Abstract
The in vitro transfection activity of a novel series of N,N′-diacyl-1,2-diaminopropyl-3-carbamoyl-(aminoethane) derivatives was evaluated against a mouse melanoma cell line at different +/- charge ratios, in the presence and absence of helper lipids. Only the unsaturated derivative N,N′-dioleoyl-1,2-diaminopropyl-3-carbamoyl-(aminoethane), (1,2lmp[5]) mediated significant increase in the reporter gene level which was significantly boosted in the presence of DOPE peaking at +/- charge ratio of 2. The electrostatic interactions between the cationic liposomes and plasmid DNA were investigated by gel electrophoresis, fluorescence spectroscopy, dynamic light scattering and electrophoretic mobility techniques. In agreement with the transfection results, 1,2lmp[5]/DOPE formulation was most efficient in associating with and retarding DNA migration. The improved association between the dioleoyl derivative and DNA was further confirmed by ethidium bromide displacement assay and particle size distribution analysis of the lipoplexes. Differential scanning calorimetry studies showed that 1,2lmp[5] was the only lipid that exhibited a main phase transition below 37 °C. Likewise, 1,2lmp[5] was the only lipid found to form all liquid expanded monolayers at 23 °C. In conclusion, the current findings suggest that high in vitro transfection activity is mediated by cationic lipids characterized by increased acyl chain fluidity and high interfacial elasticity.
Keywords: Cationic lipids; Gene delivery; lipofection; 1,2-Dialkylamido; membrane fluidity; interfacial elasticity
1. Introduction
Gene delivery for the treatment of inherited and acquired diseases has been a therapeutic challenge in the last few decades that, once solved, will revolutionize our view and methods for the treatment of diseases. In addition to plasmid DNA, the recent advances in the fields of siRNA and oligonucleotide antisense and the significant potential these new therapeutics carry [1] emphasize the necessity for establishing an effective and safe delivery platform for these nucleic acid-based molecules [2,3]. Among the various gene delivery vectors, cationic lipids represent a promising tool for transfection of somatic cells. However, the relatively low efficiency of these vectors has limited their development into a clinically useful gene medicine [4]. Libraries of cationic lipids have been synthesized and screened in a restless effort to improve the transfection efficiency of these molecules [5,6]. Creative rational designs of pH-sensitive [7,8], photosensitive [9] and reducible cationic lipids [10] were also pursued but the outcome, unfortunately, was not the hoped one.
Several groups shifted their focus toward identifying and optimizing the different steps of the lipofection process to improve transfection efficiency of these vectors. Among the different steps of the lipofection process, the first step, namely interaction of cationic lipids with DNA and formation of the lipoplexes, is considered a key step from a pharmaceutical formulation point of view and therefore was thoroughly investigated. Specifically, the structural [11] and the thermodynamic [12,13] aspects of cationic lipid/DNA interactions and the effect of various factors on these interactions as well as on the final transfection activity were investigated [14-16]. In addition, the structure of the final lipoplex has been the topic for numerous studies to identify the ‘transfection active structure’ and factors that play a role in its formation [17-25]. While the transfection competent lipoplex macrostructure is not well defined yet, the corresponding microstructure appears to be composed of a multilammellar array of alternating lipid bilayers and DNA monolayers or inverted hexagonal assemblies composed of DNA tubes surrounded by a lipid monolayer, depending on the particular cationic and helper lipid [26-28].
In general, the molecular architecture of the cationic lipid is believed to influence and modulate the various steps in the lipofection process. The contribution of each of the cationic lipid segments to the lipofection process is, as yet, inadequately understood. One reason is that, aside from studies involving synthesis of cationic lipids and/or characterization of their lipoplexes, less was invested toward identifying the physicochemical properties of cationic lipids in isolation that correlates with improved transfection activity. The structure-activity relationship studies conducted so far were mainly empirical; trying to identify the molecular requirements of cationic lipids for improved transfection activity by solely screening of the cationic lipid and monitoring their activity profiles.
Toward this end, we have launched a long term structure-activity relationship study utilizing dialkylamidopropane-based cationic lipids [29-32]. In these cationic lipids, we have systematically varied the acyl chain length, the position of the alkyl chains, 1,3-dialkylamido versus 1,2-dialkylamido, as well as degree of methyl substitution of the cationic polar head group. In the current study, we continue our investigation by introducing the asymmetric 1,2-dialkylamidopropane-based derivatives bearing a single primary amine group as the cationic head group. Their in vitro transfection and cytotoxicity profiles are reported. The electrostatic interactions of these novel cationic lipids with plasmid DNA were characterized with several techniques and the effect of the presence of helper lipid on these interactions is described. Lastly, the physicochemical properties of these lipids such as main phase transition and interfacial properties were investigated and correlated with their in vitro transfection activity.
2. Materials and Methods
2.1. Materials
Ethylenediamine (95.5 %), tris (99.8+ %), cholesterol (>99 %), ninhydrin, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolam (MTT) and o-nitrophenyl β-D-galactopyranoside were purchased from Sigma–Aldrich (St. Louis, MO). DOPE was from AVANTI Polar Lipids Inc. (Alabaster, AL). Agarose, ethidium bromide, RPMI medium, fetal bovine serum, combined penicillin-streptomycin aqueous solution (10,000 U/ml and 10,000 μg/ml, respectively), sodium pyruvate and trypsin–EDTA 1x (10 mg/ml) were from Invitrogen Life Technologies (Carlsbad, CA). Water for buffer preparation was obtained from a Barnstead NANOpure ultrapure water system (Barnstead, Dubuque, IA). The resistivity and surface tension of the ultrapure water were 16-18 × 106 Ω cm and 72.5 mN/m, respectively.
2.2. Analytical procedures
Purification of compounds by column chromatography was performed with silica gel, 70–230 mesh, 60 Å (Sigma–Aldrich) and KONTES chromatography columns (VWR). Solvents for column chromatography were obtained from commercial suppliers and were all of HPLC grade. The reaction progress was followed by thin layer chromatography (TLC) developed on 0.25 mm silica gel plates (Fisher Scientific) using the following solvent systems: (A) CHCl3/CH3OH (20:1, v/v) and (B) CHCl3/CH3OH/aqueous NH4OH (66:33:1, v/v). TLC of the intermediate compounds was visualized by iodine vapor and UV. TLC plates of the final primary cationic lipids were additionally visualized using the amine group specific reagent, ninhydrin. 1H NMR spectra were recorded on a Varian Inova 400 MHz spectrometer using TMS as an internal standard. High-resolution mass spectroscopy was performed by the Department of Chemistry at Ohio State University. Elemental analysis (C, H, N) was carried out by Robertson Microlit Laboratories Inc. (Madison, NJ).
2.3. Synthesis
2.3.1. N,N′-dilauroyl-1,2-diaminopropyl-3-carbamoyl-(aminoethane), 1,2lmp[1]
The activated N,N′-diacyl-1,2-diaminopropane-3-(4-nitrophenyl)carbonate intermediates with varying acyl chains were synthesized as described previously [29]. To a solution of N,N′-dilauroyl-1,2-diaminopropane-3-(4-nitrophenyl) carbonate (1.2 g, 1.94 mmol) in 50 ml CHCl3, ethylenediamine (0.35 g, 5.82 mmol) was added dropwise with stirring at room temperature. After 3 h, an additional 150 ml of CHCl3 was added to dissolve the precipitated crude product and the reaction mixture was washed three times with 100 ml alkaline brine. The organic layer was collected and concentrated under diminished pressure. Purification by column chromatography gradient elution with 0 to 30 % CH3OH:CHCl3) afforded the pure compound as a white powder (0.547 g, 52.2 %). Rf = 0.39 (B). Anal. calcd for C30H60N4O4: C, 66.67; H, 11.11; N, 10.37. Found: C, 65.99; H, 11.34; N, 10.17. MS (ESI) m/z: calcd mass [M+Na]+ = 563.450, found mass = 563.453. 1H NMR (400 MHz, CDCl3, 20°C, TMS), δH (ppm): 6.82–6.78 [d, 1H, -CO-NH-CH-], 6.6–6.56 [t, 1H, -CO-NH-CH2-], 5.51–5.47 [t, 1H, -OCONH-], 4.13–4.08 [m, 3H, -CH-CH2-OCON-], 3.43–3.28 [m, 2H, -NH-CH2-CH-], 3.21–3.17 [m, 2H, -OCONH-CH2-], 2.82–2.78 [m, 2H, -CH2-NH2], 2.20–2.10 [m, 4H, -CO-CH2-], 1.61–1.50 [m, 4H, -CO-CH2-CH2-], 1.40–1.18 [coherent, 32H, -(CH2)8-], 0.85–0.82 [m, 6H, -CH3].
All other derivatives were synthesized using an analogous procedure as described above for the lauroyl derivative.
2.3.2. N,N′-dimyristoyl-1,2-diaminopropyl-3-carbamoyl-(aminoethane), 1,2lmp[2]
Yield = 47.5 %. Rf = 0.42 (B). Anal. calcd for C34H68N4O4: C, 68.45; H, 11.41; N, 9.39. Found: C, 67.81; H, 11.9; N, 9.35. MS (ESI) m/z: calcd mass [M + Na]+ = 619.513, found mass = 619.519. 1H NMR (400 MHz, CDCl3, 20°C, TMS), δH (ppm): 6.74–6.70 [d, 1H, -CO-NH-CH-], 6.52–6.48 [t, 1H, -CO-NH-CH2-], 5.40–5.35 [t, 1H, -OCONH-], 4.15–4.09 [m, 3H, -CH-CH2-OCON-], 3.43–3.30 [m, 2H, -NH-CH2-CH-], 3.22–3.18 [m, 2H, -OCONH-CH2-], 2.82–2.78 [m, 2H, -CH2-NH2], 2.20–2.10 [m, 4H, -CO-CH2-], 1.60–1.45 [m, 4H, -CO-CH2-CH2-], 1.32–1.16 [coherent, 40H, -(CH2)8-], 0.85–0.80 [m, 6H, -CH3].
2.3.3. N,N′-dipalmitoyl-1,2-diaminopropyl-3-carbamoyl-(aminoethane), 1,2lmp[3]
Yield = 55.2 %. Rf = 0.48 (B). Anal. calcd for C38H76N4O4: C, 69.94; H, 11.66; N, 8.59. Found: C, 67.99; H, 12.34; N, 8.27. MS (ESI) m/z: calcd mass [M + Na]+ = 675.576, found mass = 675.577. 1H NMR (400 MHz, CDCl3, 20 °C, TMS), δH (ppm): 6.70–6.67 [d, 1H, -CO-NH-CH-], 6.50–6.46 [t, 1H, -CO-NH-CH2-], 5.42–5.38 [t, 1H, -OCONH-], 4.15–4.10 [m, 3H, -CH-CH2-OCON-], 3.43–3.32 [m, 2H, -NH-CH2-CH-], 3.22–3.18 [m, 2H, -OCONH-CH2-], 2.85–2.80 [m, 2H, -CH2-NH2], 2.20–2.12 [m, 4H, -CO-CH2-], 1.64–1.52 [m, 4H, -CO-CH2-CH2-], 1.30-1.15 [coherent, 48H, -(CH2)8-], 0.85–0.80 [m, 6H, -CH3].
2.3.4. N,N′-distearoyl-1,2-diaminopropyl-3-carbamoyl-(aminoethane), 1,2lmp[4]
Yield = 50.9 %. Rf = 0.53 (B). Anal. calcd for C42H84N4O4: C, 71.18; H, 11.86; N, 7.91. Found: C, 70.86; H, 13.5; N, 7.73. MS (ESI) m/z: calcd mass [M + Na]+ = 731.638, found mass = 731.637. 1H NMR (400 MHz, CDCl3, 20 °C, TMS), δH (ppm): 6.75–6.70 [d, 1H, -CO-NH-CH-], 6.50–6.45 [t, 1H, -CO-NH-CH2-], 5.34–5.28 [t, 1H, -OCONH-], 4.12–4.05 [m, 3H, -CH-CH2-OCON-], 3.40–3.30 [m, 2H, -NH-CH2-CH-], 3.25–3.18 [m, 2H, -OCONH-CH2-], 2.83–2.78 [m, 2H, -CH2-NH2], 2.20–2.14 [m, 4H, -CO-CH2-], 1.60–1.50 [m, 4H, -CO-CH2-CH2-], 1.25-1.05 [coherent, 56H, -(CH2)8-], 0.85–0.80 [m, 6H, -CH3].
2.3.5. N,N′-dioleoyl-1,2-diaminopropyl-3-carbamoyl-(aminoethane), 1,2lmp[5]
Yield = 51 %. Rf = 0.58 (B). Anal. calcd for C42H80N4O4: C, 71.59; H, 11.36; N, 7.95. Found: C, 70.68; H, 11.34; N, 7.76. MS (ESI) m/z: calcd mass [M + Na]+ = 727.607, found mass = 727.607. 1H NMR (400 MHz, CDCl3, 20 °C, TMS), δH (ppm): 6.90–6.85 [d, 1H, -CO-NH-CH-], 6.65–6.60 [t, 1H, -CO-NH-CH2-], 5.65–5.60 [t, 1H, -OCONH-], 5.30–5.25 [m, 4H, -CH=CH-], 4.15–4.08 [m, 3H, -CH-CH2-OCON-], 3.42–3.35 [m, 2H, -NH-CH2-CH-], 3.25–3.20 [m, 2H, -OCONH-CH2-], 2.88–2.82 [m, 2H, -CH2-NH2], 2.18–2.10 [m, 4H, -CO-CH2-], 2.00–1.88 [m, 8H, -CH2-CH=CH- CH2-], 1.60–1.50 [m, 4H, -CO-CH2-CH2-], 1.32–1.10 [coherent, 40H, -(CH2)10-], 0.85–0.77 [m, 6H, -CH3].
2.4. Plasmid DNA Preparation
A pUC19 plasmid DNA vector containing the beta-galactosidase (β-Gal) reporter gene driven by the human cytomegalovirus (CMV) immediate-early promoter was amplified in E coli DH5α competent cells and extracted by detergent alkaline lysis [33]. Purification of the plasmid was effected by gel permeation chromatography on a Sepharose 4B column eluted with 2.5 M ammonium acetate. The concentration of plasmid DNA was determined spectrophotometrically at 260 nm using the relationship 1 OD = 50 μg/ml of plasmid DNA. The purity and quality of the plasmid DNA were verified by A260/A280 ratio and gel electrophoresis. A ratio of 1.89 was found suggesting a high quality plasmid free from protein and salt contamination.
2.5. Preparation of Liposomes
The newly synthesized 1,2lmp derivatives were found to be freely soluble in a 4:1 v/v chloroform/methanol mixture. Aliquots of cationic lipid solutions prepared on a weight per volume basis, along with other lipids were transferred to 12 mm × 75 mm borosilicate glass tubes (VWR). Lipid films were deposited on the walls of the tubes by evaporating the bulk solvent with a stream of dry nitrogen gas. To remove any residual solvents, the tubes were vacuum desiccated for an additional 4 hours. MLVs were prepared by hydrating the dry lipid films with 40 mM tris buffer pH 7.2 at 90 °C for one hour with occasional vortexing. The tubes were carefully sealed to prevent any loss of water during this process. The prepared MLVs were finally downsized by a 5-minute bath sonication. For transfection experiments, cationic lipid/DOPE and cationic lipid/DOPE/cholesterol molar ratios were fixed at 6:4 and 6:4:2, respectively.
2.6. Cell Culture and Transfection Assays
B16-F0 mouse melanoma cells (ATCC, CRL-6322) were cultured at 37 °C and 5% CO2 in RPMI media supplemented with 10 % fetal bovine serum, 1 mM sodium pyruvate, 50 U/ml penicillin and 50 μg/ml streptomycin. Approximately 5 × 104 cells were seeded into each well of a 48-well plate 12 h prior to transfection. Lipoplexes were prepared 30 min prior to transfection by mixing appropriate amounts of the cationic lipid liposomal dispersions with DNA in serum free media. Positive-to-negative charge ratio calculations were based on a 330 average nucleotide molecular weight. Each well was transfected with 250 μl of lipoplex dispersion corresponding to 1 μg of plasmid DNA. The transfection protocol and β-galactosidase assay followed herein are described in detail elsewhere [30].
2.7. In vitro Cytotoxicity Assay
Viability of B16-F0 mouse melanoma cells cultured and treated with lipoplexes in a similar fashion as described above for in vitro transfection experiments was evaluated by a modified MTT based cytotoxicity assay [34]. Briefly, after 44 h of transfection, 50 μl of MTT solution in PBS (5 mg/ml) were added to each well and cells were incubated for an additional 4 h at 37 °C. The media was discarded and the formed purple formazan crystals were dissolved with 250 μl of DMSO. The plates were gently agitated for 20 minutes before the absorption was read at 630 nm using Universal Microplate reader (Elx 800 UV, Bio-Tek Instruments Inc.). The absorption obtained for cells treated with DNA alone was taken to be 100%.
2.8. Agarose Gel Electrophoresis
The electrophoretic casting apparatus and power source utilized in this assay were obtained from BioRad®. The gel mold was filled to an approximate thickness of 5.0 mm with a standard Low-EEO agarose (0.8%) melted in a 16 mM TAE buffer pH 7.2, containing Ethidium bromide (0.5 μg/mL) for DNA band visualization.
Lipoplexes at +/- charge ratios of 0.5, 1, 2, 4, 6 and 8 were prepared in microfuge tubes by mixing 0.2 μg of DNA with the appropriate amount of the cationic lipid aqueous dispersions. One microliter of a 0.25 % bromophenol solution (30.0 % glycerol, 0.25 % bromophenol blue, 0.25 % xylene cyanol FF) was added to each tube and the total volume was brought to 11 μl with 40 mM tris buffer pH 7.4. Lipoplexes were briefly centrifuged at a speed of 14,000 rpm, and then allowed to equilibrate at ambient temperatures for 30 minutes. Naked DNA was loaded on the outermost lanes as a control. All samples were electrophoresed by applying a voltage of 5 V per centimeter length of casted gel for 35 minutes. The migration of lipoplexes was visualized using a KODAK Gel Logic 200 gel Imaging System equipped with a 590 nm filter (Eastman Kodak Co., Rochester, NY).
2.9. Ethidium Bromide Displacement Assay
Plasmid DNA (22.5 μg equivalent to 68.2 nmoles of negative charge) and ethidium bromide (2 nmoles) were added to a quartz cuvette and diluted to 3 ml with 40 mM tris buffer pH 7.2. Cationic liposomes prepared in the same buffer at 2 mM concentration were added in aliquots to the DNA/Eth-Br complex under constant stirring at 23 °C. The fluorescence of ethidium bromide was recorded with a Cary Eclipse fluorescence spectrophotometer (Varian Inc., CA) at an excitation wavelength of 515 nm (slit width 5 nm) and an emission wavelength of 595–605 nm (slit width 2.5 nm), with a 5-min interval between additions. A negative control lacking the fluorescence probe was run side by side to the original experiment to correct for light scattering effects. The fluorescence signal of ethidium bromide solution was subtracted from all measurements and the relative ethidium bromide displacement was calculated as described previously [29].
2.10. Dynamic Light Scattering
Hydrodynamic particle size distribution of 1,2lmp aqueous dispersions and their lipoplexes were analyzed by dynamic light scattering using a Malvern Zetasizer Nano ZS particle sizer (Malvern Instruments Inc., MA). The instrument was validated with polystyrene microspheres provided by the manufacturer. Lipoplexes were prepared in 0.2 μm filtered 40 mM tris buffer pH 7.2 by mixing cationic liposomes with plasmid DNA in a similar fashion to the in vitro transfection studies. All samples were allowed to equilibrate at 23 °C for 5 minutes, after which the samples were illuminated with a 532 nm laser beam and the scattered light was collected in a backscatter detection mode at an angle of 173°. The data presented are the intensity-based mean diameter (Zaverage) and polydispersity index (PI) obtained by a cumulant fit of the correlation function.
2.11. Differential Scanning Calorimetry
Lipid samples that were dried extensively under vacuum for 48 h were weighed (2-4 mg) directly into pre-weighed aluminum hermetic pans using a sartorius semi-micro balance (readability = 0.1 ± 0.03 mg). Aliquots of 40 mM tris buffer pH 7.2, calculated to achieve water/lipid molar ratio in the range of 35 - 40, were added to the lipid and the pan was immediately sealed. The lipids were hydrated in the DSC furnace at 90 °C for 1 h, after which time, heat/cool scans were recorded between -50 and 90 °C at a scan rate of 1 °C/min using a Q100 differential scanning calorimeter (TA instruments, DE) with an empty sealed pan as a reference. All samples were heat-cooled in the specified range for at least 4 consecutive cycles to ensure complete annealing of the sample and reproducibility of the scans. Maximum peak temperatures (Tm) and main phase transition enthalpies were determined using the TA universal analysis software (version 3.9A) provided by the manufacturer.
2.12. Monolayer Studies
The interfacial properties of the 1,2lmp amphiphilic derivatives in isolation were investigated using the Langmuir film balance technique. A computer controlled KSV minitrough film balance (KSV instruments LTD, Finland) equipped with two hydrophilic polyacetal made barriers and a platinum Wilhelmy plate was used to construct the surface pressure/area isotherms of monolayers of the cationic derivatives at the air/water interface. All experiments were conducted with a trough made of solid PTFE (1364 × 75 mm) filled with 140 ml of 40 mM tris buffer pH 7.2, as the subphase solution. Compression isotherms of 1,2lmp derivatives were constructed at a constant subphase temperature of 23 °C maintained by the aid of an external water bath circulator. Cationic lipid solutions at 0.75 μM concentration were prepared in chloroform/methanol (4:1) solvent mixture, stored at −20 °C and used within two days of preparation. Each experiment was repeated 3-4 times in an open-air vibration-free environment to ensure isotherm reproducibility. After each experiment, the trough and glasswares were exhaustively washed with purified water.
In a typical experiment, 30 μL aliquots of 1,2lmp lipid-spreading solutions were applied carefully drop by drop on the surface of the aqueous subphase using a Hamilton glass micro syringe. After an initial waiting period of 10 minutes, to ensure chloroform evaporation, the barriers were closed at a constant speed of 9.99 mm/minute. Analysis of the constructed π/A isotherms was carried out as described elsewhere [29,31].
3. Results
3.1. In vitro Transfection Activity and Toxicity
The ability of the five newly synthesized 1,2lmp cationic lipids (Figure 1) to mediate gene transfer in vitro was tested in B16-F0 mouse melanoma cell line. All saturated derivatives failed to mediate cell transfection in the absence or presence of helper lipids (not shown). Contrary to that, lipoplexes formulated with the dioleoyl derivative 1,2lmp[5] elicited a significant expression of the reporter gene which was further boosted in the presence of DOPE, peaking at 2:1 +/- charge ratio to levels comparable to those achieved by the commercially available cationic lipid DOTAP (Figure 2). Inclusion of cholesterol in the formulation or increasing the +/- charge ratio 1.2lmp[5]/DOPE reduced the transfection activity of this formulation.
Figure 1.
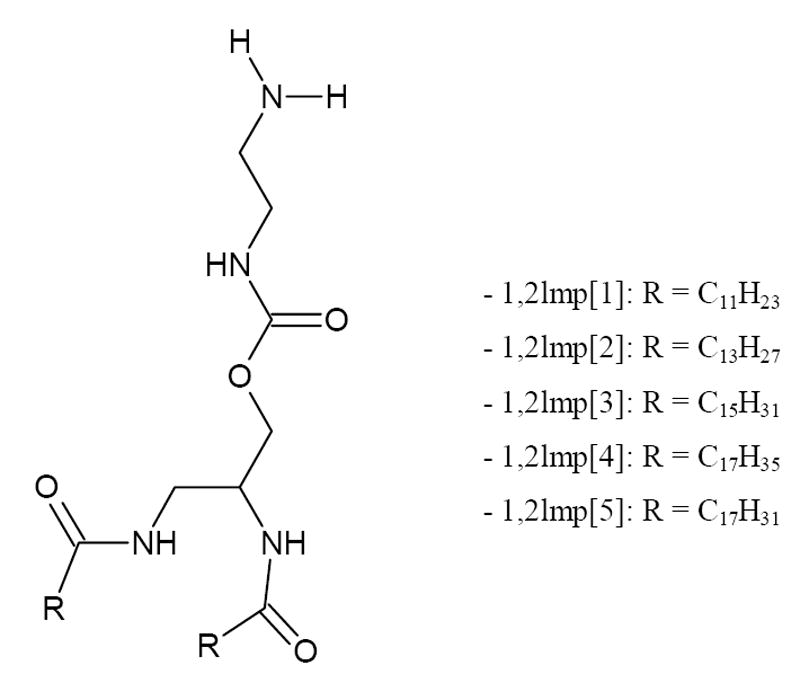
General structure of 1,2lmp derivatives.
Figure 2.
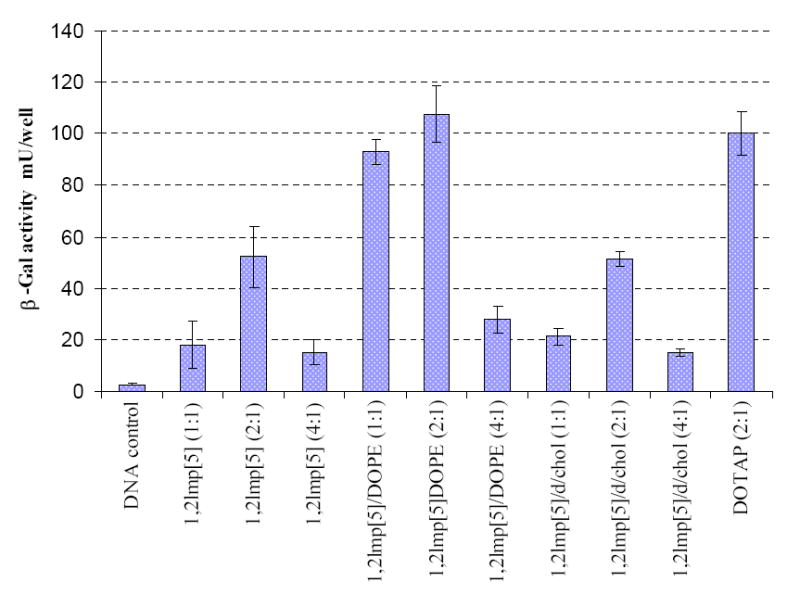
β-Gal gene expression in B16-F0 mouse melanoma cells transfected with various formulations of 1,2lmp[5]. The data shown present the average of four wells treated at the same day. The experiment was repeated 4 times and day-to-day variations were found to be within one fold of the presented data. β-galactosidase levels are expressed as mU/well, where each well is equivalent to 150 µl of cell lysate. Numbers in parenthesis denote +/- charge ratios.
As shown in Figure 3, formulations of 1,2lmp[5] were in general a bit less tolerated than DOTAP with a cell survival rate ≥65 % compared to 79 % for cells treated with DOTAP. On the other hand, in spite of their inefficiency to elicit any significant transfection activity, the saturated derivatives 1,2lmp[1 to 4] displayed comparable cytotoxicity profiles to those of 1,2lmp[5] (data not shown).
Figure 3.
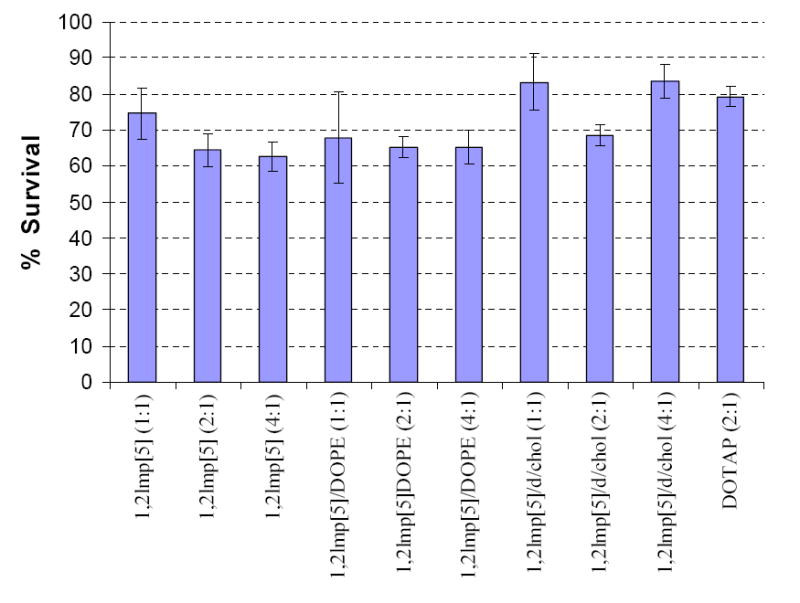
Percentage survival of B16-F0 mouse melanoma cells transfected with various formulations of 1,2lmp[5]. Data presented are the average of 4 wells treated at the same day. Results were calculated taking cells treated with DNA alone to be 100 %.
3.2. Agarose Gel Electrophoresis
As a first step to understand the transfection profile of the various formulations of 1,2lmp derivatives, their ability to complex plasmid DNA was investigated by gel electrophoresis retardation assay. All five lipids retarded the migration of plasmid DNA in the absence of the helper lipids in a similar fashion. As shown in Figure 4, complexation and complete retardation of plasmid DNA was evident at +/- charge ratio >2. The association of 1,2lmp[1] with DNA was practically unaffected in the presence of DOPE and cholesterol, while the complexation efficiency of 1,2lmp[5] slightly improved with DOPE but remained unaffected in the presence of cholesterol. Contrary to that, complexation of 1,2lmp[2] and 1,2lmp[3] was adversely affected by the inclusion of DOPE, whereas the 1,2lmp[4]/DOPE formulation failed to fully complex with plasmid DNA even at +/- charge ratios of 8. Inclusion of both DOPE and cholesterol in the formulations resulted in further reduction of the complexation efficiency of these latter lipids with DNA.
Figure 4. Representative gel electrophoresis pictures of lipoplexes prepared with various 1,2lmp formulations.
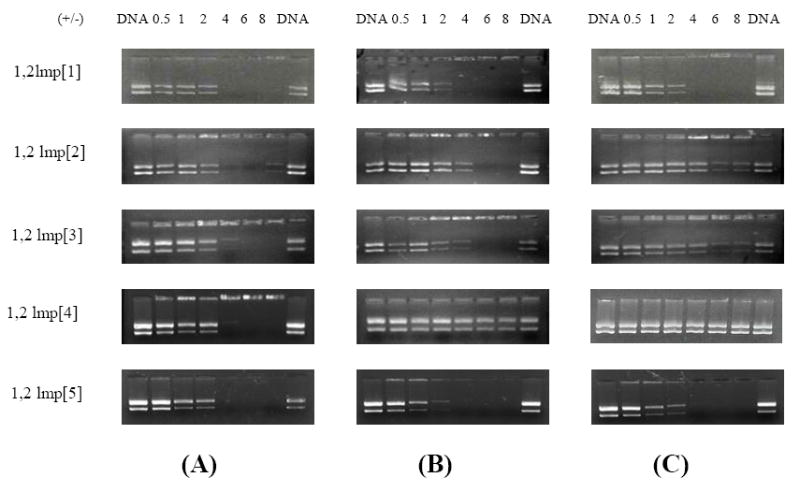
(A) Lipoplexes prepared with cationic lipids alone, (B) cationic lipid/DOPE (6:4 molar ratios) and (C) cationic lipid/DOPE/cholesterol (6:4:2 molar ratios).
3.3. Ethidium Bromide Displacement Assay
The ability of these lipids to condense plasmid DNA in 40 mM tris buffer pH 7.2 was quantitatively assessed from the reduction of emitted fluorescence of DNA intercalated ethidium bromide (Figure 5). The transfection efficient dioleoyl derivative 1,2lmp[5] could effectively induce 84 % condensation of plasmid DNA at +/- charge ratio of 1.6. The maximum condensation that could be induced before lipoplexes started to aggregate was equal to 88.5 % at an +/- charge ratio of 1.8. Contrary to the 1,2lmp[5], the first three saturated lipids, 1,2lmp[1 to 3], could only condense 45-50 % of the plasmid DNA at pH 7.2. This condensation percentage was achieved at a +/- charge ratio of 1.6, after which aggregation and precipitation of lipoplexes occurred and the titration had to be ceased. Finally, the stearoyl derivative 1,2lmp[4] could induce only 21.5 % condensation of plasmid DNA at +/- charge ratio of 1.8.
Figure 5.
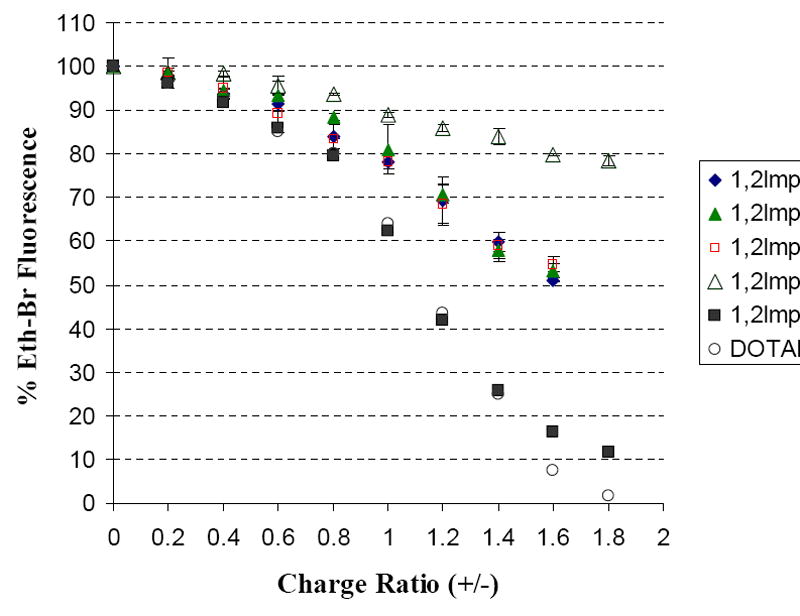
Cationic lipid induced DNA condensation at pH 7.2 monitored by the reduction in fluorescence due to the displacement of ethidium bromide from the DNA double helix.
3.4. Dynamic Light Scattering
Particle size distributions of the liposomal dispersions of 1,2lmp derivatives and their lipoplexes are summarized in Table 1. In 40 mM tris buffer pH 7.2, cationic liposomes prepared in the absence of helper lipids exhibited a mean diameter below 200 nm. Cationic liposomes composed of 1,2lmp[1]/DOPE, 1,2lmp[1]/DOPE/Chol, 1,2lmp[5]/DOPE and 1,2lmp[5]/DOPE/Chol exhibited similar size to the corresponding cationic liposomes made without helper lipids. Contrary to that, inclusion of DOPE and Cholesterol in formulations made with 1,2lmp[2], 1,2lmp[3] and 1,2lmp[4] resulted in significant increase of liposome size or aggregate formation. Similarly, aggregate formation took place when 1,2lmp[2 to 4] were complexed with DNA in the absence and presence of helper lipids, at all +/- charge ratios investigated (not shown). The aggregation behavior of these lipids correlates with their poor transfection activity.
Table 1.
Particle size distributions of 1,2lmp cationic liposomes and lipoplexes at different +/- charge ratios in 40 mM tris buffer pH 7.2a.
| Cationic Lipid | Cationic Lipid/DOPE | Cationic Lipid/DOPE/Chol. | |||||||
|---|---|---|---|---|---|---|---|---|---|
| In 40 mM tris buffer, pH 7.2 | D (nm) | PIb | D (nm) | PI | D (nm) | PI | |||
| 1,2lmp[1] | Liposomes | 166 | 0.16 | 184 | 0.29 | 167 | 0.28 | ||
| Lipoplexes (1:1) | 324 | 0.23 | 374 | 0.44 | 447 | 0.46 | |||
| Lipoplexes (2:1) | AGc | ---- | AG | ---- | AG | ---- | |||
| Lipoplexes (4:1) | AG | ---- | AG | ---- | AG | ---- | |||
| 1,2lmp[2] | Liposomes | 196 | 0.16 | 1248 | 0.94 | 490 | 0.5 | ||
| 1,2lmp[3] | Liposomes | 181 | 0.19 | AG | ---- | AG | ---- | ||
| 1,2lmp[4] | Liposomes | 169 | 0.4 | AG | ---- | AG | ---- | ||
| 1,2lmp[5] | Liposomes | 108 | 0.2 | 118 | 0.21 | 139 | 0.19 | ||
| Lipoplexes (1:1) | 162 | 0.21 | 171 | 0.21 | 192 | 0.21 | |||
| Lipoplexes (2:1) | 1501 | 0.28 | AG | ---- | AG | ---- | |||
| Lipoplexes (4:1) | 245.7 | 0.23 | 203 | 0.17 | 384 | 0.28 | |||
Experiments were done in duplicates and data points shown represent the mean diameter and polydispersity index values obtained from one experiment. Values from the other experiment were within the standard deviation of the presented results. The standard deviation is calculated from the corresponding polydispersity index using the relation: PI = (σ/Zaverage)2
PI denotes for polydispersity index.
AG denotes for aggregates with polydispersity indices = 1.
1,2lmp[2-4] resulted in visible aggregates when complexed with DNA at all +/- charge ratios, and are not included in the Table.
The dilauroyl derivative 1,2lmp[1] alone or in the presence of helper lipids interacted with DNA and formed small size lipoplexes at 1:1, but exhibited aggregate formation at charge ratio ≥ 2 (Table 1; +/- charge ratio of 6 and 8 not shown). Contrary to the other lipids, complexation of cationic liposomes made with 1,2lmp[5] alone or in the presence of helper lipids with plasmid DNA at +/- ratios of 1:1, 2:1 and 4:1, resulted in small size, large size and very small size lipoplex particles, respectively.
3.5. Differential Scanning Calorimetry
Representative DSC heating scans of hydrated bilayers of the five 1,2lmp cationic derivatives in 40 mM tris buffer pH 7.2 are presented in Figure 6 and the thermodynamic parameters extracted from these scans are summarized in Table 2. The most striking feature is that the dioleoyl derivative 1,2lmp[5] exhibited a gel-to-liquid crystalline phase transition at a significantly much higher temperature than what would be expected for a double-chained ester analog. Nevertheless, the presence of the cis double bond in the hydrocarbon chain of 1,2lmp[5] is reflected by the extensive broadening of the main phase transition peak over ~ 28 degrees. The onset point temperature of the 1,2lmp[5] phase transition, determined by the TA universal analysis software (TA instruments, DE) as the point at which a change in the slope of the curve occurs, and the mean temperature of the transition, were 10.71 °C ± 1.45 and 27.17 °C ± 0.86, respectively.
Figure 6.
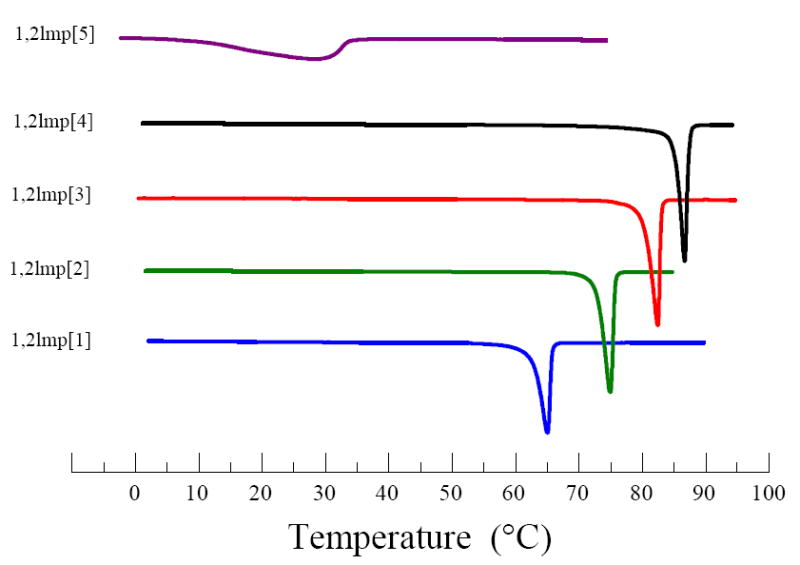
Representative heating scan of various 1,2lmp derivatives in 40 mM tris buffer pH 7.2.
Table 2.
Main phase transition parameters of 1,2lmp derivatives in 40 mM tris buffer pH 7.2
| Lipid | Tm (°C) | ΔH (KJ/mol) |
|---|---|---|
| 1,2lmp[1] | 65.27 ± 0.24 | 48.462 ± 1.22 |
| 1,2lmp[2] | 75.12 ± 0.16 | 59.189 ± 1.00 |
| 1,2lmp[3] | 82.40 ± 0.10 | 69.495 ± 1.01 |
| 1,2lmp[4] | 86.05 ± 0.57 | 81.557 ± 0.31 |
| 1,2lmp[5] | 27.17 ± 0.86 | 28.739 ± 0.04 |
The four saturated derivatives, 1,2lmp[1 to 4] exhibited sharp close to symmetric phase transitions with mean temperatures notably higher than physiological temperature (Figure 6). These phase transition temperatures appear to increase steadily but disproportionally with acyl chain length. More specifically, the difference in the main phase transition temperature was initially 10 degrees between 1,2lmp[1] and 1,2lmp[2]. Increasing the chain length by an additional 2 carbons resulted in only a 7.2 degree increase in the phase transition temperature. Further increase in the chain length from 16 to 18 carbons was accompanied by a smaller increase of the phase transition temperature of 3.35 degrees. Unlike the main phase transition temperature, the transition enthalpy of bilayers prepared with the saturated 1,2lmp derivatives increases in a linear fashion as the length of the hydrocarbon acyl chain length is increased (Figure 7 and Table 2). On average, increasing the acyl chain length of saturated 1,2lmp derivatives by 2 carbons is accompanied by 11.03 ± 0.92 kJ/mole.
Figure 7.
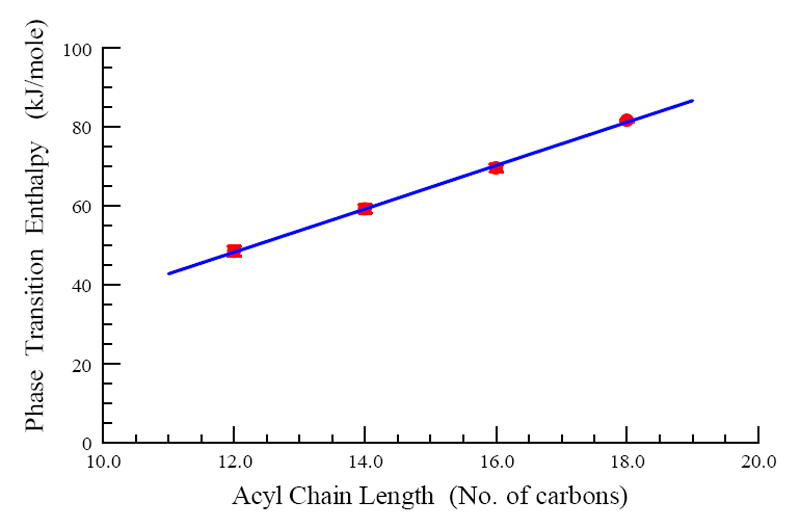
Main phase transition enthalpies of various 1,2lmp derivatives in 40 mM tris buffer pH 7.2 as a function of chain length. The data presented are mean values and standard deviations obtained from triplicate experiments.
3.6. Monolayer Studies
Representative π/A isotherms of the five derivatives as well as the first derivative of the surface pressure as a function of the mean molecular area are presented in Figure 8 and the monolayer parameters of 1,2lmp cationic lipids are summarized in Table 3.
Figure 8.
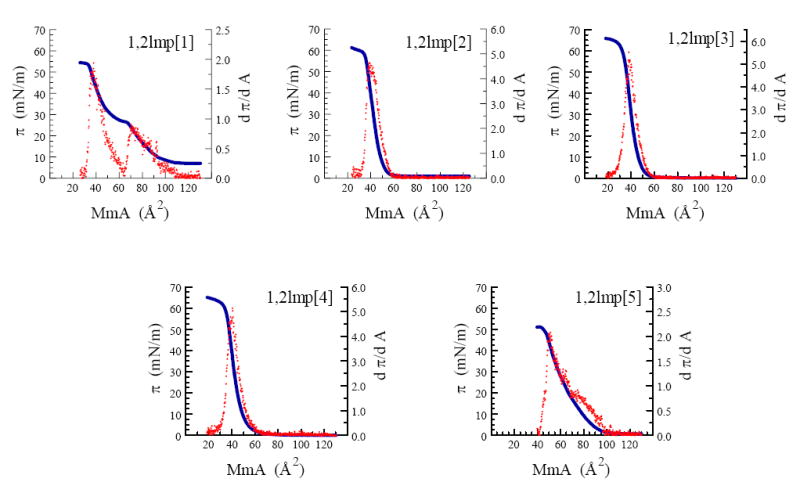
Surface pressure (π, —) and first derivative of surface pressure (dπ/dA, ˙) as a function of mean molecular area of various 1,2lmp derivatives at 23 °C.
Table 3.
Monolayer parameters of 1,2lmp amphiphilic seriesa.
| MmA (Ǻ2) | πc (mN/m) | K (mN/m) | dπ/dA | Phase state | |
|---|---|---|---|---|---|
| 1,2lmp[1] | 37.98 ± 0.33
70.87 ± 1.46b |
47.23 ± 1.35
23.77 ± 0.45 |
73.49 ± 5.97
69.1 ± 4.83 |
2.05 ± 0.16
0.96 ± 0.06 |
L2
L1 |
| 1,2lmp[2] | 38.06 ± 0.19 | 46.95 ± 1.32 | 205.3 ± 6.8 | 4.66 ± 0.23 | L2 |
| 1,2lmp[3] | 37.64 ± 0.56 | 43.10 ± 2.69 | 208.6 ± 9.51 | 5.27 ± 0.29 | L2 |
| 1,2lmp[4] | 38.90 ± 0.68 | 44.81 ± 1.11 | 198.8 ± 10.97 | 4.91 ± 0.27 | L2 |
| 1,3lmp[5] | 51.35 ± 0.23 | 41.17 ± 0.70 | 104.5 ± 8.26 | 1.66 ± 0.17 | L1 |
L1 denotes liquid-expanded phase state.
L2 denotes liquid-condensed phase state.
MmA denotes mean molecular area expressed in Ǻ2 per molecule.
measured in 40 mM tris buffer pH 7.2, 23 °C. All parameters are ‘onset’ transition values.
phase transition denoted by a maximum in the compression isotherm.
Monolayers of the shortest derivative, 1,2lmp[1], exhibited a typical liquid-expanded to liquid-condensed transition in its compression isotherm at a molecular area and surface pressure of 70.87 Å2 and 23.77 mN/m, respectively and a monolayer collapse at a molecular area and surface pressure of 37.98 Å2 and 47.23 mN/m, respectively. Increasing the chain length of 1,2lmp by only 2 carbons dramatically affected the surface behavior of these lipids. At 23 °C, the surface pressure of 1,2lmp[2] monomolecular films remained relatively zero until a mean molecular area of 60 Å2 was reached. Further reduction of the molecular area resulted in a steep increase in the surface pressure until a monolayer collapse at 38.06 Å2 mean molecular area and 46.95 mN/m surface pressure was reached. The palmitoyl 1,2lmp[3] and the stearoyl 1,2lmp[4] derivatives exhibited an all liquid-condensed state with similar lift off and collapse mean molecular area as 1,2lmp[2]. Unlike the saturated derivatives, the transfection efficient lipid 1,2lmp[5] was the only lipid that formed all liquid-expanded monolayers at 23 °C. In addition, the molecular surface area of 1,2lmp[5] at the monolayer collapse was ~ 13 Å2 larger than that of the saturated analogues.
4. Discussion
The work presented herein is primarily focused on the effect of acyl chain lenth on the physicochemical and biological properties of N,N′-diacyl-1,2-diaminopropyl-3-carbamoyl-(aminoethane) analogs. The in vitro transfection activity of the five 1,2lmp derivatives was evaluated in B16-F0 mouse melanoma cell line. As Figure 2 shows, the dioleoyl derivative 1,2lmp[5] mediated significant in vitro transfection in this cell line with maximum expression of the reporter gene elicited by the 1,2lmp[5]/DOPE formulation at a +/- charge ratio of 2. In sharp contrast, none of the saturated derivatives mediated transfection activity in the absence or presence of helper lipids at all +/- charge ratios.
The electrostatic interactions between these novel cationic lipids and plasmid DNA were thoroughly investigated by a variety of techniques in an attempt to interpret their distinctive in vitro transfection profiles. In agreement with the in vitro transfection results, the DNA complexation efficiency of the various lipid formulations studied using a gel electrophoresis retardation assay shows that the optimum formulation 1,2lmp[5]/DOPE was the most efficient formulation in complexing and retarding plasmid DNA (Figure 4). Interestingly, inclusion of the helper lipid DOPE in the aqueous dispersion of the saturated derivatives either didn’t improve their DNA complexation efficiency, as in the case of 1,2lmp[1], or impeded their association of DNA. Moreover, the ethidium bromide displacement assay confirmed the DNA-compaction superiority of 1,2lmp[5] (Figures 5). This transfection active lipid condensed plasmid DNA at least by 45 % more than the saturated derivatives.
Formation of lipoplexes was further studied by means of dynamic light scattering technique, with particular emphasis on evaluating the effects of helper lipid DOPE in formulations composed of the shortest derivative 1,2lmp[1] and the unsaturated derivative 1,2lmp[5], since all other derivatives formed visible aggregates in the presence of DOPE and/or DNA. As shown in Table 1, the striking difference between these two lipids is that the size of 1,2lmp[1] lipoplexes monotonously increases with increasing +/- charge ratio, whereas the size of the 1,2lmp[5] lipoplexes increases at 2:1 but it again decreases at 200-400 nm at a +/- charge ratio ≥ 4, in the presence or absence of helper lipids.
Interfacial compressibility moduli (K) values and the rate of change of surface pressure with respect to the change of monolayer surface, dπ/dA, were calculated at 23 °C from monolayers assembled on 40 mM tris buffer pH 7.2. At monolayer collapse, 1,2lmp[1] and 1,2lmp[5] monolayers had K values of 73.49 mN/m and 104.5 mN/m, respectively, suggesting a higher in-plane elasticity of the 1,2lmp[1], although it does exist at a liquid-condensed state (see also Figure 6), as compared to the liquid-expanded state exhibited by the dioleoyl derivative. This result is quite unusual, but nonetheless the interfacial compressibility of the five analogues is in the order of 1,2lmp[1] > 1,2lmp[5] ≫ 1,2lmp[2] ≥ 1,2lmp[3] ≥ 1,2lmp [4], at 23 °C.
The observation that numerous lipid mixing and fusogenic events take place during the processes of liposome preparation, lipoplex formation and endosomal escape [11,19] points the attention toward the importance of the physicochemical properties of cationic lipids in isolation, such as phase state and interfacial elasticity. Several reports [35-37], including studies of our own [31,32] had established a correlation between membrane fluidity and improved in vitro transfection activity.
In the present study, assessment of membrane fluidity by means of differential scanning calorimetry indicated that the transfection efficient dioleoyl primary derivative 1,2lmp[5] was the only lipid to exhibit a phase transition below the physiological temperature (Figure 6). The current findings suggest that the bilayer of the dioleoyl derivative 1,2lmp[5] is in a gel-fluid coexisting phase state at room temperature while it exhibits an all liquid crystalline state at the physiological temperature. The transfection-deficient saturated 1,2lmp derivatives, on the other hand, are in a solid gel phase at 37 °C. In agreement with the differential scanning calorimetry data, monolayer studies showed that 1,2lmp[5] was the only lipid that exhibited an all liquid-expanded phase state when spread on 40 mM tris buffer pH 7.2 at 23 °C (Figures 8).
Using an amide bond to link the acyl chains to the polar region of 1,2-dialkylamidopropane-based derivatives increased the rigidity of these lipids as compared to their chain equivalent phospholipids and cationic lipids containing ester bonds instead. The increased stiffness of these lipids is indicated by the elevated phase transition temperatures and enthalpies of 1,2lmp cationic lipids (Table 2). More specifically, the average energy per CH2 group was calculated to be 2.619 KJ/mol as compared to 2.1 KJ/mol for diacyl phosphadityl cholines [38]. As suggested earlier, this could be in part due to the introduction of hydrogen bond functionalities in the polar part of this class of lipids [29] and to a smaller part to an increased restriction of acyl chain mobility due to the higher rotational energy barrier of amide bonds as compared to ester bonds. The rotational energy barrier of the amide bond in formamide, for example, is 18-19 kcal/mol while that for C-O is 3.9 kcal/mol in formic acid and 4.75 kcal/mol in methyl formate [39]. The kink in the unsaturated oleoyl chain introduced by the cis configuration of the double bond prevented tight packing of these molecules in monolayers or bilayers. Thereby, only the dioleoyl derivative possessed the acyl chain fluidity required for high in vitro transfection activity.
In summary, the current findings suggest that efficient association and encapsulation of plasmid DNA by cationic lipids are required for improved in vitro transfection activity. These processes can be only promoted by cationic lipids that are characterized by acyl chain fluidity indicated by a (1) main phase transition temperature of bilayers below 37 °C and (2) liquid-expanded state and higher interfacial elasticity of their corresponding monolayers.
Acknowledgments
The authors gratefully acknowledge Dr. Christopher M. Hadad in the Department of Chemistry, Ohio State University, for performing the mass spectroscopy analysis. This work was supported in part by a grant from the National Institutes of Health, EB004863 and by the Arnold & Marie Schwartz College of Pharmacy and Health Sciences.
Abbreviations
- DOPE
Dioleoylphosphatidylethanoleamine
- lmp
lipid monovalent primary
- 1,2lmp[1]
dilauroyl derivative
- 1,2lmp[2]
dimyristoyl derivative
- 1,2lmp[3]
dipalmitoyl derivative
- 1,2lmp[4]
distearoyl derivative
- 1,2lmp[5]
dioleoyl derivative
- MmA
mean molecular area
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zhou D, He QS, Wang C, Zhang J, Wong-Staal F. Curr Top Med Chem. 2006;6:901–911. doi: 10.2174/156802606777303630. [DOI] [PubMed] [Google Scholar]
- 2.Li SD, Huang L. Mol Pharm. 2006;3:579–588. doi: 10.1021/mp060039w. [DOI] [PubMed] [Google Scholar]
- 3.Spagnou S, Miller AD, Keller M. Biochemistry. 2004;43:13348–13356. doi: 10.1021/bi048950a. [DOI] [PubMed] [Google Scholar]
- 4.Kodama K, Katayama Y, Shoji Y, Nakashima H. Curr Med Chem. 2006;13:2155–2161. doi: 10.2174/092986706777935276. [DOI] [PubMed] [Google Scholar]
- 5.Ilies MA, Seitz WA, Balban AT. Curr Pharm Design. 2002;8:2441–2473. doi: 10.2174/1381612023392748. [DOI] [PubMed] [Google Scholar]
- 6.Miller AD. Chem Int Ed. 1998;37:1768–1785. [Google Scholar]
- 7.Benatti CR, Ruysschaert J, Lamy MT. Chem Phys Lipids. 2004;131:197–204. doi: 10.1016/j.chemphyslip.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 8.Singh RS, Goncalves C, Sandrin P, Pichon C, Midoux P, Chaudhuri A. Chem Biol. 2004;11:713–723. doi: 10.1016/j.chembiol.2004.03.026. [DOI] [PubMed] [Google Scholar]
- 9.Nagasaki T, Taniguchi A, Tamagaki S. Bioconjug Chem. 2003;14:513–516. doi: 10.1021/bc0256603. [DOI] [PubMed] [Google Scholar]
- 10.Wetzer B, Byk G, Frederic M, Airiau M, Blanche F, Pitard B, Scherman D. Biochem J. 2002;356:747–756. doi: 10.1042/0264-6021:3560747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huebner S, Battersby BJ, Grimm R, Cevc G. Biophys J. 1999;76:3158–3166. doi: 10.1016/S0006-3495(99)77467-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lobo BA, Davis A, Koe G, Smith JG, Middaugh CR. Arch Biochem Biophys. 2002;386:95–105. doi: 10.1006/abbi.2000.2196. [DOI] [PubMed] [Google Scholar]
- 13.Patel MM, Anchordoquy TJ. Biophys J. 2005;88:2089–2103. doi: 10.1529/biophysj.104.052100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferrari ME, Rusalov D, Enas J, Wheeler CJ. Nucleic Acids Res. 2001;29:1539–1548. doi: 10.1093/nar/29.7.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kennedy MT, Pozharski EV, Rakhmanova VA, MacDonald RC. Biophys J. 2000;78:1620–1633. doi: 10.1016/S0006-3495(00)76714-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matulis D, Rouzina I, Bloomfield VA. J Am Chem Soc. 2002;124:7331–7342. doi: 10.1021/ja0124055. [DOI] [PubMed] [Google Scholar]
- 17.Caracciolo G, Caminiti R. Chem Phys Letters. 2005;411:327–332. [Google Scholar]
- 18.Cherezov V, Qiu H, Pector V, Vandenbranden M, Ruysschaert JM, Caffrey M. Biophys J. 2002;82:3105–3117. doi: 10.1016/S0006-3495(02)75652-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gershon H, Ghirlando R, Guttman SB, Minsky A. Biochemistry. 1993;32:7143–7151. doi: 10.1021/bi00079a011. [DOI] [PubMed] [Google Scholar]
- 20.Koltover I, Salditt T, Rädler JO, Safinya CR. Science. 1998;281:78–81. doi: 10.1126/science.281.5373.78. [DOI] [PubMed] [Google Scholar]
- 21.Lin AJ, Slack NL, Ahmad A, George CX, Samuel CE, Safinya CR. Biophys J. 2003;84:3307–3316. doi: 10.1016/S0006-3495(03)70055-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pitard B, Oudrhiri N, Vigneron JP, Hauchecorne M, Aguerre O, Toury R, Airiau M, Ramasawmy R, Scherman D, Crouzet J, Lehn JM, Lehn P. Proc Natl Acad Sci USA. 1999;96:2621–2626. doi: 10.1073/pnas.96.6.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Radler JO, Koltover I, Salditt T, Safinya CR. Science. 1997;275:810–814. doi: 10.1126/science.275.5301.810. [DOI] [PubMed] [Google Scholar]
- 24.Sternberg B, Sorgi FL, Huang L. FEBS Lett. 1994;356:361–366. doi: 10.1016/0014-5793(94)01315-2. [DOI] [PubMed] [Google Scholar]
- 25.Wiethoff CM, Gill ML, Koe GS, Koe JG, Middaugh CR. J Biol Chem. 2002;277:44980–44987. doi: 10.1074/jbc.M207758200. [DOI] [PubMed] [Google Scholar]
- 26.Koltover I, Salditt T, Safinya CR. Biophys J. 1999;77:915–924. doi: 10.1016/S0006-3495(99)76942-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koynova R, Wang L, Tarahovsky Y, MacDonald RC. Bioconjug Chem. 2005;16:1335–1339. doi: 10.1021/bc050226x. [DOI] [PubMed] [Google Scholar]
- 28.Koynova R, Wang L, MacDonald RC. Proc Natl Acad Sci U S A. 2006;103:14373–14378. doi: 10.1073/pnas.0603085103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aljaberi A, Cheng P, Savva M. Chem Phys Lipids. 2005;133:135–149. doi: 10.1016/j.chemphyslip.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 30.Sheikh M, Feig J, Gee B, Li S, Savva M. Chem Phys Lipids. 2003;124:49–61. doi: 10.1016/s0009-3084(03)00033-1. [DOI] [PubMed] [Google Scholar]
- 31.Savva M, Aljaberi A, Feig J, Stolz DB. Colloids Surf B. 2005;43:43–56. doi: 10.1016/j.colsurfb.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 32.Savva M, Chen P, Aljaberi A, Selvi B, Spelios M. Bioconjug Chem. 2005;16:1411–1422. doi: 10.1021/bc050138c. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J, Russel DW. Molecular cloning. A laboratory manual. Cold Spring Harbor Laboratory Press; New York: 2001. [Google Scholar]
- 34.Savva M, Duda E, Huang L. Int J Pharm. 1999;184:45–51. doi: 10.1016/s0378-5173(99)00092-7. [DOI] [PubMed] [Google Scholar]
- 35.Akao T, Nakayama T, Takeshia K, Ito A. Mol Biol Int. 1994;34:915–920. [PubMed] [Google Scholar]
- 36.Wang J, Guo X, Xu Y, Barron L, Szoka FC., Jr J Med Chem. 1998;41:2207–2215. doi: 10.1021/jm950802i. [DOI] [PubMed] [Google Scholar]
- 37.Singh RS, Mukherjee K, Banerjee R, Chaudhuri A, Hait SK, Moulik SP, Ramadas Y, Vijayalakshmi A, Rao NM. Chem Eur J. 2002;8:900–909. doi: 10.1002/1521-3765(20020215)8:4<900::aid-chem900>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 38.Isrealachvili J. Intermolecular and Surface Forces. Second Edition. Academic Press; NY: 1992. p. 355. [Google Scholar]
- 39.Wiberg KB, Laidig KE. J Am Chem Soc. 1987;109:5935–5943. [Google Scholar]


