Abstract
Background and Aims
Chronic lymphocytic leukaemia (CLL) is a frequent non‐Hodgkin lymphoma characterised by a heterogeneous clinical course. Assessment of cell cycle phase kinetics might be important for prediction of clinical behaviour and prognosis.
Methods
Distribution of neoplastic cells in CLL within the cell cycle was evaluated by determining the labelling indices (LI, i.e. percentage of positive cells) of markers specific for late G1‐phase (cyclin E), S‐phase (cyclin A), and G2/M‐phase (cyclin B1), and Mcm2, a novel marker of proliferative potential, in a large cohort of patients (n = 79) using tissue microarray (TMA) technology. Utilising a combination of these markers, an algorithm was developed—subtracting the combined LIs of cyclin E, cyclin A and cyclin B1 from the LI of Mcm2—to determine the percentage of tumour cells residing in early G1‐phase, which is probably a critical state for the malignant potential of CLL.
Results
27.11% of cells had acquired proliferative potential as indicated by expression of Mcm2. Only a small number of cells were found to be in late G1‐phase (7.16%), S‐phase (3.31%) or G2/M‐phase (0.98%), while 15.66% of cells were considered to be in early G1‐phase.
Conclusion
Cell cycle phase distribution can easily be assessed by immunohistochemistry in routinely processed paraffin‐embedded specimens. A large number of neoplastic cells in CLL have proliferative potential, with a significant sub‐population residing in early G1‐phase. Estimates of these cells may identify cases likely to exhibit a more aggressive biological behaviour and adverse clinical course.
Keywords: chronic lymphocytic leukaemia, cell cycle, minichromosome maintenance protein, cyclins, tissue microarray
Chronic lymphocytic leukaemia (CLL) represents the most frequent type of leukaemia; its worldwide incidence ranges between 0.5 and 5.5 cases per 100 000 habitants per year.1 Although the overall median survival is approximately 10 years, the clinical course and prognosis of CLL are extremely heterogeneous. While some patients never require treatment and have a survival similar to that of healthy age‐matched individuals, others have a poor prognosis and an early treatment requirement.2 The proliferation of cells is a highly regulated process that depends on the precise duplication of DNA in each cell cycle. The licensing of replication origins and therefore the initiation of DNA replication is achieved by loading of the minichromosome maintenance (MCM) proteins 2–7 onto the DNA at the beginning of the G1‐phase of the cell cycle.3 Expression of MCM proteins is only observed in cycling cells; there is no expression in quiescent and differentiating cells.4
The standard marker for assessment of proliferation by immunohistochemistry has so far been Ki‐67.5,6 However, antibodies for detection of MCM proteins in routinely processed tissue specimens have been found superior to Ki‐67 in defining the proliferative compartments in both normal and abnormal tissues.7,8,9,10,11 MCM proteins are—unlike Ki‐67—expressed throughout the whole cell cycle.4,12 It has been shown that MCM proteins are expressed in Ki‐67 negative cells, which are therefore non‐proliferating but have already acquired “proliferative potential”.13,14 The progression of cells through the cell cycle is promoted by the oscillatory expression of cyclins: cyclin E in late G1‐phase, cyclin A in S‐phase, and cyclin B1 in G2‐phase and M‐phase.15,16,17,18
Relatively little attention has been paid so far to the analysis of cell cycle phase distribution in CLL.19 This is remarkable as it has been hypothesised that being in early G1‐phase instead of a G0 state could allow the neoplastic cells in CLL to be more responsive to external stimuli to further progress in the cell cycle.20 This is probably due to the fact that there is no specific marker available which allows estimation of the number of cells residing in early G1‐phase as opposed to G0‐phase; even the standard proliferation marker Ki‐67 is not expressed in early G1‐phase.21
We have now developed an approach to determine the number of cells in early G1‐phase by assessing the number of cells in late G1‐, S‐, G2‐ and M‐phase utilising phase specific markers and subtracting them from the number of Mcm2‐expressing cells.
The aim of this study was to critically investigate if these expression profiles can provide new insights into proliferative state and biology of CLL. Ultimately, it has to be determined whether determination of cell cycle phase parameters is of prognostic significance in CLL.
Materials and methods
Tissue microarrays
Tissue microarrays were constructed as described previously.22 Three TMAs were constructed, each containing tumour samples from different institutions (Basel, Bologna, Innsbruck). The TMAs contained a total of 79 cases of CLL. All cases included in this analysis had been classified according to the World Health Organization classification.23 All samples had been obtained at the time of diagnosis, before any treatment had been given. Samples from lymph nodes had been used; no bone marrow biopsies were included. Clinical data had been obtained by reviewing the charts. Retrieval of tissue and clinical data was performed according to the regulations of the local institutional review board and data safety laws.
Immunohistochemistry and cell cycle phase analysis
Sections (4 μm) of the TMA blocks were cut to adhesive‐coated slides (Instrumedics Inc, Hackensack, New Jersey, USA) and stained by standard procedures utilising an avidin–biotin peroxidase method with diaminobenzidine chromatogen. After antigen retrieval (microwave oven for 30 min at 320 W or pressure cooker for 5 min), immunohistochemistry was carried out in a NEXES immunostainer (Ventana, Tucson, Arizona, USA). The following primary antibodies were used: Ki‐67 (mouse monoclonal, clone MIB1, Dako, Hamburg, Germany; final dilution 1:50), BM28 for detection of Mcm2 (mouse monoclonal, clone 46, BD Biosciences, San José, California, USA; final dilution 1:3000), cyclin E (mouse monoclonal, clone 13A3, Novocastra, Newcastle, UK, final dilution 1:5), cyclin B1 (mouse monoclonal, clone 7A9, Novocastra; final dilution 1:5) and cyclin A (mouse monoclonal, clone 6E6, Novocastra; final dilution 1:100). Dilutions of primary antibodies had been established using adequate controls. Negative controls were obtained by omitting the primary antibody. Only cases containing clearly recognisable tumour tissue were analysed. At least 10% of cases were re‐evaluated by a second observer. For Ki‐67, Mcm2, cyclin E and cyclin A, only nuclear staining was considered to be specific, while for cyclin B1, cells with nuclear and/or cytoplasmic staining were regarded to be positive for this marker. One hundred cells were assessed in each tumour core and the percentage of labelled cells (labelling index (LI)) was calculated. If a biopsy core contained less than 100 cells, as many neoplastic cells as possible were evaluated. The percentage of cells in early G1‐phase was derived by subtracting the combined LIs of cyclin E, cyclin A and cyclin B1 from the LI of Mcm2. Cells which were negative for Mcm2 were considered to reside in G0.24 Descriptive statistics were performed using SPSS V.10.0 (SPSS, Chicago, Illinois, USA).
Results
Clinical data
The gender was known for 51 patients; 70.6% of these were male and 29.4% were female. Clinical data concerning the age of patients at the time of diagnosis were available in 35 cases. The mean age at time of diagnosis was 65 years (range 38–87 years).
Immunohistochemistry
Investigation of protein expression was informative for Ki‐67 in 70.9% (56/79), Mcm2 in 69.6% (55/79), cyclin A in 74.7% (59/79) and for cyclin B1 as well as cyclin E in 73.4% (58/79) of all cases. Data for all parameters were available in 51 of 79 cases (64.6%). Non‐informative cases were attributable to the array technology, in particular missing samples.
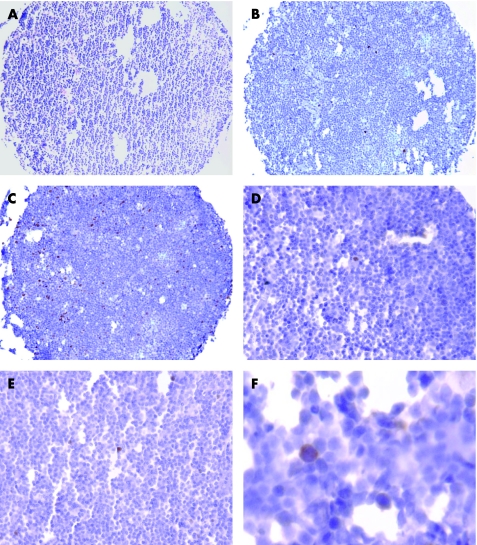
All markers—with the exception of cyclin B1—showed staining restricted to the nuclei. For cyclin B1, nuclear and/or cytoplasmic staining was encountered (fig 1). Non‐neoplastic lymphoid tissue, which was used as a positive control and for establishing the appropriate staining protocols, showed expression of all markers mainly in the germinal centres harbouring proliferating cells. Identical patterns have been described for Ki‐67 and Mcm2 as well as for cyclin B1 and cyclin A.25,26,27
Figure 1 H&E stain of a core biopsy (A). Immunohistochemical staining of Ki‐67 (B), Mcm2 (C), cyclin E (D), cyclin A (E), and cyclin B1 (F).
Cell cycle phase analysis
The number of cells expressing Ki‐67 was low, with a mean value of 5.73% (range 0–44%; SD 7.67). The average LI of Mcm2 was in excess of the LI of Ki‐67 with a mean value of 27.11% (range 2–62%; SD 9.49). Mcm2 expression always exceeded Ki‐67 expression except in one case, where it was equal (LI 44%).
The mean LI was 7.16% (range 0–34%; SD 7.72) for cyclin E, 3.31% (range 0–22%; SD 3.56) for cyclin A and 0.98% (range 0–8%; SD 1.61) for cyclin B1. The mean number of cells residing in early G1‐phase was calculated by the number of cells expressing cyclin E, A or B1 subtracted from the number of Mcm2 positive cells. There were five cases in which the combined LI for cyclin E, A and B1 exceeded the LI for Mcm2 (mean 5.6%; range 2–11%). The average of cells considered to reside in early G1‐phase was therefore 15.66%. Figure 2A shows the distribution of cells within the various cell cycle phases.
Figure 2 (A) Distribution of cells within the cell cycle in chronic lymphocytic leukaemia (CLL). Most cells are resting in G0‐phase, but a large number of cells has already entered the cell cycle and resides in early G1‐phase. Only few cells are found in late G1‐, S‐, G2/M‐phase. (B) Resting cells, cells with proliferative potential and actively proliferating cells in CLL. Most cells are resting in G0‐phase as indicated by lack of Ki‐67 and Mcm2 expression. A large number of cells has already acquired proliferative potential and shows expression of Mcm2, while only a small percentage of cells is actively proliferating as indicated by simultaneous expression of Mcm2 and Ki‐67.
Discussion
In this study, we used tissue microarray technology and immunohistochemistry to evaluate the distribution of neoplastic cells within the cell cycle by applying cell cycle phase specific markers in a large series of CLL. TMAs are highly efficient tools for the investigation of large series of neoplasms, including lymphoma with defined clinicopathological characteristics.28,29,30 Analysis of the cell cycle by applying antibodies specifically detecting different cell cycle phases has been proposed as a novel method for cell cycle assessment which may be of diagnostic and prognostic value.24
We were able to show that a large number of neoplastic cells in CLL have already acquired a “proliferative potential” as indicated by expression of Mcm2 (fig 2B). The number of cells with proliferative potential, i.e. expression of Mcm2 (mean 27.11%) was far in excess of the number of actively proliferating cells expressing Ki‐67 (mean 5.73%). This result is supported by a previous study which reports marked over‐expression of Mcm2 in contrast to Ki‐67 in low‐grade non‐Hodgkin lymphoma.25 In this aforementioned study, both Ki‐67 and Mcm2 expression was high in high‐grade lymphomas. Therefore we speculate that high‐grade lymphomas have fewer cells resting in G1‐phase than low‐grade lymphomas, especially CLL; however, this needs to be investigated further.
Furthermore, we were able to show that a significant sub‐population (15.66%) of neoplastic cells had entered the G1‐phase. This might be crucial as cells resting in G1‐phase are thought to be more prone to external stimuli to further progress into the cell cycle and therefore proliferate than quiescent cells in G0‐phase. As expected, only a few neoplastic cells had made further progression into the cell cycle, i.e. late G1‐phase (7.16%), S‐phase (3.31%) and G2/M‐phase (0.98%). These findings are supported by previous studies which showed that only a small number of cells in CLL are actively proliferating.31,32 It would be interesting to see if other low‐grade lymphomas show a similar percentage of cells with proliferative potential in G1‐phase or if other mechanisms such as prevention of apoptosis in follicular lymphoma are responsible for their growth potential. An immunohistochemical method to estimate cell cycle phase distribution within routinely processed formalin‐fixed, paraffin‐embedded specimens as shown here has the advantage that it can easily be performed in a routine diagnostic setting, whereas other methods (e.g. flow cytometry, autoradiographic 3H‐thymidine labelling) require either fresh material or sophisticated cost‐intensive technology.33
The validity of our approach is supported by a previous study which showed cell cycle analysis results obtained by immunohistochemistry to be comparable to those obtained by flow cytometry utilising a set of markers similar to ours.7 There were 5 out of 51 evaluable cases in our study in which the combined LI for cyclin E, A and B1 exceeded the LI for Mcm2; this was probably attributable to tumour heterogeneity and a small overlap of protein expression at the transgression point of the various phases.
Take‐home messages
Cell cycle phase distribution of neoplastic cells can be assessed by a panel of immunohistochemical markers in routinely processed tissue specimens.
Cells which have acquired a proliferative potential and are therefore licensed to replicate their DNA can be identified by antibodies against Mcm2.
In CLL, a significant sub‐population of cells resides in early G1‐phase; in this state cells may be more prone to external proliferation stimuli as opposed to cells in G0‐phase.
Cell cycle assessment in neoplastic disease may be useful to predict clinical course and outcome.
It has been speculated that CLL is composed of neoplastic cells arrested in G0‐ and/or early G1‐phase.20 Utilising Mcm2 in combination with other phase specific markers allows cells in early G1‐phase to be distinguished from those arrested in G0. The combined evaluation of a set of markers as used in this study might be a predictor of disease progression. In view of the cell cycle inhibitors that are currently entering clinical trials, these molecules might also represent potential therapeutic targets.34,35 Due to the lack of clinical data we had no information about other prognostic parameters. Further studies are now required to validate our data. Prospective studies are now especially called for to assess the significance of cell cycle phase analysis in conjunction with established prognostic parameters to predict clinical course and outcome in CLL.
Acknowledgements
The authors thank A Vielberth, C Oed, H Weisskopf and R Epper for excellent technical assistance.
Abbreviations
CLL - chronic lymphocytic leukaemia
LI - labelling index/indices
MCM - minichromosome maintenance
TMA - tissue microarray
Footnotes
Funding: This study was supported by a grant of the Dr Mildred Scheel Stiftung/Deutsche Krebshilfe to EC Obermann.
Competing interests: None.
References
- 1.Redaelli A, Laskin B L, Stephens J M.et al The clinical and epidemiological burden of chronic lymphocytic leukaemia. Eur J Cancer Care 200413279–287. [DOI] [PubMed] [Google Scholar]
- 2.Gentile M, Mauro F R, Guarini A.et al New developments in the diagnosis, prognosis and treatment of chronic lymphocytic leukemia. Curr Opin Oncol 200517597–604. [DOI] [PubMed] [Google Scholar]
- 3.Blow J J, Hodgson B. Replication licensing—defining the proliferative state? Trends Cell Biol 20021272–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Todorov I T, Werness B A, Wang H Q.et al HsMCM2/BM28: a novel proliferation marker for human tumors and normal tissues. Lab Invest 19987873–78. [PubMed] [Google Scholar]
- 5.Burger P C, Shibata T, Kleihues P. The use of the monoclonal antibody Ki‐67 in the identification of proliferating cells: application to surgical neuropathology. Am J Surg Pathol 198610611–617. [DOI] [PubMed] [Google Scholar]
- 6.Brown D C, Gatter K C. Ki67 protein: the immaculate deception? Histopathology 2002402–11. [DOI] [PubMed] [Google Scholar]
- 7.Scott I S, Morris L S, Bird K.et al A novel immunohistochemical method to estimate cell‐cycle phase distribution in archival tissue: implications for the prediction of outcome in colorectal cancer. J Pathol 2003201187–197. [DOI] [PubMed] [Google Scholar]
- 8.Williams G H, Romanowski P, Morris L.et al Improved cervical smear assessment using antibodies against proteins that regulate DNA replication. Proc Natl Acad Sci USA 19989514932–14937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freeman A, Morris L S, Mills A D.et al Minichromosome maintenance proteins as biological markers of dysplasia and malignancy. Clin Cancer Res 199952121–2132. [PubMed] [Google Scholar]
- 10.Chatrath P, Scott I S, Morris L S.et al Aberrant expression of minichromosome maintenance protein‐2 and Ki67 in laryngeal squamous epithelial lesions. Br J Cancer 2003891048–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xue W C, Khoo U S, Ngan H Y.et al Minichromosome maintenance protein 7 expression in gestational trophoblastic disease: correlation with Ki67, PCNA and clinicopathological parameters. Histopathology 200343485–490. [DOI] [PubMed] [Google Scholar]
- 12.Musahl C, Holthoff H P, Lesch R.et al Stability of the replicative Mcm3 protein in proliferating and differentiating human cells. Exp Cell Res 1998241260–264. [DOI] [PubMed] [Google Scholar]
- 13.Stoeber K, Tlsty T D, Happerfield L.et al DNA replication licensing and human cell proliferation. J Cell Sci 2001114(Pt 11)2027–2041. [DOI] [PubMed] [Google Scholar]
- 14.Lea N C, Orr S J, Stoeber K.et al Commitment point during G0→G1 that controls entry into the cell cycle. Mol Cell Biol 2003232351–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hunter T, Pines J. Cyclins and cancer. Cell 1991661071–1074. [DOI] [PubMed] [Google Scholar]
- 16.Koff A, Cross F, Fisher A.et al Human cyclin E, a new cyclin that interacts with two members of the CDC2 gene family. Cell 1991661217–1228. [DOI] [PubMed] [Google Scholar]
- 17.Pines J, Hunter T. Cyclins A and B1 in the human cell cycle. Ciba Found Symp 1992170187–196. [PubMed] [Google Scholar]
- 18.Nasmyth K. Viewpoint: putting the cell cycle in order. Science 19962741643–1645. [DOI] [PubMed] [Google Scholar]
- 19.Wolowiec D, Ciszak L, Kosmaczewska A.et al Cell cycle regulatory proteins and apoptosis in B‐cell chronic lymphocytic leukemia. Haematologica 2001861296–1304. [PubMed] [Google Scholar]
- 20.Delmer A, Ajchenbaum‐Cymbalista F, Tang R.et al Overexpression of cyclin D2 in chronic B‐cell malignancies. Blood 1995852870–2876. [PubMed] [Google Scholar]
- 21.Heidebrecht H J, Buck F, Endl E.et al Ki‐Mcm6, a new monoclonal antibody specific to Mcm6: comparison of the distribution profile of Mcm6 and the Ki‐67 antigen. Lab Invest 2001811163–1165. [DOI] [PubMed] [Google Scholar]
- 22.Tzankov A, Pehrs A C, Zimpfer A.et al Prognostic significance of CD44 expression in diffuse large B cell lymphoma of activated and germinal centre B cell‐like types: a tissue microarray analysis of 90 cases. J Clin Pathol 200356747–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mueller‐Hermelink H K, Montserrat E, Cytovsky D.et al Chronic lymphocytic leukaemia/small lymphocytic lymphoma. In: Jaffe ES, Harris NL, Stein H, Vardiman JW, eds. Tumours of haematopoietic and lymphoid tissue. Lyon: IARC Press, 2001127–130.
- 24.Scott I S, Heath T M, Morris L S.et al A novel immunohistochemical method for estimating cell cycle phase distribution in ovarian serous neoplasms: implications for the histopathological assessment of paraffin‐embedded specimens. Br J Cancer 2004901583–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Obermann E C, Eward K L, Dogan A.et al DNA replication licensing in peripheral B‐cell lymphoma. J Pathol 2005205318–328. [DOI] [PubMed] [Google Scholar]
- 26.Obermann E C, Went P, Pehrs A C.et al Cyclin B1 expression is an independent prognostic marker for poor outcome in diffuse large B‐cell lymphoma. Oncol Rep 2005141461–1467. [PubMed] [Google Scholar]
- 27.Jin Y H, Park C K. Expression of cyclin B1 and cdc2 in nodal non‐Hodgkin's lymphoma and its prognostic implications. J Korean Med Sci 200217322–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schraml P, Bucher C, Bissig H.et al Cyclin E overexpression and amplification in human tumours. J Pathol 2003200375–382. [DOI] [PubMed] [Google Scholar]
- 29.Tzankov A, Zimpfer A, Pehrs A C.et al Expression of B‐cell markers in classical Hodgkin lymphoma: a tissue microarray analysis of 330 cases. Mod Pathol 2003161141–1147. [DOI] [PubMed] [Google Scholar]
- 30.Went P, Dellas T, Bourgau C.et al Expression profile and prognostic significance of CD24, p53 and p21 in lymphomas. A tissue microarray study of over 600 non‐Hodgkin lymphomas. Dtsch Med Wochenschr 20041292094–2099. [DOI] [PubMed] [Google Scholar]
- 31.Decker T, Schneller F, Hipp S.et al Cell cycle progression of chronic lymphocytic leukemia cells is controlled by cyclin D2, cyclin D3, cyclin‐dependent kinase (cdk) 4 and the cdk inhibitor p27. Leukemia 200216327–334. [DOI] [PubMed] [Google Scholar]
- 32.Mochen C, Giardini R, Costa A.et al MIB‐1 and S‐phase cell fraction predict survival in non‐Hodgkin's lymphomas. Cell Prolif 19973037–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brons P P, Raemaekers J M, Bogman M J.et al Cell cycle kinetics in malignant lymphoma studied with in vivo iododeoxyuridine administration, nuclear Ki‐67 staining, and flow cytometry. Blood 1992802336–2343. [PubMed] [Google Scholar]
- 34.Senderowicz A M, Sausville E A. Preclinical and clinical development of cyclin‐dependent kinase modulators. J Natl Cancer Inst 200092376–387. [DOI] [PubMed] [Google Scholar]
- 35.Tan A R, Headlee D, Messmann R.et al Phase I clinical and pharmacokinetic study of flavopiridol administered as a daily 1‐hour infusion in patients with advanced neoplasms. J Clin Oncol 2002204074–4082. [DOI] [PubMed] [Google Scholar]