrylate: localization of microtubules and other antigens throughout developing organs in plants of diverse taxa. Planta. 1992;187:405–413. doi: 10.1007/BF00195665. [DOI] [PubMed] [Google Scholar]
- Baskin T. I., Wilson J. E. Inhibitors of protein kinases and phosphatases alter root morphology and disorganize cortical microtubules. Plant Physiol. 1997;113:493–502. doi: 10.1104/pp.113.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballut L., Marchadier B., Baguet A., Tomasetto C., Seraphin B., Le Hir H. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat. Struct. Mol. Biol. 2005;12:861–869. doi: 10.1038/nsmb990. [DOI] [PubMed] [Google Scholar]
- Boswell R. E., Prout M. E., Steichen J. C. Mutations in a newly identified Drosophila melanogaster gene, mago nashi, disrupt germ cell formation and result in the formation of mirror-image symmetrical double abdomen embryos. Development. 1991;113:373–384. doi: 10.1242/dev.113.1.373. [DOI] [PubMed] [Google Scholar]
- Degot S., Le Hir H., Alpy F., Kedinger V., Stoll I., Wendling C., Seraphin B., Rio M.-C., Tomasetto C. Association of the breast cancer protein MLN51 with the exon junction complex via its speckle localizer and RNA binding module. J. Biol. Chem. 2004;279:33702–33715. doi: 10.1074/jbc.M402754200. [DOI] [PubMed] [Google Scholar]
- Fire A., Xu. S., Montgomery M. K., Kostas S. A., Driver S. E., Mello C. C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Gehring N. H., Neu-Yilik G., Schell T., Hentze M. W., Kulozik A. E. Y14 and hUpf3b form an NMD-activating complex. Mol. Cell. 2003;11:939–949. doi: 10.1016/s1097-2765(03)00142-4. [DOI] [PubMed] [Google Scholar]
- Hachet O., Ephrussi A. Splicing of oskar RNA in the nucleus is coupled to its cytoplasmic localization. Nature. 2004;428:959–963. doi: 10.1038/nature02521. [DOI] [PubMed] [Google Scholar]
- Hardin S., Wolniak S. M. Molecular cloning and characterization of maize ZmMEK1, a protein kinase with a catalytic domain homologous to mitogen- and stress-activated protein kinase kinases. Planta. 1998;206:577–584. doi: 10.1007/s004250050435. [DOI] [PubMed] [Google Scholar]
- Hardin S., Wolniak S. M. Expression of the mitogen-activated proteinkinasekinase ZmMEK1 in the primary root of maize. Planta. 2001;213:916–926. doi: 10.1007/s004250100564. [DOI] [PubMed] [Google Scholar]
- Hart P. E., Wolniak S. M. Spermiogenesis in Marsilea vestita: a temporal correlation between centrin expression and blepharoplast differentiation. Cell Motil. Cytoskelet. 1998;41:39–48. doi: 10.1002/(SICI)1097-0169(1998)41:1<39::AID-CM3>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Hart P. E., Wolniak S. M. Molecular cloning of a centrin homolog from Marsilea vestita and evidence for its translational control during spermiogenesis. Biochem. Cell Biol. 1999;77:101–108. doi: 10.1139/o99-013. [DOI] [PubMed] [Google Scholar]
- Hepler P. K. The blepharoplast of Marsilea: its de novo formation and spindle association. J. Cell Sci. 1976;21:361–390. doi: 10.1242/jcs.21.2.361. [DOI] [PubMed] [Google Scholar]
- Hyams J. S., Vondy K. P., Luba A., Bell P. R. Structural and macromolecular events associated with basal body morphogenesis in Marsilea. J. Submicrosc. Cytol. 1983;15:133–138. [Google Scholar]
- Kataoka N., Diem M. D., Kim V. K., Yong J., Dreyfuss G. Magoh, a human homolog of Drosophila mago nashi protein, is a component of the splicing-dependent exon-exon junction complex. EMBO J. 2001;20:6424–6433. doi: 10.1093/emboj/20.22.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim V. N., Yong J., Kataoka N., Abel L., Diem M. D., Dreyfuss G. The Y14 protein communicates to the cytoplasm the position of exon-exon junctions. EMBO J. 2001;20:2062–2068. doi: 10.1093/emboj/20.8.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klink V. P., Wolniak S. M. The utility of RNAi in the study of the plant cytoskeleton. J. Plant Growth Regul. 2000;19:371–384. doi: 10.1007/s003440000043. [DOI] [PubMed] [Google Scholar]
- Klink V. P., Wolniak S. M. RNAi treatments reveal that centrin is necessary for the formation of the motile apparatus in spermatids of Marsilea. Mol. Biol. Cell. 2001;12:761–776. doi: 10.1091/mbc.12.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klink V. P., Wolniak S. M. Changes in the abundance and distribution of conserved centrosomal, cytoskeletal and ciliary proteins during spermiogenesis in Marsilea vestita. Cell Motil. Cytoskeleton. 2003;56:57–73. doi: 10.1002/cm.10134. [DOI] [PubMed] [Google Scholar]
- Lamond A. I., Spector D. L. Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 2003;4:605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- Leatherman J. L., Jongens T. A. Transcriptional silencing and translational control: key features of early germline development. Bioessays. 2003;25:326–335. doi: 10.1002/bies.10247. [DOI] [PubMed] [Google Scholar]
- Le Hir H., Izaurralde E., Maquat L. E., Moore M. J. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000;19:6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H., Gatfield D., Braun I. C., Forler D., Izaurralde E. The protein Mago provides a link between splicing and mRNA localization. EMBO Rep. 2001a;2:1119–1124. doi: 10.1093/embo-reports/kve245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H., Gatfield D., Izaurralde E., Moore M. J. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 2001b;20:4987–4997. doi: 10.1093/emboj/20.17.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M. J., Reed R. Splicing is required for rapid and efficient mRNA export in metazoans. Proc. Natl. Acad. Sci. USA. 1999;96:14937–14942. doi: 10.1073/pnas.96.26.14937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J., Shu M. D., Steitz J. A. Communication of the position of exon-exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science. 2001;293:1836–1839. doi: 10.1126/science.1062786. [DOI] [PubMed] [Google Scholar]
- Micklem D. R., Dasgurpta R., Elliott H., Gergely F., Davidson C., Brand A., Gonzalez-Reyes A., St Johnston D. The mago nashi gene is required for the polarisation of the oocyte and the formation of perpendicular axes in Drosophila. Curr. Biol. 1997;7:468–478. doi: 10.1016/s0960-9822(06)00218-1. [DOI] [PubMed] [Google Scholar]
- Mizukami I., Gall J. Centriole Replication II. Sperm formation in the fern Marsilea and the cycad, Zamia. J. Cell Biol. 1966;29:97–111. doi: 10.1083/jcb.29.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myles D. G., Hepler P. K. Spermiogenesis in the fern Marsilea vestita: microtubules, nuclear shaping, and cytomorphogenesis. J. Cell Sci. 1977;23:57–83. doi: 10.1242/jcs.23.1.57. [DOI] [PubMed] [Google Scholar]
- Myles D. G., Hepler P. K. Shaping of the sperm nucleus in Marsilea: a distinction between factors responsible for shape generation and shape determination. Dev. Biol. 1982;90:238–252. doi: 10.1016/0012-1606(82)90373-6. [DOI] [PubMed] [Google Scholar]
- Newmark P. A., Boswell R. E. The mago nashi locus encodes an essential product required for germ plasm assembly in Drosophila. Development. 1994;120:1303–1313. doi: 10.1242/dev.120.5.1303. [DOI] [PubMed] [Google Scholar]
- Newmark P. A., Mohr S. E., Gong L., Boswell R. E. Mago nashi mediates the posterior follicle cell-to-oocyte signal to organize axis formation in Drosophila. Development. 1997;124:3197–3207. doi: 10.1242/dev.124.16.3197. [DOI] [PubMed] [Google Scholar]
- Nott A., Meislin S. H., Moore M. J. A quantitative analysis of intron effects on mammalian gene expression. RNA. 2003;9:607–617. doi: 10.1261/rna.5250403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott A., Le Hir H., Moore M. J. Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Genes Dev. 2004;18:210–222. doi: 10.1101/gad.1163204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien T. P., McCully M. E. Principles and Selected Methods, Melbourne. Australia: Termarcarphi Pty. Ltd; 1981. The Study of Plant Structure. [Google Scholar]
- Palacios I. M., St. Johnston D. Kinesin light chain-independent function of the Kinesin heavy chain in cytoplasmic streaming and posterior localisation in the Drosophila oocyte. Development. 2002;129:5473–5485. doi: 10.1242/dev.00119. [DOI] [PubMed] [Google Scholar]
- Palacios I. M., Gatfield D., St. Johnston D., Izaurralde E. An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature. 2004;427:753–757. doi: 10.1038/nature02351. [DOI] [PubMed] [Google Scholar]
- Pennell R. I., Vondy K. P., Bell P. R., Hyams J. S. Composition and function of the blepharoplast of Marsilea vestita. Eur. J. Cell Biol. 1988;46:51–60. [Google Scholar]
- Sharp L. W. Spermatogenesis in Marsilia. Bot. Gaz. 1914;58:419–432. [Google Scholar]
- Sheth U., Parker R. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell. 2006;125:1095–1109. doi: 10.1016/j.cell.2006.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya T., Tange T. Ø., Sonenberg N., Moore M. J. eIF4AIII binds spliced mRNA in the exon junction complex and is essential for nonsense-mediated decay. Nat. Struct. Mol. Biol. 2004;11:346–351. doi: 10.1038/nsmb750. [DOI] [PubMed] [Google Scholar]
- Smith D., Johnson K. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Steel J. H., Gordon L., O'D. McGee J. In Situ Hybridization; Principles and Practice. In: Polak J. K., O'D. McGee J., editors. Oxford, United Kingdom: Oxford University Press; 1998. pp. 35–69. [Google Scholar]
- Tange T. Ø., Shibuya T., Jurica M. S., Moore M. J. Biochemical analysis of the EJC reveals two new factors and a stable tetrameric protein core. RNA. 2005;11:1869–1883. doi: 10.1261/rna.2155905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tange T. Ø., Nott A., Moore M. J. The ever-increasing complexities of the exon junction complex. Curr. Opin. Cell Biol. 2004;16:279–284. doi: 10.1016/j.ceb.2004.03.012. [DOI] [PubMed] [Google Scholar]
- Tsai C. W., Wolniak S. M. Cell cycle arrest allows centrin translation but not basal body formation during spermiogenesis in Marsilea. J. Cell Sci. 2001;114:4265–4272. doi: 10.1242/jcs.114.23.4265. [DOI] [PubMed] [Google Scholar]
- Tsai C.-W., van der Weele C. M., Wolniak S. M. Differential segregation and modification of mRNA during spermiogenesis in Marsilea vestita. Dev. Biol. 2004;269:319–330. doi: 10.1016/j.ydbio.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Wiegand H. L., Lu S., Cullen B. R. Exon junction complexes mediate the enhancing effect of splicing on mRNA expression. Proc. Natl. Acad. Sci. USA. 2003;100:11327–11332. doi: 10.1073/pnas.1934877100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolniak S. M. Organelle distribution and apportionment during meiosis in microsporocytes of Ginkgo biloba L. Am. J. Bot. 1976;63:251–258. [Google Scholar]
- Wolniak S. M., Klink V. P., Hart P. E., Tsai C.–W. Control of development and motility in the spermatozoids of lower plants. Gravit. Space Biol. Bull. 2000;13:85–93. [PubMed] [Google Scholar]
- Zhou Z., Luo M. J., Straesser K., Katahira J., Hurt E., Reed R. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature. 2000;407:401–405. doi: 10.1038/35030160. [DOI] [PubMed] [Google Scholar]