Abstract
Human interleukin 4 (IL-4) binds to its cellular receptor with a Kd in the subnanomolar range, similar to many other 4-helix-bundle proteins interacting with members of the hematopoietin (cytokine) receptor superfamily. In the IL-4 system this interaction is predominantly determined by the extracellular domain (IL4-BP) of the receptor α chain (Kd ≈ 150 pM). Now a high-resolution mutational and kinetic analysis has revealed that the high-affinity binding of IL-4 originates from a continuous patch of a few mostly polar or charged amino acid side chains located on helices A and C. The binding epitope comprises (i) a set of side chains determining the dissociation rate (koff) and (ii) a partially overlapping set determining the association rate constant (kon) of the IL-4/IL4-BP complex. The koff epitope is assembled from two juxtaposed main determinants (Glu-9 and Arg-88) surrounded by five side chains (Ile-5, Thr-13, Arg-53, Asn-89, and Trp-91) of lower importance. The cumulative increase in koff after alanine substitution is 105-fold for the central mixed-charge pair and 3 × 103-fold for the satellites. The kon epitope is formed by five positively charged residues on helix C (Lys-77, Arg-81, Lys-84, Arg-85, and Arg-88) and two neighboring residues on helix A (Glu-9 and Thr-13). The cumulative loss in kon of the alanine variants is only about 10-fold. These results provide the basis for an understanding of molecular recognition in cytokine receptor complexes and for an IL-4 antagonist design.
Keywords: receptor recognition, mutational analysis, biosensor, functional epitope, electrostatic steering
Interleukin 4 (IL-4) is a pleiotropic cytokine which plays a key regulatory role in the immune system (1). The most notable functions of IL-4 include the development of T-helper cells to a type 2 cytokine-producing phenotype (2) and the class switching to IgE (3). A dysregulation of IL-4 function appears to contribute to type 1 hypersensitivity reactions, such as allergies and asthma (4). Accordingly, inhibitors of IL-4 activity have therapeutic potential (5, 6).
IL-4, as many other cytokines, evokes a cellular response by promoting the formation of a heterodimeric receptor complex in the plasma membrane (7–9). The high-affinity binding of human IL-4 to its cellular receptor (10) is mediated nearly exclusively by the receptor α chain (11). The affinity is only marginally increased if both the α chain and γc, the second functional receptor chain, are present (7). Antagonistic IL-4 variants retain the high-affinity binding to the α chain but are deficient in γc binding (12, 13).
IL-4 is one of the short-chain 4-helix bundle cytokines (14). Its three-dimensional structure has been established in solution (15–17) and in crystals (18–20). Residues important for α-chain binding have been located on the antiparallel helices A and C (11, 21). The present detailed analysis of this discontinuous epitope addresses several questions relevant for an understanding of molecular recognition in cytokine receptor complexes and in particular for a rational IL-4 antagonist design.
(i) A large planar interface of ≈500 Å2 has been modelled for the human IL-4/IL4-BP complex (22, 23), similar to and only slightly smaller than that of the complex of human growth hormone (hGH) and its binding protein (hGHbp) (24). We wanted to know if only a few residues in the IL-4 structural epitope are critical in binding as established for the hGH epitope (25). (ii) Transient protein–protein interfaces have been discussed to be dominated by hydrophobic interactions similar to the interior of a protein (26, 27). The importance of charged IL-4 residues for binding (28) might be at variance with such a postulate. It was therefore interesting to identify and analyze the complete set of functional residues and their binding increments in the IL-4 epitope. (iii) A conspicuous feature of human IL-4 is a highly positive net charge and in particular a cluster of basic residues in helix C. This patch of positive charges has been surmised to be important for complex formation, since a surplus of negatively charged side chains is found in the modeled interface of IL4-BP. “Electrostatic steering” has been invoked to explain the fast association rate for IL-4/IL4-BP complex formation (29).
Here we have studied the effects of systematic alanine and glutamine substitutions (alanine scanning) as well as of charged residue substitutions (charge scanning) in IL-4 variants on the kinetics of IL4-BP interaction. The alanine variants were expected to exhibit a loss in binding energy according to the contribution of the original side chain atoms beyond the β carbon. In hGH (25) the sum of reductions in free energy caused by alanine substitutions was comparable to the total free energy of binding between hGH and hGHbp. For barnase/barstar association (30), however, side-chain contributions were not always additive due to cooperative interactions. Nevertheless, the mutational and kinetic analysis of IL-4/IL4-BP binding should identify the functional residues, and it might give a first estimation of their quantitative contributions to total binding free energy. The charge variants of IL-4 were studied, since they might exhibit loss of binding due to an electrostatic mismatch, and variants altered in their binding affinity thus could be affected at any of the side chains buried at the interface. The charge variants might in addition show a pronounced alteration in their association rate constant with IL4-BP, if electrostatic steering augments complex formation.
The kinetics of IL-4 variant binding were measured by means of the BIAcore system with a recombinant IL4-BP immobilized at the biosensor matrix (28). In addition, equilibrium binding of informative IL-4 variants was determined for T cells, to learn whether biosensor data give information relevant for the binding of the IL-4 variants to the receptor of whole cells. Finally, the results of the physical binding and kinetic measurements were compared with the proliferative activity of IL-4 variants for activated T cells.
MATERIALS AND METHODS
The human IL-4 variants were produced in Escherichia coli and purified to apparent chromatographic homogeneity (11). The freeze-dried proteins were dissolved in water at 300–600 μM concentration and stored at −20°C. The concentration was measured photospectrometrically (28) using an extinction coefficient ɛ278 = 8860 M−1·cm−1 for all variants but IL-4[W91A], for which ɛ278 = 3360 M−1·cm−1 was used.
The IL-4 cDNA was mutated by inserting synthetic double-stranded DNA cassettes between engineered restriction endonuclease cutting sites. Some cDNAs could not be cloned (e.g., K12A). Some variants could not be refolded and purified (e.g., D87A).
The BIA2000 system (Pharmacia Biosensor) was used to measure the rate constants for association and dissociation of the complex between IL-4 variants and IL4-BP as well as the equilibrium binding, as described (28). Briefly, a recombinant IL4-BP[C182A, Q206C] was biotinylated at Cys-206 and immobilized at the streptavidin-coated matrix of biosensor chip CM5 to a density of 50–70 resonance units corresponding to 50–70 pg of protein per mm2. The IL-4 variants were diluted with HBS running buffer (10 mM Hepes, pH 7.4/150 mM NaCl/3.4 mM EDTA/0.005% surfactant P20) to 1 or 10 μM and applied at concentrations between 2.5 and 1000 nM. Flow rates were set to 50 μl·min−1 at 25°C, flow path was 1-2-3-4, and data were recorded at 2.5 Hz. Sensograms were evaluated on the basis of a 1:1 association model according to fitting routines 2 or 3 provided by the BIA evaluation 2.1 software (Pharmacia Biosensor). The mean standard deviations were 15% for on-rate constants and 25% for the off-rate constants using the results of 12 to 24 independent measurements at different analyte concentrations.
Competitive radioligand binding to activated T-cells was performed as described (12), applying 100–300 pM 125I-labeled IL-4 with a specific activity of 2.2 × 106 dpm·pmol−1. Competing IL-4 variants were applied in serial 3-fold dilutions. Measurements were repeated at least twice. T cell proliferation was measured in triplicate by the incorporation of [3H]thymidine in the presence of serial 2-fold dilutions of the IL-4 variants (11).
Mouse IL-4 was produced in SF-9 cells and purified to apparent homogeneity (31). Protein was measured by the BCA protein assay (Pierce).
RESULTS
The high-affinity binding site of human IL-4 for IL4-BP has been located on helices A and C during previous studies on binding-deficient IL-4 variants (11, 21) and on IL-4 epitopes recognized by neutralizing monoclonal antibodies (32). Now, “loss-of-function” variants were produced at all surface positions of helices A and C by alanine or glutamine substitutions (alanine scanning). In addition, a collection of previously constructed glutamine variants, covering all conserved charged residues of IL-4, was reanalyzed. Finally, a charge scanning mutational analysis was performed to identify the complete IL-4BP-binding interface. All IL-4 proteins were purified to apparent homogeneity and submitted to biosensor-based binding experiments. A recombinant IL4-BP biotinylated at Cys-206 and immobilized at the streptavidin-coated matrix of the biosensor was used as receptor α-chain analogue (28). Sensograms recorded at different concentrations of IL-4 variants were evaluated for the kinetic constants of the association (kon and koff) as compiled in Table 1.
Table 1.
Rate constants for association (kon) and dissociation (koff) of the complex between IL-4 variants and IL4-BP immobilized at a biosensor matrix
| IL-4 variant | kon × 10−6 M−1·s−1 | koff × 103 s−1 | IL-4 variant | kon × 10−6 M−1·s−1 | koff × 103 s−1 |
|---|---|---|---|---|---|
| IL4 | 13 | 2.1 | |||
| I5A | 12 | 14 | I5R | 14 | 8.7 |
| T6A | 14 | 1.9 | T6D | 8.8 | 15 |
| Q8A | 16 | 2.5 | Q8R | 11 | 1.9 |
| E9Q | 8.7 | 270 | E9K | ND | ND |
| E9A | ND | ND | |||
| I11A | 11 | 2 | |||
| K12S | 12 | 1.9 | K12E | 7.3 | 1.5 |
| T13A | 8.4 | 7.1 | T13D | 7.6 | 0.85 |
| N15A | 15 | 2.3 | N15D | 12 | 1.7 |
| S16A | 16 | 1.9 | S16D | 11 | 1.5 |
| E19A | 18 | 1.7 | E19R | 12 | 1.6 |
| R53Q | 11 | 7.3 | |||
| K77A | 10 | 2.1 | K77E | 4.4 | 2 |
| Q78A | 11 | 2.2 | Q78E | 11 | 2.7 |
| R81A | 7.7 | 2.8 | R81E | 3.2 | 6.1 |
| F82A | 15 | 2.1 | F82D | 12 | 0.73 |
| K84A | 10 | 2.9 | K84D | 2.4 | 9.3 |
| R85A | 8.1 | 2.7 | R85E | 3.6 | 4.6 |
| R88Q | 7.3 | 140 | R88D | ND | ND |
| R88A | 8.3 | 760 | |||
| N89A | 12 | 27 | N89R | ND | ND |
| W91A | 11 | 6.1 | W91D | 5.8 | 8.5 |
The association and dissociation of IL-4 variants and IL4-BP immobilized at a biosensor matrix was measured in real time by means of a BIA2000 system. The rate constants represent mean values of 12–24 independent measurements performed with at least three different analyte concentrations. The mean standard deviation was 15% for the kon and 25% for the koff values. ND, not determined (only unspecific binding was found).
The association rate constants were found to be unaffected or only slightly altered in all analyzed variants. A 1.5- to 2-fold reduction in on-rate occurred in alanine or glutamine variants E9Q, T13A, K77A, R81A, K84A, R85A, R88A/Q, and W91A (Table 1). With the exception of T13A, only substitutions of charged, mostly basic, residues produced a significant on-rate effect. The on-rate of variant E19A was consistently found to be faster than that of IL-4.
Charge variants at the basic positions of helix C exhibited a larger decrease in on-rate than the corresponding alanine variants. The replacement of a basic residue by an acidic residue of opposite charge in variants K77E, R81E, K84D, and R85D was paralleled by a 3- to 5-fold reduced on-rate. Charge variants T6D, K12D, and T13D on helix A showed an about 2-fold lower kon than IL-4.
These results indicated that the rate constant of 1.3 × 107 M−1·s−1 of association between IL-4 and IL4-BP is remarkably insensitive to amino acid substitutions, even when large alterations in size or charge had been introduced. The sensitivity showed a positional effect, with the largest changes found among the basic side chains on helix C. Substitutions leading to a loss of one positive charge had in most instances smaller effects (1.5- to 2-fold) than those leading to a loss of two positive charges (3- to 5-fold). This suggested that a loss of charges, usually positive charges, of certain side chains at specific positions of the protein surface slows down the rate of IL-4/IL4-BP association.
In previous experiments, a 4- to 5-fold decrease in the kon of IL-4 had been observed when the salt concentration was increased form 150 mM to 0.7 M NaCl (28). This negative salt effect was found to be only 2- to 3-fold for the kon of variants R81E, K84D, and R85D (data not shown). In none of the variants was the off-rate constant altered by salt. Thus, both high ionic strength of the medium and the loss of charges in the IL-4 protein weakened electrostatic steering during IL-4/IL4-BP association. It is possible that a kon in the range of 1 to 2 × 106 M−1·s−1 is a limit value attainable in the absence of electrostatic attraction (see ref. 33).
Large changes in dissociation rate constant were observed in a subset of the alanine or glutamine variants (Table 1). By far the most sensitive positions were those of Glu-9 and Arg-88. The off-rate constant was 70- to 130-fold faster when the charges of these side chains had been removed by glutamine substitution. The increase in koff was 300-fold when the whole side chain of Arg-88 had been replaced by that of alanine. In two further alanine variants the off-rate constant was increased 14-fold (N89A) and 7-fold (I5A). A 3-fold higher off-rate was found in variants T13A, R53Q, and W91A. A marginal but significant increase in koff occurred in IL-4 variants R81A, K84A, and R85A. R53Q was the only variant with an amino acid substitution located on helix B. An estimate of the cumulative effect of the substitutions in all these variants yielded an increase in koff by a factor of about 1 × 109.
The analyzed charge variants exhibited a pattern of changes in koff confirming and overlapping with that of the alanine or glutamine variants. All positions where alanine substitutions increased the koff also yielded faster dissociation rate constants after charged residue substitutions. The only exception was position Thr-13. Remarkably, variant T13D and also F82D exhibited a 3- to 4-fold reduced koff. The substituted acidic residue therefore stabilized the complex with IL4-BP. At the position of Thr-6, where alanine substitution had no effect, a faster off-rate constant was obtained in the charge variant. Variant T6D dissociated with an off-rate 8-fold faster than IL-4. The increase in off-rate observed with the charge variants R81E, K84D, R85D, and W91D was 3- to 5-fold and thus more pronounced than that of the corresponding alanine variants. The increased off-rates probably result from an electrostatic repulsion at the contact site with the receptor rather than from a loss in binding function of the original residue. These changes were generally quite low, however. From this observation it might be concluded that also in alanine or glutamine variants repulsive forces due to uncompensated charged receptor side chains appear to be negligible.
A set of further IL-4 variants with glutamine substitutions, which had been analyzed previously by competitive radioligand binding (11), showed association and dissociation rate constants with IL4-BP indistinguishable from those of IL-4. The only exception, as mentioned above, was variant R53Q.
Several IL-4 variants (E9K, E9A, R88D, and N89R) bound unspecifically to the biosensor matrix when applied at concentrations >1 μM. This prevented a measurement of the kinetic and equilibrium constants.
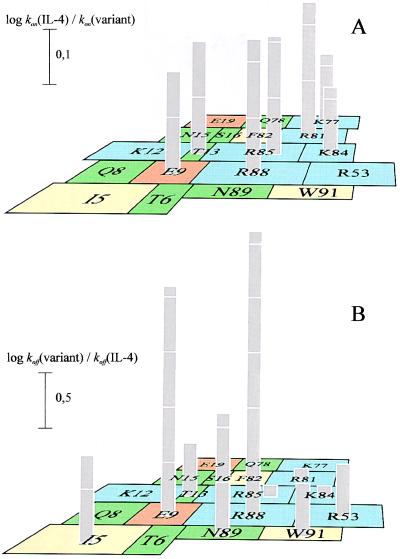
The high-resolution mutational analysis of IL-4 indicated that the side chains of Glu-9 and Arg-88, and especially their charges, provide the largest contribution to the stabilization of the complex. As illustrated for the koff epitope in Fig. 1B (see also Fig. 2), both side chains are juxtaposed on the surface of IL-4, where they form a pair of oppositely charged binding determinants. This mixed-charge pair is surrounded by five side chains of lower functional importance—i.e., Ile-5, Thr-13, Arg-53, Asn-89, and Trp-91. The off-rate determinants are not the same as the on-rate determinants. The kon epitope is shifted toward helix C and comprises most of the basic side chains in that surface (Fig. 1A). This positively charged epitope might be attracted by the negatively charged surface of the receptor, thereby increasing the association rate constant by the increment sensitive to ionic strength (5- to 10-fold).
Figure 1.
IL-4 epitopes determining association (A) and dissociation (B) rate constants for the IL-4/IL4-BP complex. The location of the mutated residues on helices A and C are indicated by rectangles with sizes corresponding to their accessible surface areas (17). The bars represent the change in kon or koff of IL-4 variants in relation to the kon or koff of IL-4 due to alanine or glutamine (E9, R53) substitution (see Table 1).
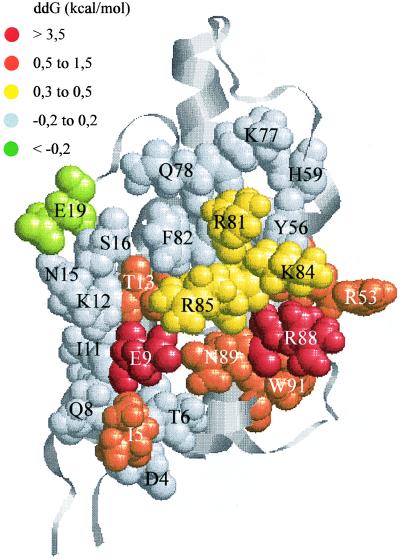
Figure 2.
Functional epitope of IL-4 determining the high-affinity binding of the receptor α chain. The 4-helix-bundle structure of IL-4 (17) is depicted as ribbons. The mutated residues are shown by space-filling models with their colors indicating the loss in binding free energy {ddG = 1.36 log(Kd[variant]/Kd[IL-4])} due to alanine or glutamine (E9, R53) substitution (see Table 2).
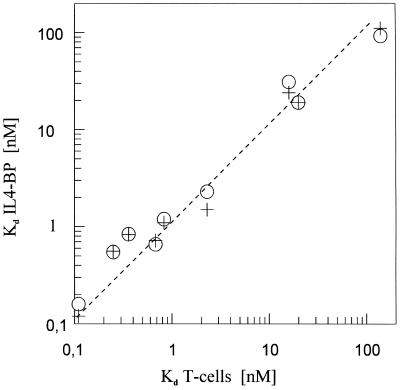
The total change in binding free energy (dG) for the interaction of IL-4 with IL4-BP is 13.3 kcal/mol (1 kcal = 4.18 kJ) as calculated from a dissociation constant of 160 pM. The loss of binding energy in alanine or glutamine variants (ddG) as calculated from the change in the dissociation constant Kd is shown in Table 2. The Kd values obtained either from the kinetic measurements (koff/kon) or from the equilibrium binding phase of the biosensor experiments, were similar to those from competitive radioligand binding to whole T cells (Fig. 3). The loss of binding free energy after alanine or glutamine substitution of seven residues (Ile-5, Glu-9, Thr-13, Arg-53, Arg-88, Asn-89, and Trp-91) amounted together to 12 kcal/mol. This accounted for 90% of the total binding energy. Another three residues (Arg-81, Lys-84, and Arg-85) when replaced by alanine produced a loss of 1.2 kcal/mol, which could account for a further 10% of the total binding energy. No significant energetic contributions could be found for other amino acid side chains. The cumulative loss of binding free energy established by the analyzed variants is probably too low, since, e.g., the Ala variant of Glu-9 could not be analyzed and interactions with the main chain cannot be studied by the present mutational approach. The high cumulative loss may indicate that the individual mutations are not completely independent.
Table 2.
Equilibrium binding of IL-4 variants to IL4-BP and to the receptor on T cells is similar but differs from their T cell proliferative activity
| IL-4 variant |
Kd (IL4-BP), nM
|
Equilib. Kd (T cells), nM | EC50 for T cell proliferation, nM | |
|---|---|---|---|---|
| koff/kon | Equilib. | |||
| IL4 | 0.16 | 0.11 | 0.12 | |
| I5A | 1.2 | 1.1 | 0.83 | 0.08 |
| T6A | 0.14 | 0.06 | 0.11 | |
| Q8A | 0.16 | 0.1 | ||
| E9Q | 31 | 24 | 16* | 3.1 |
| E9A | ND | |||
| I11A | 0.18 | |||
| K12S | 0.15 | |||
| T13A | 0.84 | 0.83 | 0.5 | 0.12 |
| N15A | 0.15 | 0.11 | ||
| S16A | 0.12 | 0.11 | ||
| E19A | 0.094 | 0.13 | ||
| R53Q | 0.66 | 0.72 | 0.68* | 0.13 |
| K77A | 0.21 | |||
| Q78A | 0.2 | |||
| R81A | 0.36 | 0.08 | ||
| F82A | 0.14 | 0.1 | ||
| K84A | 0.29 | 0.1 | ||
| R85A | 0.33 | 0.09 | ||
| R88Q | 19 | 19 | 20* | 2.5 |
| R88A | 92 | 110 | 140 | 8.1 |
| N89A | 2.3 | 1.5 | 2.3 | 0.09 |
| W91A | 0.55 | 0.56 | 0.25 | 0.09 |
The Kd values of IL-4 variants were calculated from the kinetic constants of Table 1 (Kd = koff/kon) and from equilibrium binding to biosensor-immobilized IL4-BP. The Kd values for IL-4 variants and the receptor on T cells were determined by competitive radioligand binding. The EC50 values are the concentrations effecting half-maximal proliferation of T cells. ND, not determined.
*Data from ref. 11.
Figure 3.
The dissociation constants measured for IL-4 variants and IL4-BP on biosensors are similar to those on T cells. The Kd values derived from koff/kon (○) or equilibrium binding (+) with IL4-BP during BIA2000 experiments are plotted on the y-axis against the Kd values obtained by competitive radioligand binding experiments with whole T cells on the x-axis. Data are taken from Table 2.
The location of the binding epitope is indicated in the space-filling IL-4 model presented in Fig. 2. The losses in binding affinity due to alanine substitution of the functional residues are represented by a color code. The contiguous binding epitope was considerably smaller than the total IL-4/IL4-BP interface derived from recent model-building exercises (22, 23). Furthermore, the location differed from earlier postulates (see below). The important koff epitope (residues colored in red and orange) extended perpendicular to the axis of the helix bundle and comprised two helical turns on helix A, one helical turn on helix C, and one residue on helix B. The small contribution of the kon epitope to binding is indicated by the yellow basic residues in the upper half of helix C. The functional residues in IL-4 were all polar with the exception of Ile-5 and Trp-91.
The biological activity of the IL-4 variants as determined in a T cell proliferation assay was indistinguishable from that of IL-4, with the exception of the variants affected at the key positions Glu-9 and Arg-88 (Table 2). But even with these variants the loss in biological activity (EC50) was 10 to 20 times less than the changes in binding affinity. The receptor on T cells and IL4-BP immobilized to the biosensor matrix bound the IL-4 variants with similar affinities (Fig. 3), and differences between the measurements in the two systems could not be responsible for these discrepancies. The unexpectedly low deficiencies of the variants in T cell proliferative activity, therefore, supported the earlier notion (28) that the biological activity of human IL-4 is not primarily determined by the equilibrium binding to the IL-4 receptor α chain.
IL-4 exhibits a marked species specificity with respect to both biological activity and physical receptor binding (see, e.g., ref. 10). Up to now, mouse IL-4 was considered not to cross-react with the human α chain to a measurable extent. When the functional epitope of human IL-4 was compared with the probably equivalent surface residues of mouse IL-4, however, several invariant positions were recognized. The two key residues of human IL-4 (Glu-9, Arg-88) are present in mouse IL-4. In addition, the minor determinants Arg-53 and Arg-85 are retained. Accordingly, it was a distinct possibility that mouse IL-4 binds to human IL4-BP in the micromolar range. A glycosylated mouse IL-4 expressed in Sf-9 cells bound during biosensor experiments to immobilized human IL4-BP with a 15 μM Kd at 150 mM NaCl and with a 35 μM Kd in the presence of 0.5 M NaCl (data not shown). The measured binding affinity is about 10 times weaker than estimated. This relatively small discrepancy could result from sterical hindrance at the interface between the mouse IL-4 and the human IL4-BP. It may be predicted that the interaction of bovine or porcine IL-4 with human IL4-BP has a still lower Kd, since the putative binding epitopes are conserved in these species and in human IL-4 with the exception of Trp-91 (Table 3).
Table 3.
Homologies between the functional epitope (koff epitope) of human IL-4 and the possibly equivalent epitopes of IL-4 from other species
| Species | Residue at position
|
||||||
|---|---|---|---|---|---|---|---|
| 5 | 9 | 13 | 53 | 88 | 89 | 91 | |
| Human | I | E | T | R | R | N | W |
| Cow | I | E | T | R | R | N | S |
| Pig | I | E | T | R | R | N | S |
| Sheep | I | E | T | R | R | N | S |
| Mouse | N | E | I | R | R | A | R |
| Rat | S | E | T | R | R | G | S |
The amino acid alignments are from ref. 34.
DISCUSSION
The present study has identified all amino acid side chains in human IL-4 that are engaged in the high-affinity binding of IL4-BP, the extracellular domain of the receptor α chain. A saturating mutational analysis of all surface residues on IL-4 helices A and C yielded all but one of the functional residues. Reanalysis of a collection of glutamine variants (11) confirmed that Arg-53 on helix B provides an additional functional side chain. The existence of further binding determinants is unlikely, but cannot be rigorously excluded at present. The variant D87A could not be renatured, probably because the original residue is structurally important. In addition, however, it may be involved in binding. Contributions of small residues—e.g., glycine or alanine—or of the backbone are also possible, but would not have been detected by the approach applied. Nevertheless, the salient features of the binding epitope could be clearly established. It is interesting to note that the receptor α-chain-binding epitope of IL-4 coincides with a B cell epitope recognized by IL-4 neutralizing monoclonal antibody 7D7 (32).
IL-4 residues buried in the postulated large flat interface with IL4-BP (22, 23) contribute very unequally to binding as assessed from the loss of binding free energy in alanine or glutamine variants. This is especially striking in the case of the four buried arginines, where the key determinant Arg-88 is located beside the minor determinant Arg-53 and also beside putative interface residues Arg-81 and Arg-85, which have very low functional importance. The three lysines at positions 12, 74, and 77 can be replaced by alanine with no or only marginal effect. Large hydrophobic side chains as those of Ile-5 or Trp-91 are of some functional importance, whereas Phe-82 can be replaced by a small alanine residue with no discernible consequences on binding. Thus, the functional epitope of IL-4 is clearly smaller than the putative structural epitope, similar to the binding in the hGH/hGHbp complex (25, 26). This leads to the question how a residue becomes functional—i.e., how it can make a positive contribution to the binding free energy. Why, e.g., is Arg-88 critical for binding but not the other arginines in the interface, although several of them appear to be engaged in ionic interactions with aspartic residues in IL4-BP (23)? Dehydration and/or mobility of side chains may be important parameters determining the binding potency of an interface residue. These questions may be answered when the orientation and the mobility of interface residues both in the complex and in the free IL-4 and IL4-BP proteins become known.
Charged and polar determinants predominate in the high-affinity binding epitope of human IL-4. The hydrophobic functional side chains of Ile-5, Thr-13, and Trp-91 are of minor importance, and single losses in binding due to alanine or even aspartate substitutions are less than 10-fold. The proposed interface of IL4-BP contains several polar and charged side chains. Ion pairs could be formed between Asp-67 in the CD loop of IL4-BP and IL-4 Arg-88, or between Lys-86 in the interdomain peptide of IL4-BP and Glu-9 of IL-4 (22). In the hGH receptor two tryptophans (Trp-104 and Trp-169) are the most important binding determinants involved in hydrophobic bonds to the hormone (35). The possibly equivalent positions in IL4-BP are Ala-66 and Tyr-124. Ala-66 is located in the polar sequence Ser-Ala-Asp-Asn. Thus, the functional IL-4/IL4-BP epitopes are probably not in accordance with the assumption that the large transient interfaces formed by the association of cytokines and cytokine receptors are mainly hydrophobic, similar to the interior of proteins (26).
Electrostatic steering likely contributes to the fast association rate constant of 1.3 × 107 M−1·s−1 between IL-4 and IL4-BP (29). All charged residues in the kon epitope of IL-4 as depicted in Fig. 1A could help to prealign the proteins by electrostatic interactions with the complementary IL4-BP interface. The association rate constant, but not the dissociation rate constant, is decreased at higher salt concentrations (28). For IL-4 the on-rate extrapolated for indefinitely high ionic strength amounts to about 2 × 106 M−1·s−1. This value is comparable to the on-rate constant predicted by Northrup and Ericson (33) for a generic protein–protein association on the basis of Brownian dynamics simulation and assuming a two-point encounter complex. The kon of charge-reversal variants is ½ to 1/5 that of IL-4, whereas the off-rates of these variants are unaltered (see Table 1). The salt effect on the association rate constant, however, is smaller than with IL-4, suggesting that a similar on-rate would be extrapolated for very high salt concentration (see also ref. 36). It is interesting to note that the kon for the association between mouse IL-4 and mouse IL4-BP is only 4 × 106 M−1·s−1 (31). Mouse IL-4 is less basic than the human IL-4; in particular, some of the basic residues on helix C are absent. Thus, electrostatic steering probably is of lower importance for the association of the mouse IL-4 receptor complex.
Some variants exhibit an up to 3-fold increased binding affinity compared with IL-4. This tighter binding is caused either by an increased on-rate (E19A) or by a decreased off-rate (T13D, F82D). Amino acid substitutions which increase the binding affinity have been detected in the hGH/hGHbp complex as well (25).
The biosensor experiments yield the energy contributions of the functional residues either by evaluating the kinetic constants (Kd = koff/kon) or by measuring the equilibrium binding at different IL-4 concentrations. The values obtained by both procedures are the same within the experimental error. When the receptor binding of variants to whole T cells is analyzed by competitive radioligand binding, the relative energy contributions of the functional side chains are similar to those obtained by means of the biosensor measurements. This indicates that the results obtained with the immobilized recombinant IL4-BP are relevant for the receptor binding on whole cells. Largely dissimilar, however, are the biological activities of the binding-deficient IL-4 variants. During a T cell proliferation assay all variants showed normal IL-4-like specific activities, with the exception of E9Q, R88Q, and R88A. But also with these variants the 10- to 20-fold loss in biological activity is much less pronounced than the more than 100-fold loss in receptor binding. Thus, the receptor-binding affinity of IL-4 variants (Kd) is not clearly correlated with their biological activity (EC50). The same holds for the correlation between koff and EC50. The koff for the IL-4/IL4-BP complex means a half-life of 6.5 min. As discussed previously (28), this is similar to the half-life of about 10 min for receptor-bound IL-4 internalization in whole cells. Possibly the biological activity of IL-4 and IL-4 variants is determined predominantly by the kon as long as the koff is in the time scale of the receptor internalization rates.
Human IL-4 has a three-dimensional structure similar to other 4-helix bundle cytokines, despite the large differences in the amino acid sequences (34). The cytokine receptors, on the other hand, exhibit related primary structures of their extracellular binding domains (14) which are probably all folded into two fibronectin type III domains. Therefore, the functional epitope of human IL-4 has been suggested to be organized like that in other 4-helical cytokines, in particular like that of hGH (see, e.g., ref. 17). The present results indicate, however, that several fundamental differences exist. Most importantly, the high-affinity binding epitope of IL-4 is located on helices A and C, and not on helix D like the high-affinity binding site of hGH. In hGH four main binding determinants exist on the same helix D (Lys-172, Thr-175, Phe-176, and Arg-178) contributing binding energies of around 2 kcal/mol. In IL-4 the binding is concentrated in two main determinants with energy contributions of 3–4 kcal/mol, and these residues exist on different helices. Finally, the interactions contributing to the hGH/hGHbp complex binding affinity are largely hydrophobic (35). In IL-4 the binding energy appears to be provided largely by charged groups. The site 2 epitope of hGH is located on helices A and C. It has apparently a much lower affinity for hGHbp, since it interacts only when the 1:1 complex with site 1 has already been formed. The functional residues in hGH site 2, as far as they have been analyzed up to now, differ considerably from the type and position of the residues identified in the high-affinity epitope of human IL-4. It will be interesting to learn whether the high-affinity binding epitopes of other short-chain 4-helical cytokines are constructed similarly to that of IL-4.
Acknowledgments
We thank C. Söder and H. Streitenberger for excellent technical assistance. This work was supported by the Deutsche Forschungsgemeinschaft, Grant Se 235/2-2, and by the Fond der Chemischen Industrie.
ABBREVIATIONS
- IL-4
interleukin 4
- IL4-BP
IL-4-binding protein
- hGH
human growth hormone
- hGHbp
hGH-binding protein
References
- 1.Paul W E. Blood. 1991;77:1859–1870. [PubMed] [Google Scholar]
- 2.Paul W E, Seder R A. Cell. 1994;76:241–251. doi: 10.1016/0092-8674(94)90332-8. [DOI] [PubMed] [Google Scholar]
- 3.Coffman R L, Lebman D A, Rothman P. Adv Immunol. 1993;54:229–270. doi: 10.1016/s0065-2776(08)60536-2. [DOI] [PubMed] [Google Scholar]
- 4.Romagnani S. Annu Rev Immunol. 1994;12:227–257. doi: 10.1146/annurev.iy.12.040194.001303. [DOI] [PubMed] [Google Scholar]
- 5.Goldstein R A, Paul W E, Metcalfe D D, Busse W W, Reece E R. Ann Intern Med. 1994;121:698–708. doi: 10.7326/0003-4819-121-9-199411010-00011. [DOI] [PubMed] [Google Scholar]
- 6.de Vries J E. J Invest Dermatol. 1994;102:141–144. doi: 10.1111/1523-1747.ep12371750. [DOI] [PubMed] [Google Scholar]
- 7.Kondo M, Takeshita T, Ishii N, Nakamura M, Watanabe S, Arai K, Sugamura K. Science. 1993;262:1874–1877. doi: 10.1126/science.8266076. [DOI] [PubMed] [Google Scholar]
- 8.Russell S M, Keegan A D, Harada N, Nakamura Y, Noguchi M, Leland P, Friedmann M C, Miyajima A, Puri R K, Paul W E, Leonard W. Science. 1993;262:1880–1883. doi: 10.1126/science.8266078. [DOI] [PubMed] [Google Scholar]
- 9.Heldin C H. Cell. 1995;80:213–223. doi: 10.1016/0092-8674(95)90404-2. [DOI] [PubMed] [Google Scholar]
- 10.Beckmann M P, Cosman D, Fanslow W, Maliszewski C R, Lyman S D. Chem Immunol. 1992;51:107–134. [PubMed] [Google Scholar]
- 11.Kruse N, Shen B J, Arnold S, Tony H P, Muller T, Sebald W. EMBO J. 1993;12:5121–5129. doi: 10.1002/j.1460-2075.1993.tb06207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kruse N, Tony H P, Sebald W. EMBO J. 1992;11:3237–3244. doi: 10.1002/j.1460-2075.1992.tb05401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duschl A. Eur J Biochem. 1995;228:305–310. [PubMed] [Google Scholar]
- 14.Bazan J F. Proc Natl Acad Sci USA. 1990;87:6934–6938. doi: 10.1073/pnas.87.18.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith L J, Redfield C, Boyd J, Lawrence G M, Edwards R G, Smith R A, Dobson C M. J Mol Biol. 1992;224:899–904. doi: 10.1016/0022-2836(92)90457-u. [DOI] [PubMed] [Google Scholar]
- 16.Powers R, Garrett D S, March C J, Frieden E A, Gronenborn A M, Clore G M. Science. 1992;256:1673–1677. doi: 10.1126/science.256.5064.1673. [DOI] [PubMed] [Google Scholar]
- 17.Müller T, Dieckmann T, Sebald W, Oschkinat H. J Mol Biol. 1994;237:423–436. doi: 10.1006/jmbi.1994.1245. [DOI] [PubMed] [Google Scholar]
- 18.Wlodawer A, Pavlovsky A, Gustchina A. FEBS Lett. 1992;309:59–64. doi: 10.1016/0014-5793(92)80739-4. [DOI] [PubMed] [Google Scholar]
- 19.Walter M R, Cook W J, Zhao B G, Cameron R P, Jr, Ealick S E, Walter R L, Jr, Reichert P, Nagabhushan T L, Trotta P P, Bugg C E. J Biol Chem. 1992;267:20371–20376. doi: 10.2210/pdb2int/pdb. [DOI] [PubMed] [Google Scholar]
- 20.Müller T, Oehlenschlager F, Buehner M. J Mol Biol. 1995;247:360–372. doi: 10.1006/jmbi.1994.0144. [DOI] [PubMed] [Google Scholar]
- 21.Ramanathan L, Ingram R, Sullivan L, Greenberg R, Reim R, Trotta P P, Le H V. Biochemistry. 1993;32:3549–3556. doi: 10.1021/bi00065a005. [DOI] [PubMed] [Google Scholar]
- 22.Bamborough P, Hedgecock C J, Richards W G. Structure. 1994;2:839–851. doi: 10.1016/s0969-2126(94)00085-9. [DOI] [PubMed] [Google Scholar]
- 23.Gustchina A, Zdanov A, Schalk Hihi C, Wlodawer A. Proteins. 1995;21:140–148. doi: 10.1002/prot.340210208. [DOI] [PubMed] [Google Scholar]
- 24.de Vos A M, Ultsch M, Kossiakoff A A. Science. 1992;255:306–312. doi: 10.1126/science.1549776. [DOI] [PubMed] [Google Scholar]
- 25.Cunningham B C, Wells J A. J Mol Biol. 1993;234:554–563. doi: 10.1006/jmbi.1993.1611. [DOI] [PubMed] [Google Scholar]
- 26.Wells J A. Proc Natl Acad Sci USA. 1996;93:1–6. doi: 10.1073/pnas.93.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones S, Thornton J M. Proc Natl Acad Sci USA. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen B J, Hage T, Sebald W. Eur J Biochem. 1996;222:491–499. doi: 10.1111/j.1432-1033.1996.0252h.x. [DOI] [PubMed] [Google Scholar]
- 29.Demchuk E, Mueller T, Oschkinat H, Sebald W, Wade R C. Protein Sci. 1994;3:920–935. doi: 10.1002/pro.5560030607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schreiber G, Fersht A R. J Mol Biol. 1995;248:478–486. doi: 10.1016/s0022-2836(95)80064-6. [DOI] [PubMed] [Google Scholar]
- 31.Grunewald S M, Kunzmann S, Schnarr B, Ezernieks J, Sebald W, Duschl A. J Biol Chem. 1997;272:1480–1483. doi: 10.1074/jbc.272.3.1480. [DOI] [PubMed] [Google Scholar]
- 32.Reusch P, Arnold S, Heusser C, Wagner K, Weston B, Sebald W. Eur J Biochem. 1994;222:491–499. doi: 10.1111/j.1432-1033.1994.tb18890.x. [DOI] [PubMed] [Google Scholar]
- 33.Northrup S H, Erickson H P. Proc Natl Acad Sci USA. 1992;89:3338–3342. doi: 10.1073/pnas.89.8.3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rozwarski D A, Gronenborn A M, Clore G M, Bazan J F, Bohm A, Wlodawer A, Hatada M, Karplus P A. Structure. 1994;2:159–173. doi: 10.1016/s0969-2126(00)00018-6. [DOI] [PubMed] [Google Scholar]
- 35.Clackson T, Wells J A. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 36.Schreiber G, Fersht A R. Nat Struct Biol. 1996;3:427–431. doi: 10.1038/nsb0596-427. [DOI] [PubMed] [Google Scholar]