Abstract
Uncoupling protein 2 (UCP2) maps to a region on distal mouse chromosome 7 that has been linked to the phenotypes of obesity and type II diabetes. We recently reported that UCP2 expression is increased by high fat feeding in adipose tissue of the A/J strain of mice, which is resistant to the development of dietary obesity. More recently, a third UCP (UCP3) was identified, which is expressed largely in skeletal muscle and brown adipose tissue. The UCP2 and UCP3 genes are located adjacent to one another on mouse chromosome 7. Thus, the roles of these UCPs in both metabolic efficiency and the linkage to obesity and diabetes syndromes is unclear. For this reason, we examined the expression of UCP2 and UCP3 in white adipose tissue and interscapular brown adipose tissue and in gastrocnemius/soleus muscle preparations from the obesity-resistant A/J and C57BL/KsJ (KsJ) strains and the obesity-prone C57BL/6J (B6) mouse strain. In both KsJ and A/J mice, UCP2 expression in white fat was increased ≈2-fold in response to 2 weeks of a high fat diet, but there was no effect of diet on UCP2 levels in B6 mice. In skeletal muscle and in brown fat, neither UCP2 nor UCP3 expression was affected by diet in A/J, B6, or KsJ mice. However, in brown fat, we observed a 2–3-fold increase in the expression of UCP1 in response to dietary fat challenge, which may be related to diet-induced elevations in plasma leptin levels. Together, these results indicate that the consumption of a high fat diet selectively regulates UCP2 expression in white fat and UCP1 expression in brown fat and that resistance to obesity is correlated with this early, selective induction of UCP1 and UCP2 and is not associated with changes in expression of UCP3.
Keywords: UCP2, UCP3, gene expression, white adipose tissue, brown adipose tissue
Obesity is a disorder of energy balance in which energy intake is greater than energy expenditure. Methods to control obesity through limiting energy intake have had modest success at best, and it is widely recognized that energy expenditure must be increased in an obese individual if long term weight loss is to be achieved. The recent discovery of several new uncoupling proteins (UCPs) provides new molecular targets for increasing energy expenditure. The UCPs are integral membrane proteins of the mitochondrial inner membrane, where they function as a proton channel or shuttle. These proteins uncouple the process of mitochondrial respiration from oxidative phosphorylation, diminishing the resulting production of ATP and instead yielding dissipative heat. The action of these proteins creates a futile cycle that decreases the metabolic efficiency of the organism. Thus, UCPs are potentially important in disorders of energy balance such as obesity and diabetes (1, 2).
The brown fat uncoupling protein UCP1 was the first uncoupling protein to be described (3, 4). UCP1 is expressed exclusively in brown adipose tissue (BAT), where its primary role appears to be thermoregulation (5, 6), although it has also been linked to regulation of body composition (7–10). BAT containing functional UCP1 is present in neonatal humans (7), but it is still controversial whether there are sufficient quantities of BAT in adult humans to have a role in nutrient partitioning (8). Recently, two additional UCPs were identified that are more broadly expressed in metabolically active tissues of both humans and other animals: UCP2 (2) and UCP3 (9, 10). Interesting differences exist among these three UCPs in terms of both tissue distribution and physiological regulation. For example, whereas UCP1 is found exclusively in BAT, UCP2 is found in a variety of tissues including white adipose tissue (WAT), BAT, muscle tissue, and immune system tissue. UCP3 is expressed mainly in skeletal muscle but also in BAT tissue of rodents and to a lesser extent in cardiac muscle (9, 10). There is an important distinguishing feature of UCP2: its expression is modulated by diet and is not regulated by the sympathetic nervous system (2). Specifically, expression of UCP2 can be increased by dietary fat consumption (2). Although initial reports concluded that UCP3 was not regulated by cold stimulation and the sympathetic nervous system (9), more recent studies suggest that UCP3 may indeed be regulated in a similar fashion to UCP1. For example, like UCP1, UCP3 mRNA levels also are increased by thyroid hormone (T3) and by treatment with the β3AR agonist CL316,243 (ref. 11 and unpublished data).
Consumption of a high fat diet is associated with an increase in UCP2 expression in adipose tissue of the obesity-resistant A/J strain of mice but not in the obesity- and diabetes-prone B6 strain (2). Furthermore, a locus on human chromosome 11q13, which is syntenic to the UCP2 locus on mouse chromosome 7, recently was linked to metabolic rate in humans (12). Chromosomal mapping studies now place the human and mouse UCP2 and UCP3 genes in close proximity to each other on human chromosome 11 and mouse chromosome 7, respectively (11). Therefore, both UCP2 and UCP3 represent candidate genes for the quantitative trait locus of diet-induced obesity and diabetes in mice (2, 13) and in humans. In the work described here, we report that dietary fat specifically increases UCP2 expression in WAT, but not in skeletal muscle or BAT, in two obesity- and diabetes-resistant mouse strains (A/J and KsJ) with no effect on UCP2 in any tissue of the B6 mouse. Unlike UCP2, consumption of a high fat diet has no affect on UCP3 expression in WAT, skeletal muscle, or BAT.
MATERIALS AND METHODS
Animals.
Male mice (20 mice) from each of the strains B6, A/J, and KsJ were obtained from The Jackson Laboratory at 4 weeks of age. The animals were housed five per cage in a temperature-controlled room with a 12-hr light/dark cycle (lights off at 17:00 hr). Food and water was available ad libitum. Mice were fed one of two diets as follows: 10 mice from each strain were placed on a low fat/low sucrose diet or a high fat/low sucrose diet. The composition of these diets has been described (14). After 2 weeks on these diets, animals were anesthetized and killed by decapitation, and tissues were collected rapidly, cleaned, and homogenized in 4 M guanidine isothiocyanate buffer for preparation of total cellular RNA (15). Another cohort of animals was raised on these diets for 5 months for measurements of body weight, fasting plasma glucose, and insulin levels as described (16).
Genomic Mapping of Mouse UCP3 Relative to UCP2.
The chromosomal position of UCP3 was determined by linkage mapping of restriction fragment length polymorphisms in an interspecific cross as described for mouse UCP2 (2). UCP3 was found to cosegregate with UCP2 in this analysis (M. Seldin and C. Warden, personal communication). We cloned a mouse UCP3 cDNA containing all seven exons from a mouse muscle cDNA library constructed in λ gt11 (CLONTECH library ML3006b). The sequence of this UCP3 cDNA was deposited in the GenBank database under accession no. AF032902 (D.S., C. Fleury, F. Bouillaud and D.R., unpublished data). The organization of the UCP3/UCP2 locus was studied by using PCR analysis of genomic DNA from B6 mice or 129/SvJ mice. The same data were obtained from both sources of genomic DNA. The sense primer (5′-CGGGAATCTCCGTTTTGAAC→3′) used in the PCR encompassed the stop codon of UCP3, which is 264 bp upstream from the end of exon 7 of UCP3, according to the sequence of the mouse UCP3 cDNA. The reverse primer (5′-ATGAATGGACTCCACTGAGC→3′) corresponded to positions −7083 to −7063 of the UCP2 gene (relative to the UCP2 transcriptional start site). A 1.2-kb PCR product was obtained, and this product was sequenced.
Isolation and Analysis of RNA.
Total cellular RNA was prepared by the cesium chloride gradient method as detailed (15). For Northern blot hybridization, RNA was denatured by the glyoxal procedure, fractionated through 1.2% agarose gels, and blotted onto Biotrans nylon membranes (ICN) as described (17). DNA fragments that were used as hybridization probes were obtained from the following sources. We used a 950-bp PCR product of the mouse UCP2 cDNA and a 244-bp fragment of mouse UCP3 encoding the last 70 amino acids of the protein. This UCP3 probe does not cross-hybridize with UCP2. For UCP1, a 300-bp BglII fragment was used, kindly provided by Leslie P. Kozak (The Jackson Laboratory, Bar Harbor, ME) (18). A rat cDNA probe for cyclophilin was used as an internal hybridization/quantitation standard (19). Radiolabeled probes were prepared by random primer labeling (MultiPrime, Amersham) of the purified DNA fragments in the presence of [32P]dCTP to a specific activity of ≈2 × 109 dpm/μg DNA. Blots were hybridized and washed as described in other reports (17, 19). The intensity of hybridization signals was quantified by Phosphorimager (ImageQuant/Storm, Foster City, CA) and normalized to the values for cyclophilin. Permanent images of the blots were made by exposure to Kodak BioMax film.
RESULTS AND DISCUSSION
In our report of the cloning and functional activity of the UCP2 gene (2), we noted that, within a week of feeding a high fat diet, A/J mice displayed increased expression of UCP2 in WAT, whereas B6 mice lacked this response to diet for at least 5 weeks. Because we had identified a genetic linkage in the vicinity of the UCP2 locus on mouse chromosome 7 that is associated with the development of severe obesity and diabetes in the B6 strain, we hypothesized that increased expression of UCP2 in adipose tissue of A/J mice may prevent obesity by expending calories as heat instead of increasing the adipose tissue mass.
The UCP2 and UCP3 genes have been mapped to human chromosome 11q13 by analysis of radiation hybrids (11, 20). In mice, UCP2 and UCP3 also cosegregate in mapping studies of mouse chromosome 7 (ref. 11 and M. F. Seldin, personal communication). To question the relative organization of UCP2 and UCP3 genes, we used sequence data from a mouse UCP3 cDNA and a mouse UCP2 genomic clone (S.R., D.S., C. Pecqueur, F. Bouillaud and D.R., unpublished data). PCR analysis of genomic DNA was carried out by using material from two different strains of mice, the 129/SvJ and B6. The use of a sense primer corresponding to the 3′ end of mouse UCP3 cDNA and a reverse primer corresponding to basepair positions −7083 and −7063 of a mouse UCP2 genomic clone generated a 1.2-kb PCR product. Therefore, we conclude that the mouse UCP3 gene is 5′ to the mouse UCP2 gene and also that the distance between the 3′ extremity of UCP3 exon 7 and the transcriptional start site of UCP2 is very close to 8 kb (Fig. 1). In humans, the UCP3 gene is also 5′ to the UCP2 gene (unpublished data). Other experiments based on the hypothesis that the UCP3 gene was downstream of the UCP2 gene failed (data not shown). This close proximity, high sequence homology, and similar gene structure strongly suggest that the organization of the UCP3/2 locus is a result of a gene duplication event.
Figure 1.
Genomic organization of the mouse UCP3 and UCP2 genes. The mouse UCP3 gene is 5′ relative to the UCP2 gene. The distance between the two genes is close to 8 kb. This organization and the calculation of the distance between the two genes was predicted from PCR experiments on genomic DNA from two different strains of mice (see Materials and Methods). Roman numerals correspond to exons. The +1 position refers to the transcriptional start site of the mouse UCP2 gene determined from primer extension experiments (S.R., C. Pecqueur, and D.R., unpublished data). For UCP2 genomic structure, a genomic clone referred to as clone mmU2L2 was isolated from a mouse 129/SvJ genomic library in the λ FIXIT vector (Stratagene library 946306) and entirely sequenced. The mmU2L2 clone contains 7.1 kb of DNA upstream from the transcriptional start site (S.R. and D.R., unpublished data).
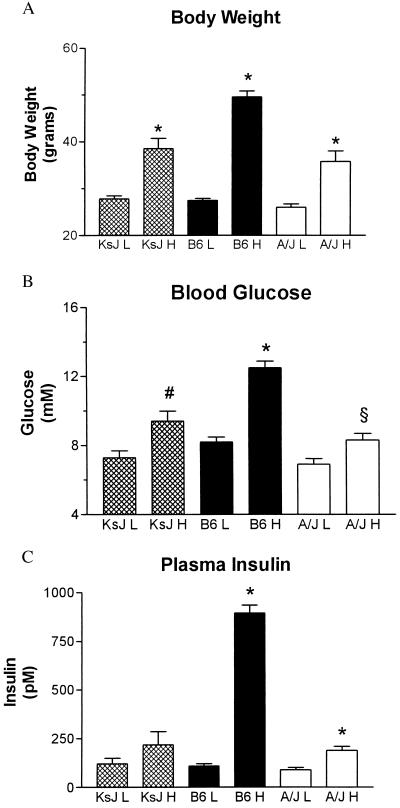
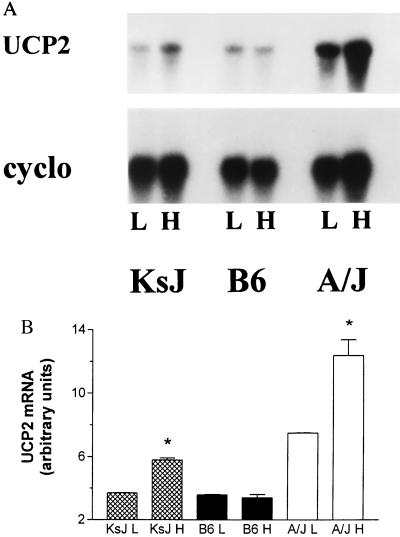
Because the UCP2 and UCP3 genes are physically so close to each other, classical genetic linkage studies cannot determine which of these genes is associated with the obesity and diabetes phenotypes we have described. Therefore, we attempted to ascertain whether a high fat diet has similar effects on the expression of both genes. To begin to address this question, we examined the effect of high fat feeding on the expression of UCP1, UCP2, and UCP3 in adipose tissues and skeletal muscle of A/J, B6, and KsJ mice. As reported (16), KsJ mice raised on a high fat diet are resistant to the development of obesity, hyperglycemia, and hyperinsulinemia (Fig. 2). Despite the fact that the ob and db mutations display similar phenotypes of severe obesity and diabetes when placed on either the B6 or the KsJ strain (21, 22), these two strains show notably different responses to consumption of a high fat diet. In this regard, the KsJ phenotype is similar to the obesity-resistant phenotype of the A/J strain. This is particularly interesting because Naggert and colleagues (23) have reported that KsJ mice are essentially a recombinant congenic of B6 and DBA mice. KsJ mice possess 84% genetic identity with the B6 strain, with the remaining genetic material derived from DBA. Most relevant to our studies is their finding that the distal region of mouse chromosome 7 in KsJ mice (in the region of UCP2 and UCP3) appear to contain DBA alleles (23). Fig. 3 shows that, similar to our earlier report (2), UCP2 mRNA levels in epididymal WAT of A/J mice are elevated 1.7 ± 0.3-fold in response to a high fat diet, whereas there is no effect of diet on the expression of UCP2 in B6 mice. In KsJ mice, UCP2 expression also is increased by 1.6 ± 0.1-fold by the high fat diet. Thus, our data suggest that diet-induced elevations of UCP2 are common to at least two obesity-resistant mouse strains. Fig. 3 also illustrates that UCP2 mRNA levels tend to be lower in B6 mice than in A/J mice (≈3-fold higher in A/J/mice, n = 4) even in low fat feeding conditions.
Figure 2.
Effect of a high fat diet on obesity and symptoms of type II diabetes in obesity-resistant and obesity-prone strains of mice. Twenty mice from each of the three strains, A/J, B6, and KsJ, were obtained from The Jackson Laboratory at 3–4 weeks of age and placed on either a low fat diet or a high fat diet as described by Surwit (14). Body weight (A), plasma levels of glucose (B), and insulin (C) after an 8-hr fast were determined after 5 months. One-way ANOVA was performed for each parameter. Body weights, glucose, and insulin values of the low fat-fed animals were not different from each other (P > 0.05). For all three measures, the values for B6 H mice were greater than those for either the KsJ H or the A/J H mice (P < 0.001), whereas those for KsJ H and A/J H mice were not different from each other (P > 0.05). Other statistical comparisons between values for high fat and low fat fed animals are as follows: ∗, P < 0.001; #, P < 0.005; §, P < 0.02.
Figure 3.
Expression of UCP2 in WAT of KsJ, B6, and A/J mice. After 2 weeks of either a low fat or a high fat diet, total cellular RNA was prepared from epididymal WAT and examined for the expression of UCP2 by Northern blot hybridization. The blots subsequently were probed with cyclophilin as a control. (A) Representative Northern blot. (B)Average of results for four mice of each strain. ∗, P < 0.01.
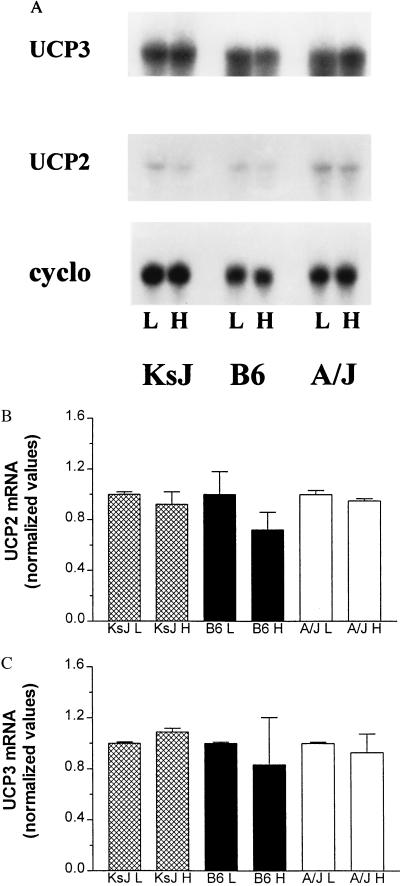
It has been reported that UCP3 mRNA levels are expressed most abundantly in skeletal muscle as well as in interscapular BAT (IBAT) (9–11). Therefore, we examined the effect of a high fat diet on the expression of both UCP3 and UCP2 in a gastrocnemius muscle/soleus muscle preparation from A/J, B6, and KsJ mice (Fig. 4). In contrast to the results obtained for UCP2 in WAT, there was no change in UCP3 or UCP2 mRNA in skeletal muscle from any of these strains as a consequence of high fat feeding. In fact, there were no detectable differences in the expression of UCP2 and UCP3 in muscle between these obesity-prone or -resistant strains under any dietary condition. UCP3 mRNA levels also were not detected in WAT in any strain consuming either diet, even after very long exposure times (data not shown).
Figure 4.
Expression of UCP2 and UCP3 in skeletal muscle of KsJ, B6, and A/J mice. After 2 weeks of either a low fat or a high fat diet, total cellular RNA was prepared from gastrocnemius and soleus muscle and examined for the expression of UCP2, UCP3, and cyclophilin by Northern blot hybridization. (A) Representative Northern blots. (B) Average of results for UCP2 from four mice of each strain. (C) Average of results for UCP3 from four mice of each strain. There were no statistically significant differences among any of the values by ANOVA or paired t tests.
Finally, we examined the expression of all three UCPs in IBAT. This tissue has been implicated in the control of body composition (24) and maintenance of body temperature in response to cold challenge (1, 6). Until the recent identification of UCP2 and UCP3, it had been assumed that all of these physiological functions in IBAT are dependent on UCP1. However, the observation that targeted disruption of UCP1 led to mice that were cold-intolerant but not obese and that displayed a 5-fold increase in the expression of UCP2 in IBAT has forced the reevaluation of the function of individual UCPs in IBAT (as well as in other tissues). Somewhat to our surprise, we found that neither UCP2 nor UCP3 expression in IBAT was affected by diet in either A/J or B6 mice (Fig. 5). Instead, we observed a 2- to 3-fold increase in the expression of UCP1 in response to dietary fat challenge. The A/J mice exhibited significantly greater stimulation of UCP1 expression than the B6 mice. These increases in UCP1 mRNA and the differences observed between the strains could be related to several factors. These factors include our observation that high fat feeding leads to a greater increase in plasma leptin levels in A/J mice than in B6 mice (25), which, in turn, could stimulate differentially sympathetic outflow to IBAT in these strains (3, 26). In addition, the greater increase in UCP1 expression in IBAT of A/J mice compared with B6 mice could be related to our recent finding that the expression and function of the βARs in IBAT are impaired more severely in B6 mice than in A/J mice when those mice are raised on high fat diets (27).
Figure 5.
Expression of UCP1, UCP2, and UCP3 in IBAT of B6 and A/J mice. After 2 weeks of either a low fat or a high fat diet, total cellular RNA was prepared from IBAT and examined for the expression of UCP1, UCP2, UCP3, and cyclophilin by Northern blot hybridization. (A) Representative Northern blots. (B–D) Average of results for each UCP from six mice of each strain. ∗, UCP1 levels in high fat fed mice were greater than in low fat mice (B6: P < 0.03; A/J: P < 0.0001). #, UCP1 levels in A/J H mice were greater than in B6 H mice (P < 0.005).
These results lead to at least two interpretations. First, the data suggest that there are tissue-specific differences in the mechanisms regulating UCP2 expression in response to dietary fat. As a corollary, it would seem that changes in dietary fat consumption do not affect the expression of the UCP2 gene equally in fat depots, as we note in the differences between WAT and BAT. A second major point is that, despite significant sequence similarity between UCP2 and UCP3, their close physical proximity in the genome, and their broader tissue distribution compared with UCP1, the signaling mechanisms regulating the expression of UCP2 and UCP3 are quite distinct. This conclusion is underscored by the fact that hypothermia, catecholaminergic stimulation, and T3 all up-regulate UCP1 and UCP3, whereas UCP2 is not affected by these stimuli (2, 11). In addition, although it has been reported that dramatically elevated plasma leptin levels may increase UCP2 expression (28), we do not find any effects of exogenously administered leptin (25) or diet-induced elevations of leptin on UCP2 expression in A/J or B6 mice. Additional experiments will be required before definitive conclusions can be drawn concerning the relationship of mRNA levels to the functional activity of these new UCPs. Nevertheless, our data provide important genetic and developmental support for the hypothesis that UCP2 is controlled specifically by factors responding to the consumption of dietary fat and that the expression of UCP2 is linked to the development of the diabetes and obesity phenotypes that we have described (14, 29).
The discovery of UCP2 and UCP3 suggests that the uncoupling of respiration may be a general biologic phenomenon not limited to brown adipose tissue. These genes are expressed widely throughout the body and therefore may explain the proton leak reported in mitochondria (e.g., 30–32), which is associated with variations in the Standard Metabolic Rate (30) and body mass across species. Furthermore, the chromosomal region containing UCP2 and UCP3 is linked to diabetes and obesity in mice (2) and to the metabolic rate in humans (12). However, the ability of a high fat diet to induce obesity and diabetes is related to the expression of UCP1 in IBAT and UCP2 in WAT and is unrelated to differential expression of UCP2 and UCP3 in skeletal muscle or IBAT. This difference would suggest that uncoupling activity in adipose tissue, and not in muscle, is a factor in mediating the effects of fat feeding on obesity and diabetes. Such a conclusion is consistent with the developing notion that the adipocyte is a key player in the pathophysiology of these conditions.
Acknowledgments
We thank Craig Warden and Michael Seldin for communicating results of genomic mapping experiments and for helpful discussions, Kiefer Daniel for assistance with some of the mouse studies, and Sheridan Snedden for comments on the manuscript. This work was supported in part by National Institutes of Health Grant R29-46793 (S.C.), Centre National de la Recherche Scientifique (D.R.), l’Association de Recherches sur le Cancer (D.R.), and an unrestricted gift from Novo-Nordisk (Copenhagen) (S.C.). D.S. was supported by the European Network “Metabolic Integration of Energy Control.”
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: UCP, uncoupling protein; BAT, brown adipose tissue; WAT, white adipose tissue; IBAT, interscapular BAT.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF032902).
References
- 1.Himms-Hagen J. Nutr Rev. 1983;41:261–267. doi: 10.1111/j.1753-4887.1983.tb07196.x. [DOI] [PubMed] [Google Scholar]
- 2.Fleury C, Neverova M, Collins S, Raimbault S, Champigny O, Levi-Meyrueis C, Bouillaud F, Seldin M F, Surwit R S, Ricquier D, et al. Nat Genet. 1997;15:269–272. doi: 10.1038/ng0397-269. [DOI] [PubMed] [Google Scholar]
- 3.Nicholls D G, Locke R M. Physiol Rev. 1986;64:1–64. doi: 10.1152/physrev.1984.64.1.1. [DOI] [PubMed] [Google Scholar]
- 4.Ricquier D, Casteilla L, Bouillaud F. FASEB J. 1991;5:2237–2242. doi: 10.1096/fasebj.5.9.1860614. [DOI] [PubMed] [Google Scholar]
- 5.Trayhurn P, Nicholls D G. Brown Adipose Tissue. Baltimore: Edward Arnold; 1986. [Google Scholar]
- 6.Enerback S, Jacobsson A, Simpson E M, Guerra C, Yamashita H, Harper M, Kozak L P. Nature (London) 1997;387:90–94. doi: 10.1038/387090a0. [DOI] [PubMed] [Google Scholar]
- 7.Himms-Hagen J, Ricquier D. In: Handbook of Obesity. Bray G, Bouchard C, James W, editors. New York: Marcel Dekker; 1997. pp. 415–441. [Google Scholar]
- 8.Danforth E, Himms-Hagen J. Eur J Endocrinol. 1997;136:362–365. doi: 10.1530/eje.0.1360362. [DOI] [PubMed] [Google Scholar]
- 9.Boss O, Samec S, Paoloni-Giacobino A, Rossier C, Dulloo A, Seydoux J, Muzzin P, Giacobino J-P. FEBS Lett. 1997;408:39–42. doi: 10.1016/s0014-5793(97)00384-0. [DOI] [PubMed] [Google Scholar]
- 10.Vidal-Puig A, Solanes G, Grujic D, Flier J S, Lowell B B. Biochem Biophys Res Commun. 1997;235:79–82. doi: 10.1006/bbrc.1997.6740. [DOI] [PubMed] [Google Scholar]
- 11.Gong D, He Y, Karas M, Reitman M. J Biol Chem. 1997;272:24129–24132. doi: 10.1074/jbc.272.39.24129. [DOI] [PubMed] [Google Scholar]
- 12.Bouchard C, Pérusse L, Chagon Y-C, Warden C, Ricquier D. Hum Mol Genet. 1997;6:1887–1889. doi: 10.1093/hmg/6.11.1887. [DOI] [PubMed] [Google Scholar]
- 13.Warden C H, Fisler J S, Shoemaker S M, Wen P-Z, Svenson K L. J Clin Invest. 1995;95:1545–1552. doi: 10.1172/JCI117827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Surwit R S, Feinglos M N, Rodin J, Sutherland A, Petro A E, Opara E C, Kuhn C M, Rebuffe-Scrive M. Metabolism. 1995;44:645–651. doi: 10.1016/0026-0495(95)90123-x. [DOI] [PubMed] [Google Scholar]
- 15.Chirgwin J M, Przybyla A E, MacDonald R J, Rutter W J. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 16.Surwit R S, Seldin M F, Kuhn C M, Secor C, Feinglos M N. Mouse Genome. 1994;92:523–525. [Google Scholar]
- 17.Collins S, Caron M G, Lefkowitz R J. J Biol Chem. 1988;263:9067–9070. [PubMed] [Google Scholar]
- 18.Jacobsson A, Stadler U, Glotzer M A, Kozak L P. J Biol Chem. 1985;260:16250–16254. [PubMed] [Google Scholar]
- 19.Collins S, Daniel K W, Rohlfs E M, Ramkumar V, Taylor I L, Gettys T W. Mol Endocrinol. 1994;8:518–527. doi: 10.1210/mend.8.4.7914350. [DOI] [PubMed] [Google Scholar]
- 20.Solanes G, Vidal–Puig A, Grujic D, Flier J S, Lowell B B. J Biol Chem. 1997;272:25433–25436. doi: 10.1074/jbc.272.41.25433. [DOI] [PubMed] [Google Scholar]
- 21.Coleman D L, Hummel K P. Diabetologia. 1973;9:287–293. doi: 10.1007/BF01221856. [DOI] [PubMed] [Google Scholar]
- 22.Coleman D L. Diabetologia. 1978;14:141–148. doi: 10.1007/BF00429772. [DOI] [PubMed] [Google Scholar]
- 23.Naggert J K, Mu J L, Frankel W, Bailey D W, Paigen B. Mamm Genome. 1995;6:131–133. doi: 10.1007/BF00303258. [DOI] [PubMed] [Google Scholar]
- 24.Rothwell N J, Stock M J. Nature (London) 1979;281:31–35. doi: 10.1038/281031a0. [DOI] [PubMed] [Google Scholar]
- 25.Surwit R, Petro A, Parekh P, Collins S. Diabetes. 1997;46:1516–1520. doi: 10.2337/diab.46.9.1516. [DOI] [PubMed] [Google Scholar]
- 26.Collins S, Kuhn C M, Petro A E, Swick A G, Chrunyk B A, Surwit R S. Nature (London) 1996;380:677. doi: 10.1038/380677a0. [DOI] [PubMed] [Google Scholar]
- 27.Collins S, Daniel K W, Petro A E, Surwit R S. Endocrinology. 1997;138:405–413. doi: 10.1210/endo.138.1.4829. [DOI] [PubMed] [Google Scholar]
- 28.Zhou Y, Shimabukuro M, Koyama K, Lee Y, Wang M, Trieu F, Newgard C B, Unger R H. Proc Natl Acad Sci USA. 1997;94:6386–6390. doi: 10.1073/pnas.94.12.6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Surwit R S, Kuhn C M, Cochrane C, McCubbin J A, Feinglos M N. Diabetes. 1988;37:1163–1167. doi: 10.2337/diab.37.9.1163. [DOI] [PubMed] [Google Scholar]
- 30.Brand M, Couture P, Else P, Withers K, Hulbert A. Biochem J. 1991;275:81–86. doi: 10.1042/bj2750081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Porter R, Brand M. Nature (London) 1993;362:628–630. doi: 10.1038/362628a0. [DOI] [PubMed] [Google Scholar]
- 32.Porter R, Brand M. Am J Physiol. 1995;269:R1213–R1224. doi: 10.1152/ajpregu.1995.269.5.R1213. [DOI] [PubMed] [Google Scholar]