Abstract
Objective
To describe the prevalence of Staphylococcus warneri on the hands of nurses and the clinical relevance of this organism among neonates in the neonatal intensive care unit (NICU).
Design
Prospective cohort study that examined the microbial flora on the hands of nurses and clinical isolates recovered from neonates during a 23-month period (March 1, 2001, through January 31, 2003).
Setting
Two high-risk NICUs in New York City.
Participants
All neonates hospitalized in the NICUs for more than 24 hours and all full-time nurses from the same NICUs who volunteered to participate.
Intervention
At baseline and then every 3 months, samples for culture were obtained from each nurse’s cleaned dominant hand. Pulsed-field electrophoresis compared S. warneri isolates from neonates and staff.
Results
Samples for culture (n = 834) were obtained from the hands of 119 nurses; 520 (44%) of the 1,195 isolates of coagulase-negative staphylococci recovered were identified as S. warneri. Of the 647 clinically relevant isolates recovered from neonates, 17 (8%) of the 202 isolates that were identified to species level were S. warneri. Pulsed-field electrophoresis revealed a common strain of S. warneri that was shared among the nurses and neonates. Furthermore, 117 (23%) of 520 S. warneri isolates from nurses’ hands had minimum inhibitory concentrations for vancomycin of 4 μg/mL, which indicate decreasing susceptibility.
Conclusions
Our findings that S. warneri can be pathogenic in neonates, is a predominant species of coagulase-negative staphylococci cultured from the hands of nurses, and has decreased vancomycin susceptibility underscore the importance of continued surveillance for vancomycin resistance and pathogenicity in pediatric care settings.
Coagulase-negative staphylococci (CoNS) are the most common microorganisms that cause healthcare-associated infections in neonates hospitalized in the neonatal intensive care unit (NICU),1,2 with Staphylococcus epidermidis being the most common species recovered from clinically significant cultures.3-5 Although not a predominant pathogen, Staphylococcus warneri has been recovered from the hands of healthcare workers6,7 and hand carriage has been linked to the transmission of disease.8-10 Because of high oxacillin resistance rates among CoNS, vancomycin is the antimicrobial agent of choice. However, a trend toward decreased susceptibility to this glycopeptide has been shown among CoNS isolates.11,12
Few data are available regarding the prevalence and clinical significance of CoNS species other than S. epidermidis. We report on the prevalence of CoNS on the hands of nurses in 2 NICUs, with a focus on the epidemiology and vancomycin susceptibility of S. warneri. In addition, the clinical relevance of S. warneri among neonates in the same NICUs is explored.
Methods
Sample and Setting
This study was conducted in 2 NICUs that are part of the New York-Presbyterian Hospital in Manhattan in New York City. Data were obtained from all neonates hospitalized in the NICUs for more than 24 hours. Nurse participants were full-time employees who volunteered to participate in a larger clinical trial conducted from March 1, 2001, through January 31, 2003, that examined the relationship between hand hygiene practices and healthcare-associated infections in critically ill neonates.13 Sixty-one (77%) of 79 nurses from NICU 1, a 43-bed unit, and 58 (76%) of 76 nurses from NICU 2, a 50-bed unit, agreed to participate. Institutional review board approval was obtained from the participating institutions, and each nurse participant provided written consent before data collection. Data from 2,935 neonatal admissions and 119 nurses were included in this study.
Procedures
Patient cultures
Clinical specimens were collected from neonates by NICU staff when clinically indicated. A nurse epidemiologist hired specifically for this study conducted prospective surveillance for infections in neonates in both NICUs. Infections met Centers for Disease Control and Prevention case definitions14 and included bloodstream infections, pneumonia, conjunctivitis, skin and soft-tissue infections, and infections of the central nervous system. Samples for surveillance cultures were not obtained from infants.
Surveillance cultures from nurses’ hands
Samples for culture were collected from the dominant hand of each nurse participant at baseline and every 3 months during the 23-month study. Before sampling, participants cleansed their hands using the hand hygiene product (ie, 2% chlorhexidine gluconate or alcohol-based hand rub) available on the unit. A modified “glove-juice” technique was used for sampling, as described elsewhere.15
Microbiologic Procedures
All bacteriologic isolation, identification, antibiotic susceptibility tests, and molecular analyses were performed by the Clinical Microbiology Service of Columbia University Medical Center. As reported elsewhere,15 undiluted aliquots and diluted aliquots (10-fold and 100-fold) of samples of microbial flora were inoculated onto 5% sheep blood agar (Becton Dickinson Microbiology Systems) for determining total colony counts.
Staphylococcal isolates from cultures were speciated using the MicroScan System (Dade Behring) according to the manufacturer’s instructions. We confirmed the ability of the MicroScan System to identify S. warneri accurately to species level by 16S ribosomal RNA sequencing in 4 isolates from this study.
Antimicrobial susceptibility testing was performed using the microbroth dilution MicroScan System according to the manufacturer’s instructions. CoNS susceptibility to oxacillin was indicated by a minimum inhibitory concentration (MIC) of 0.25 μg/mL or less, and resistance was indicated by an MIC of 0.5 μg/mL or more. We used the current Clinical Laboratory Standards Institute (formerly National Committee for Clinical Laboratory Standards) guidelines16 for vancomycin MIC breakpoints of susceptible (4 μg/mL or less), intermediate (8 to 16 μg/mL), and resistant (more than 32 μg/mL). For the purposes of this study, MIC susceptibility subcategories of 2 μg/mL or less and 4 μg/mL were examined to determine the degree of decreased vancomycin resistance among S. warneri isolates. The Etest (AB Biodisk North America) was used to confirm vancomycin MICs for a random sample of S. warneri isolates (n = 24) recovered from nurses’ hands. To limit the possibility of error in susceptibility testing, all isolates from both neonates and nurses were tested in real time by the same laboratory technician. Isolates were stored by the clinical microbiology laboratory at -70°C for future analyses. For comparative purposes, the antimicrobial susceptibility patterns of unrelated S. warneri strains isolated during the study period from pediatric patients (n = 20) and adult patients (n = 37) hospitalized outside the NICU in 1 study hospital were analyzed.
Molecular Typing
The clinical isolates of S. warneri from neonates with bloodstream infection and S. warneri from the hands of nurses who provided care to these infected neonates in the same NICU underwent molecular typing. The isolates from nurses that were selected for molecular typing fulfilled the following criteria: the strain was isolated from a nurse’s culture sample within 1 month of a neonatal infection with S. warneri, and the S. warneri strains from the nurse and infected neonate had 2 or fewer differences in susceptibility to the antimicrobial agents tested. For comparative purposes, S. warneri strains from NICU nurses who did not provide nursing care to the infected neonates (n = 4), from adults with hospital-acquired infections caused by S. warneri (n = 4), and from adults in the community with hand carriage of S. warneri (n = 4) during the same period were also typed.
Bacterial genomic DNA was digested using SmaI endo-nuclease, and its genetic fingerprint determined by pulsed-field gel electrophoresis (PFGE) using the GenePath system (BioRad), as described elsewhere.17 The pattern of DNA restriction fragments was interpreted as indistinguishable, related, closely related, possibly related, or different, according to established criteria.18
Results
A total of 647 bacterial isolates were associated with clinical infection in the neonates during the 23-month study, of which 442 (68%) were from neonates in NICU 1 and 205 (32%) from neonates in NICU 2. Two hundred twenty-one (34%) of the isolates were CoNS, and 202 (91%) were identified to the species level (Table 1). Most CoNS-associated neonatal infections were caused by S. epidermidis (151 [68%]), followed by S. warneri (17 [8%]). The 17 neonatal infections caused by S. warneri included 15 bloodstream infections (13 catheter-related, 2 non-catheter related), 1 skin infection, and 1 eye infection.
Table 1.
Distribution of Coagulase-Negative Staphylococcus (CoNS) Species From Nurses’ Hands and Clinical Infections Among Neonates in 2 Neonatal Intensive Care Units (NICUs)
| No. (%) of isolates |
|||
|---|---|---|---|
| CoNS Species | From nurses | From neonates | Pa |
| S. epidermidis | 524 (44) | 151 (68) | <.001 |
| S. warneri | 520 (44) | 17 (8) | <.001 |
| S. capitis | 49 (4) | 6 (3) | .33 |
| S. hominis | 27 (2) | 16 (7) | <.001 |
| S. haemolyticus | 27 (2) | 4 (2) | .81 |
| S. simulans | 16 (1) | 5 (2) | .30 |
| S. auricularis | 15 (1) | 1 (.5) | .50 |
| Otherb | 17 (2) | 2 (1) | .75 |
| Total | 1,195 | 202 | ... |
χ2 or Fisher exact test.
Includes S. cohnii (n = 6), S. lugdunensis (n = 5), S. saprophyticus (n = 3), S. schleiferi (n = 1), S. sciuri (n = 1), and S. xylosus (n = 3).
A total of 834 samples for culture were analyzed from the nurses in NICU 1 (n = 450) and NICU 2 (n = 384). Of 1,442 bacterial isolates recovered, 1,195 (83%) were CoNS. Most CoNS isolates were S. epidermidis (524 [44%]), followed by S. warneri (520; 44%). Eleven additional CoNS species were also identified (Table 1).
Antibiotic Susceptibility
The distribution of S. warneri MICs for oxacillin and vancomycin is given in Table 2. Most NICU isolates were resistant to oxacillin; 84% of isolates from NICU nurses and 88% of S. warneri isolates from neonates had MICs of 0.5 μg/mL or higher.16 In comparison, lower rates of resistance to oxacillin were found among isolates from pediatric patients (65% of isolates) and adult patients (30% of isolates) hospitalized outside the NICU. Furthermore, 402 (77%) of the S. warneri isolates from nurses’ hands had vancomycin MICs of 2 μg/mL or less, and 117 (23%) had vancomycin MICs of 4 μg/mL. Only 1 isolate had intermediate susceptibility to vancomycin (MIC, 8 μg/mL). In contrast, 100% of the S. warneri isolates from neonates in the NICU and the isolates from pediatric and adult comparative populations had vancomycin MICs of 2 μg/mL or less.
Table 2.
Vancomycin and Oxacillin Susceptibilities of Staphylococcus warneri Isolates From Study Subjects
| No. (%) of isolates, by agent and susceptibility category |
||||||
|---|---|---|---|---|---|---|
| No. of isolates | Vancomycin MIC, μg/mLa |
Oxacillin MIC, μg/mL |
||||
| Cohort | Susceptible, ≤ 2 | Susceptible, 4 | Intermediate, 8-16 | Susceptible, <0.25 | Resistant, >0.5 | |
| Nurses in NICU | 520 | 402 (77) | 117 (23) | 1 (0.2) | 81 (16) | 438 (84) |
| Neonates in NICU | 17 | 17 (100) | 0 | 0 | 2 (12) | 15 (88) |
| Pediatric patients, other units | 20 | 20 (100) | 0 | 0 | 7 (35) | 13 (65) |
| Adult patients | 37 | 37 (100) | 0 | 0 | 26 (70) | 11 (30) |
NOTE. MIC, minimum inhibitory concentration; NICU, neonatal intensive care unit.
The χ2 was used to compare MICs among the 4 groups (P < .001).
Molecular Typing
Fifteen S. warneri isolates were recovered from 12 neonates with bloodstream infection in the NICU, 13 of which were available for PFGE. Of these isolates from neonates, 4 (31%) were indistinguishable from each other (ie, PFGE-designated clone A), 8 (61%) were related (ie, clones A1 and A2), and 1 isolate was unrelated. Eighteen isolates from nurses caring for 6 infected neonates were compared. Overall, 5 (83%) of these 6 neonates had isolates of clone A that were either indistinguishable from or related to isolates from at least 1 of their nurse caregivers. The Figure depicts the PFGE patterns of select isolates from 6 adults (different strains) and 8 study subjects with clone A.
Figure 1.
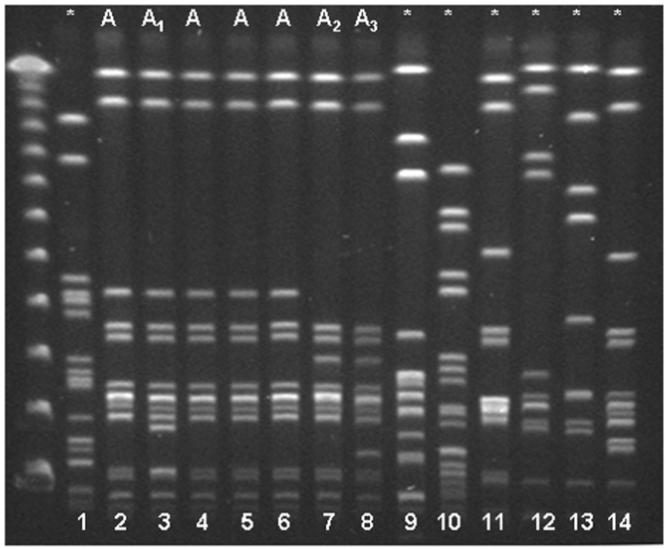
Pulsed-field gel electrophoresis patterns of Staphylococcus warneri isolates. Asterisk indicates unrelated isolates. Lanes 1-4, isolates from study neonates; lanes 5 and 6, isolates from nurses in neonatal intensive care unit 2; lanes 7 and 8, isolates from nurses in neonatal intensive care unit 1; lanes 9-11, isolates from adult patients; and lanes 12-14, isolates from adults from the community.
Discussion
CoNS in Hospitalized Neonates
To our knowledge, this is the largest study to examine the prevalence and molecular epidemiology of S. warneri among NICU staff and patients. Previous studies have assessed the relative prevalence of S. warneri compared with other CoNS species and demonstrated that, after S. epidermidis, S. warneri is usually the second most common species, responsible for 10%-50% of infections caused by CoNS.19-21 In a retrospective study at a children’s hospital, Buttery and colleagues20 reported that S. warneri was the second most common CoNS species associated with bloodstream infection. They reported no evidence of a single strain of S. warneri, but half of the isolates (6 of 12) were oxacillin resistant. In contrast, our susceptibility results show that 88% of the neonates and 84% of the NICU nurses had S. warneri strains that were oxacillin resistant. Among the isolates from neonates, no trend toward decreased resistance to vancomycin was seen; 100% of the strains had MICs of less than 2 μg/mL.
In the relatively few reports that describe the molecular typing of S. warneri associated with late-onset sepsis, small numbers of isolates have been studied. Kacica et al.21 demonstrated that 4 S. warneri isolates that caused late-onset sepsis were unrelated, whereas Raimondo et al.4 reported that during an initial 12-month study period, all S. warneri isolates (n = 8) that caused late-onset sepsis were unrelated, but during a follow-up 12-month study conducted 3 years later, most S. warneri isolates (5 of 7) were related. Our findings are similar in that almost all neonates (12 of 13) infected with S. warneri had isolates that were shown to be indistinguishable or related by PFGE.
CoNS Flora on Nurses’ Hands
As reported by others, most isolates cultured from the hands of nurses were CoNS6,7,22,23; however, the predominance of S. warneri has not been previously described. Horn et al.7 cultured samples from the skin of nurses, physicians, office workers, and students in an acute care hospital and found that 38% of CoNS isolates were S. epidermidis and 2% were S. warneri. Larson et al.6 examined the hand flora of 40 nurses and found that 131 (60.6%) of 216 isolates were staphylococcal species, of which 6.8% were Staphylococcus aureus, 31.3% were S. epidermidis, 21.4% were Staphylococcus hominis, 19.1% were S. warneri, and 21.4% were other CoNS species. Kloos et al.24 noted that S. warneri was “only occasionally” isolated from the skin of 40 adult participants. Although other studies have reported that NICU nurses are colonized with strains of CoNS shared with patients,25-27 to our knowledge none have described a predominant clone of S. warneri shared among nurses and neonates.
Several possible explanations for the high prevalence of S. warneri on nurses’ hands were considered. It is possible that prevalent species change over time or vary by geographic region. Perhaps prevalent species and skin colonization patterns emerge within specific work settings. For example, we noted in this same study that the staphylococcal species and antimicrobial resistance patterns of the hand flora of new graduate nurses changed after a few months of employment in the study unit and more closely resembled the flora of nurses who had been long-term employees.28
We also speculate that patterns of antimicrobial use, specifically vancomycin, in neonates may have facilitated S. warneri hand carriage. Of interest, we observed a trend toward reduced vancomycin susceptibility (MICs greater than 2 μg/mL) among S. warneri isolates recovered from nurses’ hands. The increased use of vancomycin in medical centers due to high oxacillin resistance may be responsible for the selection of subpopulations with decreased susceptibility to vancomycin. Center et al.29 reported that exposure to vancomycin, especially exposure for more than 10 days, and an NICU stay of more than 28 days were significantly associated with bloodstream infections caused by S. warneri strains with vancomycin MICs of greater than 2.0 μg/mL. Although none of the isolates that caused neonatal infections in our study were resistant to vancomycin, the decreased susceptibility to vancomycin seen in the S. warneri isolates among nurses’ hand flora may reflect trends in the strains that colonized the neonates. This speculation is strengthened by the observations that strains were shared among staff and neonates and that hand hygiene was suboptimal during the study.30 However, there may be other important, as yet unknown, epidemiologic or biologic factors associated with the transmission dynamics of S. warneri.
In conclusion, S. warneri can be a pathogen in neonates. Our finding that S. warneri was the predominant CoNS species cultured from the hands of nurses in the NICU is unique. The decreased vancomycin susceptibility noted among these S. warneri strains underscores the need for continued surveillance for vancomycin resistance. Future studies should further explore the dynamics of transmission of antibiotic-resistant flora among NICU patients and staff and the mechanisms of vancomycin resistance. Continued surveillance is needed to assess the potentially changing epidemiology of S. warneri to determine whether it is emerging as a healthcare-associated pathogen and whether the decreased susceptibility to vancomycin is contributing to its spread.
Acknowledgments
This study was funded by a grant from the National Institutes of Health (NINR 1 R01 NR05197, Staff Hand Hygiene and Nosocomial Infection in Neonates).
References
- 1.Gaynes RP, Edwards JR, Jarvis WR, Culver DH, Tolson JS, Martone WJ. Nosocomial infections among neonates in high-risk nurseries in the United States: National Nosocomial Infections Surveillance System. Pediatrics. 1996;98:357–361. [PubMed] [Google Scholar]
- 2.Grohskopf LA, Sinkowitz-Cochran RL, Garrett DO, et al. A national point-prevalence survey of pediatric intensive care unit-acquired infections in the United States. J Pediatr. 2002;140:432–438. doi: 10.1067/mpd.2002.122499. [DOI] [PubMed] [Google Scholar]
- 3.Fallat ME, Gallinero RN, Stover BH, Wilkerson S, Goldsmith LJ. Central venous catheter bloodstream infections in the neonatal intensive care unit. J Pediatr Surg. 1998;33:1383–1387. doi: 10.1016/s0022-3468(98)90013-6. [DOI] [PubMed] [Google Scholar]
- 4.Raimundo O, Heussler H, Bruhn JB, et al. Molecular epidemiology of coagulase-negative staphylococcal bacteraemia in a newborn intensive care unit. J Hosp Infect. 2002;51:33–42. doi: 10.1053/jhin.2002.1203. [DOI] [PubMed] [Google Scholar]
- 5.Udo EE, Jacob LE, Chugh TD. Antimicrobial resistance of coagulase-negative staphylococci from a Kuwait hospital. Microb Drug Resist. 1995;1:315–320. doi: 10.1089/mdr.1995.1.315. [DOI] [PubMed] [Google Scholar]
- 6.Larson EL, Hughes CA, Pyrek JD, Sparks SM, Cagatay EU, Bartkus JM. Changes in bacterial flora associated with skin damage on hands of health care personnel. Am J Infect Control. 1998;26:513–521. doi: 10.1016/s0196-6553(98)70025-2. [DOI] [PubMed] [Google Scholar]
- 7.Horn WA, Larson EL, McGinley KJ, Leyden JJ. Microbial flora on the hands of health care personnel: differences in composition and antibacterial resistance. Infect Control Hosp Epidemiol. 1988;9:189–193. doi: 10.1086/645831. [DOI] [PubMed] [Google Scholar]
- 8.Gupta A, Della-Latta P, Todd B, et al. Outbreak of extended-spectrum beta-lactamase-producing Klebsiella pneumoniae in a neonatal intensive care unit linked to artificial nails. Infect Control Hosp Epidemiol. 2004;25:210–215. doi: 10.1086/502380. [DOI] [PubMed] [Google Scholar]
- 9.Huang YC, Lin TY, Leu HS, Peng HL, Wu JH, Chang HY. Outbreak of Candida parapsilosis fungemia in neonatal intensive care units: clinical implications and genotyping analysis. Infection. 1999;27:97–102. doi: 10.1007/BF02560505. [DOI] [PubMed] [Google Scholar]
- 10.Foca M, Jakob K, Whitter S, et al. Endemic Pseudomonas aeruginosa infection in a neonatal intensive care unit. N Engl J Med. 2000;343:695–700. doi: 10.1056/NEJM200009073431004. [DOI] [PubMed] [Google Scholar]
- 11.Diekema DJ, Pfaller MA, Schmitz FJ, et al. Survey of infections due to Staphylococcus species: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United Status, Canada, Latin America, Europe, and the western Pacific region for the SENTRY Antimicrobial Surveillance Program, 1997-1999. Clin Infect Dis. 2001;32(Suppl 2):S114–S132. doi: 10.1086/320184. [DOI] [PubMed] [Google Scholar]
- 12.Tacconelli E, Tumbarello M, de Gaetano Donati K, et al. Glycopeptide resistance among coagulase-negative staphylococci that cause bacteremia: epidemiological and clinical findings from a case-control study. Clin Infect Dis. 2001;33:1628–1635. doi: 10.1086/323676. [DOI] [PubMed] [Google Scholar]
- 13.Larson EL, Cimiotti J, Haas J, et al. Effect of antiseptic handwashing vs alcohol sanitizer on health care-associated infections in neonatal intensive care units. Arch Pediatr Adolesc Med. 2005;159:377–383. doi: 10.1001/archpedi.159.4.377. [DOI] [PubMed] [Google Scholar]
- 14.Horan TC, Emori TG. Definitions of key terms used in the NNIS System. Am J Infect Control. 1997;25:112–116. doi: 10.1016/s0196-6553(97)90037-7. [DOI] [PubMed] [Google Scholar]
- 15.Larson E, Silberger M, Jakob K, et al. Assessment of alternative hand hygiene regimens to improve skin health among neonatal intensive care unit nurses. Heart Lung. 2000;29:136–142. [PubMed] [Google Scholar]
- 16.Clinical and Laboratory Standards Institute Performance Standards for Antimicrobial Testing 2005111–113.Clinical and Laboratory Standards Institute; 15th informational supplement Wayne, PA [Google Scholar]
- 17.Olive DM, Bean P. Principles and applications of methods for DNA-based typing of microbial organisms. J Clin Microbiol. 1999;37:1661–1669. doi: 10.1128/jcm.37.6.1661-1669.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tenover FC, Arbeit RD, Goering RV, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mehr SS, Sadowsky JL, Doyle LW, Carr J. Sepsis in neonatal intensive care in the late 1990s. J Paediatr Child Health. 2002;38:246–251. doi: 10.1046/j.1440-1754.2002.00768.x. [DOI] [PubMed] [Google Scholar]
- 20.Buttery JP, Easton M, Pearson SR, Hogg GG. Pediatric bacteremia due to Staphylococcus warneri: microbiological, epidemiological, and clinical features. J Clin Microbiol. 1997;35:2174–2177. doi: 10.1128/jcm.35.8.2174-2177.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kacica MA, Horgan MJ, Preston KE, Lepow M, Venezia RA. Relatedness of coagulase-negative staphylococci causing bacteremia in low-birth-weight infants. Infect Control Hosp Epidemiol. 1994;15:658–662. doi: 10.1086/646829. [DOI] [PubMed] [Google Scholar]
- 22.Lee YL, Cesario T, Lee R, et al. Colonization by Staphylococcus species resistant to methicillin or quinolone on hands of medical personnel in a skilled-nursing facility. Am J Infect Control. 1994;22:346–351. doi: 10.1016/0196-6553(94)90033-7. [DOI] [PubMed] [Google Scholar]
- 23.Perdreau-Remington F, Stefanik D, Peters G, et al. Methicillin-resistant Staphylococcus haemolyticus on the hands of health care workers: a route of transmission or a source? J Hosp Infect. 1995;31:195–203. doi: 10.1016/0195-6701(95)90066-7. [DOI] [PubMed] [Google Scholar]
- 24.Kloos WE, Musselwhite MS. Distribution and persistence of Staphylococcus and Micrococcus species and other aerobic bacteria on human skin. Appl Microbiol. 1975;30:381–385. doi: 10.1128/am.30.3.381-395.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patrick CH, John JF, Levkoff AH, Atkins LM. Relatedness of strains of methicillin-resistant coagulase-negative Staphylococcus colonizing hospital personnel and producing bacteremias in a neonatal intensive care unit. Pediatr Infect Dis J. 1992;11:935–940. doi: 10.1097/00006454-199211110-00006. [DOI] [PubMed] [Google Scholar]
- 26.Klingenberg C, Glad GT, Olsvik R, Flaegstad T. Rapid PCR detection of the methicillin resistance gene, mecA, on the hands of medical and non-medical personnel and healthy children and on surfaces in a neonatal intensive care unit. Scand J Infect Dis. 2001;33:494–497. doi: 10.1080/00365540110026485. [DOI] [PubMed] [Google Scholar]
- 27.Milisavljevic V, Wu F, Cimiotti J, Haas J, Della-Latta P, Larson E, Saiman L. Genetic relatedness of Staphylococcus epidermidis from infected infants and staff in the neonatal intensive care unit. Am J Infect Control. 2005;33:341–47. doi: 10.1016/j.ajic.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 28.Cimiotti JP, Wu F, Della-Latta P, Nesin M, Larson EL. Emergence of resistant staphylococci on the hands of new graduate nurses. Infect Control Hosp Epidemiol. 2004;25:431–435. doi: 10.1086/502418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Center KJ, Reboli AC, Hubler R, Rodgers GL, Long SS. Decreased vancomycin susceptibility of coagulase-negative staphylococci in a neonatal intensive care unit: evidence of spread of Staphylococcus warneri. J Clin Microbiol. 2003;41:4660–4665. doi: 10.1128/JCM.41.10.4660-4665.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cohen B, Saiman L, Cimiotti J, Larson E. Factors associated with hand hygiene practices in two neonatal intensive care units. Pediatr Infect Dis J. 2003;22:494–499. doi: 10.1097/01.inf.0000069766.86901.91. [DOI] [PMC free article] [PubMed] [Google Scholar]


