Abstract
We have developed an efficient method for the derivatization of oligosaccharides, wherein the oligosaccharide is efficiently ligated to a basic aminooxyacetyl peptide by oxime formation. The resulting glycopeptide yields much higher sensitivity in matrix-assisted laser desorption/ionization mass spectrometry than does the underivatized oligosaccharide. Digestion of the glycopeptide by a exoglycosidase array and subsequent mass spectrometric assay of the digestion products provide a sensitive and rapid way to elucidate the structure of the oligosaccharide. In addition to oligosaccharide sequencing, the ligation reaction between an oligosaccharide and an aminooxyacetyl peptide also provides a potentially very convenient and efficient way for the synthesis of glycopeptides or glycoproteins.
Keywords: carbohydrate, mass spectrometry, structure analysis, sequencing
Proteins produced by eukaryotic cells are frequently posttranslationally modified by the addition of carbohydrates. The carbohydrate moieties of the glycoproteins modulate properties such as stability, activity, and binding affinity to and specificity for other biomolecules (1–4). For this reason, it is important to be able to determine the structure of the carbohydrates present in glycoproteins. However, elucidation of the structure of carbohydrates is challenging and places heavy demands on the tools used for their analysis.
When large quantities of the oligosaccharide of interest are available, tools of choice for structure analysis include nuclear magnetic resonance (5–7) and x-ray crystallography (7). Frequently, only small quantities of oligosaccharide are available, and structure analysis is accomplished by monitoring enzymatic degradation with a glycosidase array (8–10), by permethylation analysis (11), and/or by mass spectrometry (MS) (12–14). Enzymatic analysis of oligosaccharides is distinguished by its simplicity and sensitivity. The method involves digestion of aliquots of the oligosaccharide of interest with a defined exoglycosidase array, followed by analysis of the digestion products using chromatography or by gel electrophoresis (9, 10). The sequence of the oligosaccharide and the specific linkages between the individual sugar moieties can be deduced from the pattern of observed carbohydrate fragments and a knowledge of the specificities of the components of the exoglycosidase array. Although the method in its present form is useful, it is relatively slow and requires highly purified oligosaccharide and virtually complete glycosidase digestion. Therefore, further improvement of the current method is desirable.
Matrix-assisted laser desorption/ionization MS (MALDI-MS) (15–17) and electrospray ionization MS (18, 19) have been used to determine molecular masses of oligosaccharides and to obtain sequence, branching, and linkage information (12, 20, 21). The application of MS to structure elucidation is attractive because the technique is potentially rapid and sensitive. Nevertheless, application of these mass spectrometric approaches to the structure analysis of oligosaccharides has proved challenging; difficulties originate from the intrinsic complexity of oligosaccharide structures, insufficient ability to control mass spectrometric fragmentation of oligosaccharides, and the relatively low mass spectrometric response of oligosaccharides (compared, for example, with peptides). During positive ion MALDI, peptide molecules are usually ionized relatively efficiently by protonation of their basic groups. Neutral and acidic oligosaccharides lack groups with high proton affinity and are usually ionized with lower efficiency by alkali metal cationization when a small amount of metal salt is present. The resulting limited sensitivity for the mass spectrometric measurement of oligosaccharides reduces the utility of MALDI-MS for their structural analysis.
Derivatization methods for oligosaccharides have been developed with the goal of increasing the sensitivity of the mass spectrometric analysis (22, 23). Most of these methods involve covalent attachment of an appropriate moiety by reductive amination—a procedure involving formation of a covalent linkage by the reduction of a Schiff adduct between a primary amine and the acyclic aldehyde form of the oligosaccharide. For example, attachment of a basic moiety improves the probability of protonation during the ionization step.
Here, we report a method for the microscale derivatization of oligosaccharides, wherein the oligosaccharide is efficiently ligated to an aminooxyacetyl form of a basic peptide by oxime formation¶ (see Fig. 1). The resulting glycopeptide yields much higher sensitivity in MALDI-MS than does the underivatized oligosaccharide. Digestion of the glycopeptide by a exoglycosidase array and subsequent mass spectrometric assay of the digestion products (8) provides a sensitive and rapid way to elucidate the structure of the oligosaccharide.
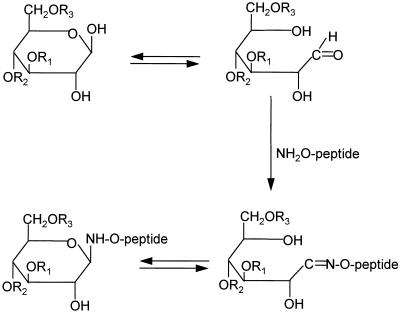
Figure 1.
Synthesis of glycopeptide from an aminooxyacetyl peptide and an oligosaccharide. R1, R2, and R3 represent sugars attached to the reducing end monosaccharide. The aminooxyacetyl peptide is NH2-O-CH2CO-KLEEQRPERVKG.
MATERIALS AND METHODS
Materials.
All oligosaccharides and exoglycosidases were purchased from Oxford GlycoSystems (Abingdon, U.K.). The procedures for preparation and storage of enzymes followed instructions from the manufacturer. HPLC grade CH3CN was purchased from Baxter Health Care (Muskegon, MI). α-Cyano-4-hydroxycinnamic acid (4-HCCA) was purchased from Sigma, and 2,5-dihydroxybenzoic acid (DHB) and 1-hydroxy isoquinoline (HIC) was from Aldrich. Aminooxyacetyl peptide (NH2-O-CH2CO-KLEEQRPERVKG) was kindly provided by Lynne E. Canne (The Scripps Research Institute, La Jolla, CA). The purity of the peptide was assayed by HPLC and MALDI-MS.
Synthesis of Glycopeptides.
The derivatization reaction was modified from the procedures for oxime formation described previously (25, 26). Thus, 2 μl (20–200 pmol) of an aqueous solution of oligosaccharide was added to 0.5 μl (18–180 pmol) of an aqueous solution of the aminooxyacetyl peptide at a molar ratio 1:0.9 (oligosaccharide/aminooxyacetyl peptide), mixed with 30 μl of CH3CN, and incubated at 37°C for 12 hr. The resulting reaction products were vacuum dried (SpeedVac; Savant Instruments) and redissolved in a suitable volume of water. The reaction products were used for glycosidase digestion and mass spectrometric analysis without further purification. Considerable care was taken to minimize the presence of aldehyde and ketone impurities in the solvents used for the derivatization reaction.
A slight excess of carbohydrate over aminooxyacetyl peptide was used during synthesis of the glycopeptides to avoid the strong mass spectrometric peak resulting from unreacted peptide. In cases where it proves desirable to use excess peptide, several alternative strategies can be applied. (i) The large unreacted peptide peak can be tolerated (or allowed to saturate) because it is in a part of the mass spectrum that does not interfere with information concerning the oligosaccharide. (ii) Unreacted peptide can be removed by reaction with activated solid beads containing aldehyde or ketone groups. (iii) Unreacted peptide ions can be discriminated against by switching the detector off during the arrival time of the unreacted peptide ions (in the same manner as is presently used for discriminating against the intense laser desorption matrix ions).
HPLC Analysis.
Reverse-phase HPLC was carried out on a Microm Ultrafast Microprotein Analyzer HPLC system (Microm Bioresource, Pleasanton, CA) with 215 nm UV detection, using C-18 analytical (5 μm, 1 × 150 mm) column. Chromatographic analysis was performed using a linear gradient from 10% to 40% of buffer B in buffer A [buffer A, 0.1% trifluoroacetic acid (TFA) in water (vol/vol); buffer B, 90% CH3CN/0.1% TFA in water (vol/vol)] in 20 min at a flowrate of 50 μl/min.
Glycosidase Digestion of Glycopeptides.
The glycopeptide was digested according to the procedures described by Sutton et al. (8), except that the amount of each reaction volume was increased five times to 5 μl. Typically, 16–40 pmol of a glycopeptide was divided into eight aliquots and mixed with reaction buffers and enzyme mixtures as given in Table 2. The resulting solutions were incubated at 37°C for about 20 hr. Two microliters of each digest was mixed with 2 μl of 4-HCCA matrix solution, and 1.5 μl of the resulting solution was loaded onto the sample probe (with a capacity for 10 samples) for MALDI time-of-flight (TOF) mass spectrometric analysis.
Table 2.
Specificities and compositions of glycosidase mixtures
| Enzyme | Source | Linkage specificity | Concentration (units/ml) | pH optimum | Exoglycosidase digests (number of 0.5 μl aliquots)
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | H | |||||
| Sialidase | Arthrobacter ureafaciens | α2–6 > α3, α8 | 1 | 5.0–5.5 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Galactosidase | Streptococcus pneumoniae | β1–4 | 0.2 | 5.5–6.5 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| GlcNAcase | Chicken liver | β1–2, α3, α4, α6 | 1 | 5.0–5.5 | 1 | 1 | 1 | 1 | ||||
| GlcNAcase | Streptococcus pneumoniae | β1–2 | 0.008 | 4.0–4.5 | 1 | 1 | ||||||
| Mannosidase | Jack bean | α1–3 | 0.1 | 4.0–4.5 | 1 | |||||||
| Mannosidase | Jack bean | α1–2, α3, α6 | 5 | 4.0–4.5 | 1 | 1 | 1 | |||||
| Mannosidase | Helix pomatia | β1–4 | 1 | 4.0–4.5 | 1 | |||||||
| Sample buffer | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||||
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||||
| 10% methanol* | 7 | 6 | 5 | 4 | 4 | 3 | 5 | 4 | ||||
Glycosidase mixtures are adapted from ref. 2.
Sample buffer includes 50 mM sodium citrate/phosphate (pH 5.0), 25 mM zinc chloride.
Mass Spectrometric Analysis.
Mass measurement was carried out on a MALDI-TOF instrument constructed at the Rockefeller University (27). The mass spectra were obtained in the positive ion mode and collected by adding individual spectra resulting from 100 laser shots.
RESULTS
Synthesis of Glycopeptides.
The synthesis of glycopeptides takes advantage of the oxime formation reaction (25, 26) between a highly reactive amine group at the N terminus of an aminooxyacetyl peptide and an aldehyde group at the reducing end of an oligosaccharide (Fig. 1). The sequence of the aminooxyacetyl peptide was chosen to contain four basic residues to increase the mass spectrometric response of the resulting glycopeptide. To optimize the reaction yields, various reaction conditions were screened, including solvent composition, pH, and temperature. A high ratio of CH3CN/water [92:8 (vol/vol)] was found to favor the synthesis of glycopeptides. No side reaction product was observed between the side chain of lysine and the oligosaccharides, suggesting that (under the conditions employed) the reaction is specific for the aminooxy group (data not shown).
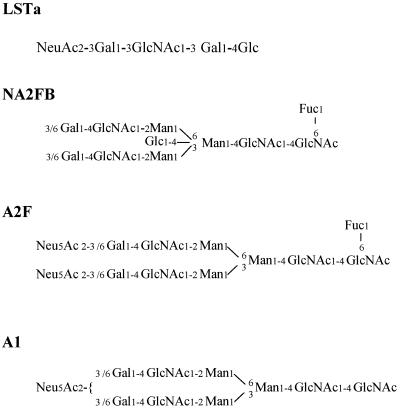
Fifteen oligosaccharides including neutral, acidic, N-linked, and O-linked oligosaccharides were successfully ligated to the aminooxyacetyl peptide. They were as follows: 3′-sialyllactose (3′-SL); 6′-sialyllactose (6′-SL); 3′-sialyl-N-acetyllactosamine (3′-SLN); 6′-sialyl-N-acetyllactosamine (SLN); 3′-sialyl-3-fucosyllactose (3′-S,3-FL); LS-tetrasaccharide a (LSTa); LS-tetrasaccharide b (LSTb); LS-tetrasaccharide c (LSTc); mono-sialylated-, galactosylated, bi-antennary (A1); di-sialylated-, galactosylated, bi-antennary, core substituted with fucose (A2F); 2′-fucosyllactose (2′FL); asialo-, agalacto-, bi-antennary (NGA2); conserved trimannosyl core (M3N2); di-sialyl-lacto-N-tetraose (DLSNT); and asialo-, agalacto-, bi-antennary, core-substituted with fucose and with bisecting GlcNAc (NA2FB) (nomenclature used in the Oxford GlycoSystems catalogue). The reaction yields of four of these oligosaccharides (Fig. 2) was analyzed by HPLC. In each case, the reaction yield was 95% or higher (see, for example Fig. 3), indicating that the reaction is quite general and efficient. The glycopeptide did not degrade after incubation at 37°C for 40 hr in aqueous buffers with pH values ranging from 4.3 to 8.0 (data not shown), suggesting the glycopeptide is quite stable. To characterize side products, the reaction mixture was separated by reverse-phase HPLC, and identities of the HPLC fractions were characterized by their molecular masses determined by MS. The major side products were formed between the peptide and aldehyde/ketone impurities, such as formaldehyde, acetylaldehyde, or acetone that were present in trace amount in the acetonitrile and/or water solvents (Fig. 3).
Figure 2.
Structures of four oligosaccharides used in the present investigation.
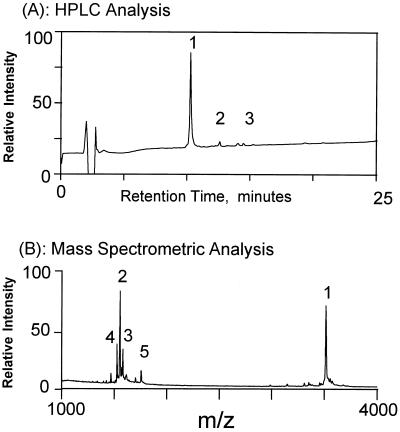
Figure 3.
(A) Reverse phase-HPLC analysis of the products formed by ligation between the oligosaccharide, NA2FB, and the aminooxyacetyl peptide. Peak 1, glycopeptide; peaks 2 and 3, side products formed between the aminooxyacetyl peptide with formaldehyde and acetone, respectively. Identities of the peaks were confirmed by mass spectrometric analysis of the HPLC fractions. The HPLC conditions are described in Materials and Methods. (B) Mass spectrometric measurement of an aliquot of the crude ligation reaction products were analyzed without further purification. Peaks 1–3 in the mass spectrum correspond to fractions 1–3 in A. Peak 4 is a fragment ion produced through cleavage of the oxime N-O bond during the MALDI-MS process. Peak 5 was not identified. Yields of the ligation reaction were calculated from the areas of the HPLC peaks.
Mass Spectrometric Sensitivities of Glycopeptides.
Sensitivities of derivatized and underivatized oligosaccharides were determined under optimized MALDI-MS conditions. We and others have observed that the composition of the matrix solution and the procedures for sample preparation significantly influence the signal response of analytes in MALDI-MS (17, 28–31). Several commonly used matrixes were evaluated for the purpose of improving the quality and sensitivity of mass spectra, including 4-HCCA (32), DHB (21), 4-hydroxy-3-methoxycinnamic acid (33), 3-hydroxypicolinic acid (34), 5-methoxysalicyclic acid (MSA) (33), DHB/MSA mixture (33), DHB/MSA/fucose mixture (33), and DHB/HIC mixture (35). We also evaluated different sample preparation procedures, including variation of the concentration of sodium chloride present in the preparation (for underivatized oligosaccharides), the evaporation environment (at atmospheric pressure or in vacuum), and the matrix-sample crystallization conditions using different organic solvents.
For the glycopeptides prepared by the present method, the strongest ion signals were observed when the sample was dried at room temperature in air using a saturated 4-HCCA solution as a matrix prepared in CH3CN/0.1% TFA in water (1:2, vol/vol). For underivatized oligosaccharides, the strongest signal was observed when the analyte was dried in vacuum with a 0.2 M DHB/0.06 M HIC (1:1, vol/vol) matrix solution prepared in CH3CN/water (1:2, vol/vol). These results are in good agreement with previous observations (35).
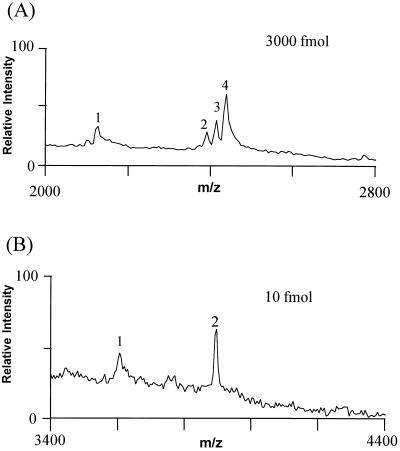
Fig. 4 compares the detection limits of the underivatized and derivatized oligosaccharide, A2F, by MALDI-MS. The derivatization increased the sensitivity of the mass measurement by 300-fold for A2F. Of four oligosaccharides tested, the derivatization increased the sensitivity of the mass spectrometric measurement between 50- and 1000-fold (Table 1). Sensitivities for the derivatized oligosaccharides were in the low femtomole range, which is in accord with our previously determined sensitivity limit for sample handling of peptides (data not shown).
Figure 4.
Comparison of the sensitivities of MALDI-MS analysis of derivatized and underivatized A2F. (A) MALDI mass spectra of underivatized A2F. The MALDI matrix solution consist of 0.2 M DHB/0.06 M HIC [1:1, vol/vol in CH3CN/water (1:2, vol/vol)] with 10 mM NaCl (to increase the ionization yield). The matrix sample mixture was dried under vacuum on the sample probe. In addition to the (M+H)+ ions (peak 2), an (M+Na)+ ion (peak 3) and an (M+2Na-H)+ (peak 4) were observed (B) Derivatized A2F. The glycopeptide was mixed with matrix solution [4-HCCA in CH3CN/0.1% TFA in water (1:2, vol/vol)], loaded on the probe, and dried at room temperature in air. The protonated glycopeptide ion (M+H)+ is labeled “2.” In A and B, peak 1 corresponds to, respectively, the sodium cationized and protonated molecule with the loss of one sialic acid.
Table 1.
Sensitivity enhancement of oligosaccharide measurement by MALDI-MS after peptide derivatization
| Carbohydrate | Underivatized, fmol | Derivatized, fmol | Signal enhancement, fold |
|---|---|---|---|
| A1 | 600 | 10 | ×60 |
| A2F | 3,000 | 10 | ×300 |
| LSTa | 10,000 | 10 | ×1000 |
| NA2FB | 100–300 | 2 | ×50–150 |
The sensitivity of MALDI-TOF mass spectrometric analysis usually deteriorates sharply when the molecular mass of an analyte falls below 800 Da (unpublished observation). Derivatization of an oligosaccharide with an aminooxyacetyl peptide not only attaches a basic peptide to the oligosaccharide but also increases its mass by 1526 Da [mass of the aminooxyacetyl peptide minus water (18 Da)] to a range optimal for MALDI-MS analysis. This advantage becomes particularly evident when analyzing small oligosaccharides or fragments of oligosaccharides from exoglycosidase digestion (see below). We note that no special effort was made to optimize the peptide for the present purposes, although the particular peptide used gave a strong mass spectrometric response (since it contains four basic groups). The peptide could also be designed to contain a fluorescent group or tryptophan that would make it compatible with UV/fluorescence detection.
Ladder Sequencing of Derivatized Oligosaccharides.
In addition to MS, there are several other approaches available for sequence analysis of oligosaccharides (6, 9, 11). One of the commonly used methods involves sequential removal of monosaccharide residues from oligosaccharides using exoglycosidases with high specificities. Dwek and coworkers (9) have refined this approach by using exoglycosidase arrays. The labeled oligosaccharide is aliquoted and digested with an exoglycosidase array to produce a list of fragments of the original oligosaccharide. The resulting glycoconjugate fragments are pooled and analyzed by column chromatography. Sequence and linkage information are deduced from the behavior of the glycoconjugate fragments in column chromatography and a knowledge of the enzyme array. More recently, Sutton et al. (8) explored the use of MALDI-MS for the analysis of carbohydrate fragments produced by enzyme-array digestion, wherein oligosaccharide structures were deduced from the molecular masses of the digestion products, and the composition and specificities of the exoglycosidase array used.
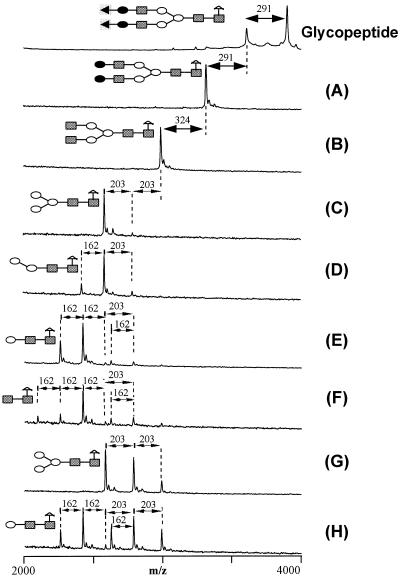
Here, we show use of the derivatized oligosaccharides for sequence analysis by applying exoglycosidase digestion (Table 2) in combination with MALDI-TOF MS. The oligosaccharide A2F (Fig. 4) was analyzed to demonstrate the methodology. Derivatized A2F was subjected to digestion without prior separation of the side-reaction products. The molecular masses of the starting material and digestion products were measured by MALDI-TOF MS (Fig. 5). Structural information was obtained by taking into account (i) the molecular mass of the starting glycopeptide, (ii) the molecular masses of the glycoconjugate fragments, and (iii) the composition and specificities of the exoglycosidases (Table 2). The type of monosaccharide was determined by molecular mass differences between adjacent peaks in the mass spectra; 203-, 291-, and 162-Da losses indicate the presence of N-acetylglucosamine, sialic acid, and hexose, respectively. The identities of the monosaccharides and the linkage types were determined using the specificities and composition of the glycosidases array (8, 9). Thus, for example, comparison of the mass spectrum obtained from the starting glycopeptide (top of Fig. 5) with that obtained from the glycopeptide treated with exoglycosidase A—i.e., sialidase, see Table 2 (Fig. 5A)—reveals that the oligosaccharide contains up to two sialic acid residues linked α2–6, α3, or α8 to an unspecified monosaccharide. The identities of these unspecified monosaccharides are revealed by treatment of the glycopeptide with exoglycosidase mixture B, which contains sialidase and galactosidase (Table 2). The resulting spectrum shows an additional loss of 324 Da (i.e., 2 × 162 Da) indicating the presence of two galactose residues. The specificity of the galactosidase reveals that the galactose linkages are Galβ1–4GlcNAc or Galβ1–4GalNAc. Continuing the analysis in a similar manner, mass spectra were compared after treatment of the glycopeptide with six additional exoglycosidase mixtures (Table 2). It is seen that structural information about A2F was readily deduced for all residues beyond the core trisaccharide (although we note that the deduced linkage information may be limited by the specificities of the available exoglycosidases).
Figure 5.
Ladder sequencing of the A2F glycopeptide. A solution of the A2F glycopeptide was divided into eight aliquots and digested by the exoglycosidase array given in Table 2. Mass differences between some of the digestion product peaks are indicated. Symbolic structures are shown for the carbohydrate portion of the starting glycopeptide and for the smallest glycopeptide fragments resulting from the series of exoglycosidase digestions (A–H). Monosaccharides are represented by symbols: ▪, N-acetylglucose; •, galactose; ○, mannose; ◂, sialic acid; ▵, fucose. For clarity, the peptide portion of the glycopeptide is omitted from the symbolic structures.
CONCLUSION
In summary, conditions for obtaining high levels of ligation between oligosaccharides and an aminooxyacetyl peptide was successfully developed. The resulting glycopeptide can be measured by MALDI-TOF MS with 50 to 1000 times more sensitivity than the underivatized oligosaccharide. The glycopeptide provides a very good reagent for structure analysis using exoglycosidase digestion. The sensitivity enhancement simplifies sample handling and is very helpful when dealing with low abundance oligosaccharides from biological sources.
MALDI-MS provides several advantages for assaying the digestion products from the enzyme array compared with the widely used chromatographic or gel electrophoretic methods (9, 10). First, MALDI-MS is much faster than the column chromatography or gel electrophoresis currently used in commercial oligosaccharide-analysis instruments (9, 10). Analysis of eight digestion samples by MALDI-MS can be completed in 0.5 hr. Second, the mass spectrometric approach is tolerant to heterogeneity of the oligosaccharides. Because MALDI-MS has much higher resolution than column chromatography or gel electrophoresis, it is less likely that impure oligosaccharides will produce confusion in the data analysis. By contrast, current commercial methods require a relatively high homogeneity of starting oligosaccharide. Third, incomplete glycosidase digestion is tolerated here because the structural information is obtained from molecular mass differences (and specificities and composition of the glycosidase mixture). Indeed, incomplete digestion can in principle give more structural information than complete digestion since more fragments are produced in an incomplete digestion. Finally, the sensitivity of MALDI-MS is very high—i.e., subpicomole amounts of glycopeptides can be easily analyzed.
In addition to oligosaccharide sequencing, the ligation reaction between an oligosaccharide and an aminooxyacetyl peptide also provides a potentially convenient and efficient way for the synthesis of glycopeptides or glycoproteins. An aminooxy group can be readily incorporated at the side chain of a natural or unnatural amino acid residue, which can be introduced into peptides during solid-phase peptide synthesis. Next, an oligosaccharide moiety can be ligated to the aminooxyacetyl peptide through the oxime formation reaction as described in this paper. Although the resulting glycopeptide does not have a natural linkage, it can be designed to provide a reasonably close approximation. A combination of glycopeptide synthesis with protein ligation techniques (25, 26) may also allow for the synthesis of artificial glycoproteins.
Acknowledgments
We thank Dr. Lynne E. Canne for the synthesis of the aminooxyacetyl peptide and Dr. Tom W. Muir for insightful discussions. This work was supported in part by Grant RR00862 from the National Institutes of Health.
ABBREVIATIONS
- 4-HCCA
α-cyano-4-hydroxycinnamic acid
- DHB
2,5-dihydroxybenzoic acid
- HIC
1-hydroxy isocarbostyril
- MALDI
matrix-assisted laser desorption/ionization
- MS
mass spectrometry
- TOF
time-of-flight
- TFA
trifluoroacetic acid
- LSTa
LS-tetrasaccharide a
- NA2FB
asialo-, agalacto-, bi-antennary, core-substituted with fucose and with bisecting GlcNAc
- A2F
di-sialylated-, galactosylated, bi-antennary, core substituted with fucose
- A1
mono-sialylated-, galactosylated, bi-antennary
Footnotes
During preparation of this manuscript, a related method was described for the preparation of glycopeptides (24).
References
- 1.Rademacher T W, Parekh R B, Dwek R A. Annu Rev Biochem. 1988;57:785–838. doi: 10.1146/annurev.bi.57.070188.004033. [DOI] [PubMed] [Google Scholar]
- 2.Varki A. Glycobiology. 1993;3:97–130. doi: 10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cumming D A. Glycobiology. 1991;1:115–130. doi: 10.1093/glycob/1.2.115. [DOI] [PubMed] [Google Scholar]
- 4.Woods R J, Edge C J, Dwek R A. Nature Struct Biol. 1994;1:499–501. doi: 10.1038/nsb0894-499. [DOI] [PubMed] [Google Scholar]
- 5.Green E D, Adelt G, Baenziger J U, Wilson S, van Halbeek H. J Biol Chem. 1988;263:18253–18268. [PubMed] [Google Scholar]
- 6.Koerner T A W, Prestegard J H, Yu R K. Methods Enzymol. 1987;138:38–59. doi: 10.1016/0076-6879(87)38006-1. [DOI] [PubMed] [Google Scholar]
- 7.Woods R J. Curr Opin Struct Biol. 1995;5:591–598. doi: 10.1016/0959-440x(95)80049-2. [DOI] [PubMed] [Google Scholar]
- 8.Sutton C W, O’Neill J A, Cottrell J S. Anal Biochem. 1994;218:34–46. doi: 10.1006/abio.1994.1138. [DOI] [PubMed] [Google Scholar]
- 9.Edge C J, Rademacher T W, Wormald M R, Parekh R B, Butters T D, Wing D R, Dwek R A. Proc Natl Acad Sci USA. 1992;89:6338–6342. doi: 10.1073/pnas.89.14.6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Starr C, Masada R I, Hague C, Skop E, Klock J. J Chromatogr. 1996;720:295–321. doi: 10.1016/0021-9673(95)00749-0. [DOI] [PubMed] [Google Scholar]
- 11.Geyer R, Geyer H. Methods Enzymol. 1995;230:86–108. doi: 10.1016/0076-6879(94)30009-7. [DOI] [PubMed] [Google Scholar]
- 12.Reinhold V N, Reinhold B B, Costello C E. Anal Chem. 1995;67:1772–1784. doi: 10.1021/ac00107a005. [DOI] [PubMed] [Google Scholar]
- 13.Philips N J, Apicella M A, Griffiss J M, Gibson B W. Biochemistry. 1993;32:2003–2012. doi: 10.1021/bi00059a017. [DOI] [PubMed] [Google Scholar]
- 14.Burlingame A L, Boyd R K, Gaskell S J. Anal Chem. 1994;66:634–683. doi: 10.1021/ac00084a024. [DOI] [PubMed] [Google Scholar]
- 15.Hillenkamp F, Karas M, Beavis R C, Chait B T. Anal Chem. 1991;63:1193–1199. doi: 10.1021/ac00024a002. [DOI] [PubMed] [Google Scholar]
- 16.Chait B T, Kent S B H. Science. 1992;257:1885–1894. doi: 10.1126/science.1411504. [DOI] [PubMed] [Google Scholar]
- 17.Karas M, Hillenkamp F. Anal Chem. 1988;60:2299–2303. doi: 10.1021/ac00171a028. [DOI] [PubMed] [Google Scholar]
- 18.Fenn J B, Mann M, Meng C K, Wong S F, Whitehouse C M. Science. 1989;246:64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 19.Smith R D, Loo J A, Edmonds C G, Barinaga C J, Udseth H R. Anal Chem. 1990;62:882–899. doi: 10.1021/ac00208a002. [DOI] [PubMed] [Google Scholar]
- 20.Linsley K, Chan S Y, Chan S, Reinhold B B, Reinhold V N. Anal Biochem. 1994;219:207–217. doi: 10.1006/abio.1994.1259. [DOI] [PubMed] [Google Scholar]
- 21.Stahl B, Steup N, Karas M, Hillenkamp F. Anal Chem. 1991;63:1463–1466. [Google Scholar]
- 22.Whittal R M, Palcic M M, Hindsgaul O, Li L. Anal Chem. 1995;67:3509–3514. doi: 10.1021/ac00115a020. [DOI] [PubMed] [Google Scholar]
- 23.Yoshino K, Takao T, Murata H, Shimonishi Y. Anal Chem. 1995;67:4028–4031. doi: 10.1021/ac00117a034. [DOI] [PubMed] [Google Scholar]
- 24.Cervigni S E, Dumy P, Mutter M. Angew Chem Int Ed Engl. 1996;35:1230–1232. [Google Scholar]
- 25.Rose K. J Am Chem Soc. 1994;116:30–33. [Google Scholar]
- 26.Canne L E, Ferre-D’Amare A R, Burley S K, Kent S B H. J Am Chem Soc. 1995;117:2998–3007. [Google Scholar]
- 27.Beavis R C, Chait B T. Proc Natl Acad Sci USA. 1990;87:6873–6877. doi: 10.1073/pnas.87.17.6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cohen S L, Chait B T. Anal Chem. 1996;68:31–37. doi: 10.1021/ac9507956. [DOI] [PubMed] [Google Scholar]
- 29.Harvey D J. J Mass Spectrom. 1995;30:1311–13245. [Google Scholar]
- 30.Harvey D J, Rudd P M, Bateman R H, Bordoli R S, Howes K, Hoyes J B, Vickers R G. Org Mass Spectrom. 1994;29:753–765. [Google Scholar]
- 31.Harvey D J. Rapid Commun Mass Spectrom. 1993;7:614–619. doi: 10.1002/rcm.1290070712. [DOI] [PubMed] [Google Scholar]
- 32.Beavis R C, Chaudhary T, Chait B T. Org Mass Spectrom. 1992;27:156–158. [Google Scholar]
- 33.Gusev A I, Wilkinson W R, Proctor A, Hercules D M. Anal Chem. 1995;67:1031–1041. [Google Scholar]
- 34.Wu K J, Shaler T A, Becker C H. Anal Chem. 1994;66:1637–1645. doi: 10.1021/ac00082a007. [DOI] [PubMed] [Google Scholar]
- 35.Mohr M D, Bornsen K O, Widmer H M. Rapid Commun Mass Spectrom. 1995;9:809–814. doi: 10.1002/rcm.1290090919. [DOI] [PubMed] [Google Scholar]