GNAS1 encodes the α subunit of the Gs protein (Gsα), which couples hormonal stimulation of multiple cell surface receptors to the activation of adenylate cyclase. GNAS1 maps to the chromosome 20q13 region and is associated with several human endocrine disorders, including Albright hereditary osteodystrophy (AHO) (also termed pseudopseudohypoparathyroidism). AHO is characterized by short stature, subcutaneous calcification, and brachydactyly and is caused by mutations that inactivate Gsα (1–3). GNAS1 mutations also cause pseudohypoparathyroidism type IA (PHPIA), a disease that displays the features of AHO but, in addition, is characterized by resistance to the peripheral action of multiple hormones, including parathyroid hormone.
The clue to how mutations that inactivate Gsα can result in different clinical manifestations comes from examination of the parental origin of the gene defect in the two diseases. Transmission of Gsα mutations through the maternal germline results in PHPIA, whereas inheritance from the father causes AHO (4). Variation of phenotype depending on parent of origin of the mutation is due to genomic imprinting. This epigenetic phenomenon establishes, at a limited number of defined loci, a difference in transcriptional activity between paternal and maternal alleles (5, 6). In the case of Gsα, it might have been predicted that the gene is expressed from the maternal allele, since maternal transmission of a mutation causes the more severe clinical manifestations. Inactivating mutations of the maternal allele would result, therefore, in absence of or greatly reduced expression of Gsα, whereas paternal mutations should have almost no effect on expression levels since the paternal copy of Gsα has been transcriptionally silenced by the imprinting mechanism. Unfortunately, the data contradict this hypothesis: levels of Gsα expression and activity are decreased by roughly 50% in the readily accessible tissues of both AHO and PHPIA individuals. A likely explanation for this conundrum is that there is predominant expression of the maternal Gsα only in certain tissues.
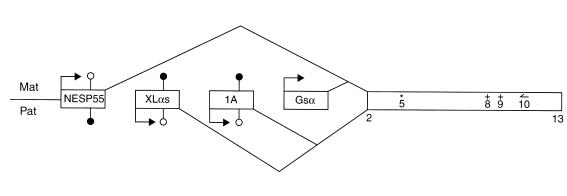
The expression and imprinting of GNAS1 is quite complex. GNAS1 has four alternative first exons that correspond to different promoters and that splice to a common set of downstream exons (3, 7–9) (Figure 1). Gsα is encoded by exon 1. The XLαs and NESP55 transcripts encode a Golgi-specific form of Gsα and a chromogranin-like protein, respectively, while the exon 1A transcript is apparently not translated. Adding to the complexity in organization of GNAS1 is the observation that, while there is maternal expression of NESP55 as well as Gsα, the exon 1A and XLαs transcripts display paternal allele–specific expression (Figure 1). These opposite patterns of imprinting have been demonstrated for all four transcripts in mice (9, 10) but not, until now, in humans for exon 1A and Gsα. In a recent article in the JCI, Liu et al. (11) establish that the exon 1A transcript displays paternal expression in humans. In addition, they show that absence of methylation of the paternal exon 1A allele is associated with yet another disease, pseudohypoparathyroidism type IB (PHPIB) (11). This disorder is characterized by renal resistance to parathyroid hormone without any other endocrine abnormalities.
Figure 1.
Organization and expression of GNAS1. Four first exons (distinct promoters) are alternatively spliced to exons 2–13 that are shared by all four transcripts. The NESP55 and Gsα transcripts are expressed from the maternal allele, while the XLαs and exon 1A transcripts display paternal expression. Note that imprinting of GNAS1 is tissue-specific and that the pattern of allelic expression shown here represents that seen in the pituitary. Lollipops correspond to CpG dinucleotides where the C is subject to modification by the addition of a methyl group at the 5′ position. A filled lollipop indicates methylation of a given CpG site, while an open lollipop indicates that the corresponding site on the other allele is not modified. RT-PCR using a primer () in exon 10 generates a product that spans a polymorphism (*) in exon 5 allows the assessment of allelic expression of all four transcripts in pituitary. RT-PCR with the exon 10–specific primer also permits the assay of sites of mutation in exons 8 and 9 (+) for the analysis of Gsα imprinting in pituitary somatotroph tumors.
Liu et al. (11) first demonstrate paternal-specific expression of the exon 1A transcript in normal peripheral blood and then confirm the previous observation made in mice that the exon 1A promoter is differentially methylated. In both humans and mice, therefore, several CpG dinucleotides in the maternal exon 1A are modified by methylation whereas the corresponding paternal sites are not (Figure 1). This difference is not evident, however, in the DNA of any of the 13 PHPIB patients examined. In these individuals, exon 1A is unmethylated on both the maternal and the paternal allele. The sites of differential methylation in the XLαs and NESP55 promoters have also been assayed in the PHPIB DNA samples, and abnormalities are detected only in a minority of patient samples. These findings strongly suggest that PHPIB results from a loss of imprinting of GNAS1 exon 1A. The link between this loss of imprinting and transcriptional changes of GNAS1 is not clear at this time, but Liu et al. (11) propose that abnormal methylation could lead to decreased Gsα expression in renal proximal tubules. For this scenario to hold true, there must be reciprocal regulation such that maternal-specific methylation (silencing) of exon 1A occurs coordinately with maternal-specific transcription of Gsα in some tissues including renal proximal tubules. Reciprocal promoter regulation is a common feature of other imprinted gene pairs, including IGF2 and H19 (12), and, in this context, Liu et al. (11) present a plausible model to explain the putative coordinate regulation of exon 1A and Gsα. The model invokes the tissue-specific imprinting of Gsα, a matter that has proved very difficult to demonstrate in humans.
In a recent issue of the JCI, Hayward et al. (13) come tantalizingly close to directly demonstrating maternal allele–specific expression of Gsα in the pituitary and add yet another twist to the involvement of GNAS1 in other human disease states. Hayward et al. studied the mRNAs transcribed from each of the exons NESP55, XLαs, 1A, and 1 in normal pituitary. Using RT-PCR with an upstream primer specific for each of these promoters and a downstream primer in exon 10, Hayward et al. could amplify each of the four transcripts (Figure 1) and could distinguish paternal from maternal allele expression on the basis of a single nucleotide polymorphism in exon 5 (13). In a pituitary DNA sample informative for this polymorphism, the XLαs and exon 1A transcripts both contain the A at exon 5, while NESP55 and Gsα both display the G at this position. Similar results are obtained for three other samples. These findings confirm that the patterns of monoallelic expression of XLαs and exon 1A are opposite that of NESP55 and strongly imply that, in normal pituitary, Gsα, like NESP55, is expressed exclusively or predominantly from the maternal allele.
Hayward et al. also studied the allelic expression in growth hormone–secreting pituitary adenomas (13). GNAS1 is referred to as the gsp oncogene in somatotroph adenomas, and 40% of such tumors bear a heterozygous Gsα mutation. The latter class of tumor, termed gsp+, displays constitutive activation of Gsα. The molecular basis of tumorigenesis in cases where there is no mutation of Gsα (gsp–) has not been elucidated. Using the RT-PCR approach described above to detect Gsα mutations in exons 8 and 9 (Figure 1), Hayward et al. show that there is mutation of the maternal Gsα allele in 21 of 22 of the gsp+ tumors (13). This finding is significant since it would explain that the observed overexpression of the mutant Gsα in gsp+ tumors simply reflects the predominant expression of the maternal allele. The authors suggest that gsp+ adenomas differ from normal pituitary in that the tumors display some Gsα expression from the (nonmutated) paternal allele. Hence, the development of gsp+ tumors involves relaxation of imprinting of Gsα as well as mutation of the maternal allele. This suggestion is further strengthened by the observation that there is loss of imprinting in the gsp-somatotroph adenomas, i.e., biallelic expression of Gsα, in the majority of gsp– tumors examined here. Loss of imprinting of Gsα may therefore represent a secondary feature in the progression of both gsp+ and gsp– somatotroph tumors. A caveat is that allelic expression of Gsα is assessed in a sample of whole pituitary, so it remains formally possible that there is biallelic expression in the normal pituitary somatotroph.
In summary, Liu et al. (11) show that there is an imprinting defect of GNAS1 in PHPIB, while Hayward et al. (13) suggest that loss of imprinting of Gsα contributes to the development of a markedly different clinical disorder, somatotroph adenomas. Liu et al. propose that the differential methylation at exon 1A is critical for Gsα imprinting and that loss of methylation of the maternal exon 1A GNAS1 allele results in PHPIB due to decreased expression of Gsα in renal proximal tubules (11). Hayward et al., on the other hand, suggest that there is relaxation of imprinting in the somatotroph adenomas resulting in activation of the normally silent paternal Gsα allele (13). To help confirm and to bridge these hypotheses, it would be important to assay the status of DNA methylation at exon 1A in somatotroph adenomas. It might be predicted that both the maternal and (to varying degrees) the paternal exon 1A promoters will prove to be methylated in these tumors. Such a finding might help to resolve some of the complexity associated with the regulation of imprinting of GNAS1 and its involvement in multiple human disease states.
Acknowledgments
The author thanks B. Graveley for help and comments.
References
- 1.Patten JL, et al. Mutation in the gene encoding the stimulatory G protein of adenylate cyclase in Albright’s hereditary osteodystrophy. N Engl J Med. 1990;322:1412–1419. doi: 10.1056/NEJM199005173222002. [DOI] [PubMed] [Google Scholar]
- 2.Weinstein LS, et al. Mutations of the Gs alpha-subunit gene in Albright hereditary osteodystrophy detected by denaturing gradient gel electrophoresis. Proc Natl Acad Sci USA. 1990;87:8287–8290. doi: 10.1073/pnas.87.21.8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weinstein LS, Yu S. The role of genomic imprinting of Galpha in the pathogenesis of Albright hereditary osteodystrophy. Trends Endocrinol Metab. 1999;10:81–85. doi: 10.1016/s1043-2760(98)00124-6. [DOI] [PubMed] [Google Scholar]
- 4.Davies SJ, Hughes HE. Imprinting in Albright’s hereditary osteodystrophy. J Med Genet. 1993;30:101–103. doi: 10.1136/jmg.30.2.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pfeifer K. Mechanisms of genomic imprinting. Am J Hum Genet. 2000;67:777–787. doi: 10.1086/303101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Preece MA, Moore GE. Genomic imprinting, uniparental disomy and foetal growth. Trends Endocrinol Metab. 2000;11:270–275. doi: 10.1016/s1043-2760(00)00277-0. [DOI] [PubMed] [Google Scholar]
- 7.Hayward BE, Moran V, Strain L, Bonthron DT. Bidirectional imprinting of a single gene: GNAS1 encodes maternally, paternally, and biallelically derived proteins. Proc Natl Acad Sci USA. 1998;95:15475–15480. doi: 10.1073/pnas.95.26.15475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayward BE, et al. The human GNAS1 gene is imprinted and encodes distinct paternally and biallelically expressed G proteins. Proc Natl Acad Sci USA. 1998;95:10038–10043. doi: 10.1073/pnas.95.17.10038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu J, Yu S, Litman D, Chen W, Weinstein LS. Identification of a methylation imprint mark within the mouse Gnas locus. Mol Cell Biol. 2000;20:5808–5817. doi: 10.1128/mcb.20.16.5808-5817.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peters J, et al. A cluster of oppositely imprinted transcripts at the Gnas locus in the distal imprinting region of mouse chromosome 2. Proc Natl Acad Sci USA. 1999;96:3830–3835. doi: 10.1073/pnas.96.7.3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu J, et al. A GNAS1 imprinting defect in pseudohypoparathyroidism type IB. J Clin Invest. 2000;106:1167–1174. doi: 10.1172/JCI10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ben-Porath I, Cedar H. Imprinting: focusing on the center. Curr Opin Genet Dev. 2000;10:550–554. doi: 10.1016/s0959-437x(00)00126-x. [DOI] [PubMed] [Google Scholar]
- 13.Hayward BE, et al. Imprinting of the Gsα gene GNAS1 in the pathogenesis of acromegaly. J Clin Invest. 2001;107:R31–R36. doi: 10.1172/JCI11887. [DOI] [PMC free article] [PubMed] [Google Scholar]