Abstract
Deregulation of the evolutionarily conserved Notch signaling is highly correlated with oncogenesis. Intracellular activated Notch (ICN) is a protooncogene linked to the transcription activation of a number of cellular genes involved in cell cycle regulation, differentiation, and proliferation. Stability of ICN is tightly regulated by the Sel10-mediated ubiquitin–proteasome pathway. Sel10 can function as a negative regulator of Notch and exhibits activities of a tumor-suppressor protein. This article shows that the Kaposi's sarcoma-associated herpesvirus (KSHV) latency-associated nuclear antigen (LANA) directly interacts with Sel10 and forms a complex in KSHV-infected cells. This results in suppression of ICN ubiquitination and degradation. The carboxyl terminus of LANA interacts with the F-box and WD40 domains of Sel10 and competes with ICN for binding to Sel10. This elevated level of ICN is also critical for maintaining the enhanced proliferation of KSHV-infected tumor cells. These findings describe a mechanism by which the KSHV-encoded LANA protein regulates ubiquitination of ICN mediated by the F-box component of the E3 ligase Sel10, leading to proliferation of the virus-infected cells.
Kaposi's sarcoma-associated herpesvirus (KSHV) is a human DNA tumor virus and is etiologically linked to Kaposi's sarcoma, the most common cancer in AIDS patients, primary effusion lymphomas, and multicentric Castleman disease (1). KSHV belongs to the γ-herpesvirus subfamily and can establish lifelong persistence in the host after primary infection (2). The diversity of the KSHV genes allows the virus to manipulate its human host using a battery of distinct strategies (3). The effects of these processes can promote cell proliferation, leading to tumor development.
Establishment of viral latency in the host is a common feature of γ-herpesviruses and KSHV-mediated pathogenesis (4). The reduced viral gene expression pattern of latency minimizes the number of viral epitopes that are presented by infected cells to cytotoxic T lymphocytes and so contributes to the ability of the virus to escape immune surveillance and long-term persistent infection (5, 6). Cells latently infected by KSHV express a limited number of viral genes, including latency-associated nuclear antigen (LANA) (7). Several important functions have been highlighted for LANA, including maintenance of the viral episome (8–10). LANA interacts with the tumor suppressors p53 and pRb, leading to the inhibition of apoptosis and cell cycle deregulation mediated by these tumor suppressors (11, 12). Additionally, LANA has been shown to regulate various other cellular pathways, including the Wnt signaling pathway stabilizing β-catenin (13, 14). LANA was also shown to transactivate the telomerase reverse-transcriptase promoter, which contributes to the oncogenic phenotype (15). It has also been shown that LANA can induce chromosome instability in KSHV-infected B cells (16). Recently, we showed that accumulation of the intracellular domain of Notch1 (ICN) is linked to increased proliferation of KSHV-infected cells (17). The enhanced levels of ICN promote the survival of KSHV-infected primary B cells (17, 18).
Notch signaling is an evolutionarily conserved pathway controlling diverse aspects of development and tissue homeostasis (19, 20). Deregulation of Notch signaling has been implicated in development of cancer with ICN associated with a subset of human T cell lymphoma and tumorigenesis (21–23). Interestingly, coexpression of LANA and ICN significantly increases the half-life of ICN (17). This raised the possibility that LANA can play a role in ICN stability. In this study, we sought to explore the mechanism leading to accumulation of ICN and abnormal Notch signaling in KSHV-infected cells.
Several studies have shown that ICN stability is regulated by the ubiquitin–proteasome pathway (24–26). An F-box protein Sel10 (also known as Fbw7, hCdc4), which belongs to Cdc4 family of proteins, has an essential role in ICN stability (24–26). The WD40 domain of Sel10 binds to ICN, and the F-box domain interacts with and recruits the E3 ligase complex containing Skp1, Cul1, and Rbx1 (24–26). Under physiological conditions, ICN degradation is precisely regulated by this protein complex which is maintained at a level critical for cell survival in the absence of any pathology. However, elevation of ICN levels in KSHV-infected tumor cells implies a role in pathogenesis induced by the virus. In this article, we tested the hypothesis that KSHV LANA may target the Sel10-mediated proteosome–ubiquitination pathway, resulting in ICN stabilization and maintenance of the proliferative state in KSHV-infected cells.
Results
LANA Directly Targets the F-Box Protein Sel10 in KSHV-Infected Pleural Effusion Lymphoma Cells.
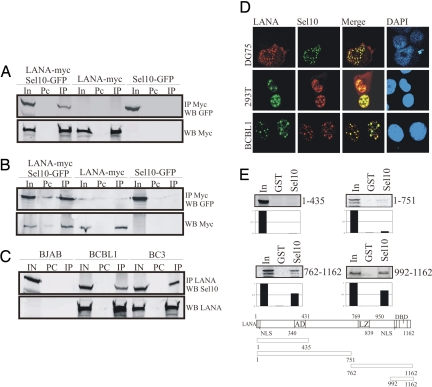
LANA is one of the predominant latent proteins expressed in the majority of KSHV-positive tumor cells. Therefore, profound cellular changes are likely to be linked to LANA expression. In our previous report, we showed that LANA can prolong the half-life of ICN when coexpressed in transfected human cells (17). We therefore hypothesized that the KSHV-encoded LANA protein may interact with ICN and directly occupy the Sel10-binding sites on ICN, preventing the SCF (composed of Skp1, Cul-1, ROC1, and F-box protein) E3 ligase from binding and ubiquitinating ICN. Thus, ICN is stabilized in LANA-expressing KSHV-positive cells. However, we were unable to detect a direct association between LANA and ICN complexed in cells (data not shown). This led us to investigate whether LANA may target Sel10 and compete with ICN for Sel10 binding. Therefore, less Sel10 will be available for binding to ICN in KSHV-positive cells, resulting in a reduction in ubiquitination of ICN by Sel10 and an increase in the stability of ICN. We performed immunoprecipitation assays in 293T and DG75 cells by transient transfection and convincingly showed that GFP-tagged Sel10 can be coprecipitated with Myc-tagged LANA when cotransfected in these cells (Fig. 1 A and B). In addition, we further verified this association between endogenous LANA and Sel10 in BCBL1 and BC3 cells latently infected with KSHV (Fig. 1C). In this experiment, immunoprecipitation of LANA by using a rabbit polyclonal antibody resulted in efficient coprecipitation of Sel10 (Fig. 1C). As expected, Sel10 was detected only in the cell lysate of the KSHV-negative BJAB cell line and not in the immunoprecipitation lane (Fig. 1C). Reverse-immunoprecipitation experiments using transfected 293T and DG75 cells or KSHV-infected BCBL1 and BC3 cells showed that LANA protein was specifically coprecipitated with Sel10 by using anti Sel10 antibodies [supporting information (SI) Fig. 6].
Fig. 1.
LANA associates with Sel10. (A and B) Immunoprecipitation analysis with mouse anti-Myc antibody or rabbit anti-GFP antibody showed that Sel10-GFP was directly immunoprecipitated with LANA-myc in 293T (A) and DG75 (B) cells. Fifteen million cells were cotransfected with 10 μg of Sel10-GFP and 10 μg of LANA-myc expression vectors or transfected with these vectors, respectively, 24 h after transfection; cell lysates were use for immunoprecipitate (IP) analysis. In, 5% total cell lysate input; Pc, preclear. (C) Endogenous LANA interacts with Sel10 in KSHV-infected primary effusion lymphoma cells BCBL1 and BC3. Immunoprecipitation analysis with polyclonal rabbit LANA antiserum showed that Sel10 was directly immunoprecipitated with LANA in BCBL1 and BC3 cells. For this experiment, 30 million cells were harvested for preparation of cell lysates, which were use for IP analysis. BJAB cells, which are KSHV-negative were used as a control. IN, 5% total lysate input; PC, preclear; IP, immunoprecipitate. (D) Immunofluorescence analysis showed that Sel10 was localized to the same nuclear compartment as LANA in different cells. (Top and Middle) DG75 and 293T cells were cotransfected with 10 μg each of Sel10-myc (Myc tagged) and pCDNA-LANA expression vectors, respectively; 24 h after transfection, cells were harvested for immunofluorescence analysis. Rabbit anti-LANA polyclonal antibody and mouse anti-Myc monoclonal antibody and corresponding secondary antibodies were used for this assay. (Bottom) BCBL1 cells were also used for immunofluorescence analysis. Human anti-LANA serum and rabbit anti-Sel10 polyclonal antibody and corresponding secondary antibodies were used for this assay. (E Top and Middle) C terminus LANA associates with Sel10 in vitro. The 35S-labeled products were incubated with GST as well as GST-Sel10 fusion protein. The pulldown products were electrophoresed on 10% SDS/PAGE gel, dried, and exposed to a PhosphorImager. Input controls of 10% of total LANA translation products. (Bottom) A schematic for the LANA clones used is given. NLS, nuclear localization signal; AD, acidic domain; LZ, leucine zipper; DBD, DNA-binding domain.
To corroborate the findings above, we further performed immunofluorescence assays to determine whether the association of LANA and Sel10 occurred in similar compartments in the nucleus or whether there was a change in LANA or Sel10 localization when both were coexpressed in cells. First, we cotransfected DG75 and 293T cells with Sel10 and LANA expression constructs. Twenty-four hours after transfection, cells were harvested, fixed, and probed with mouse monoclonal antibody against the myc epitope tagged to Sel10 and rabbit antiserum with specificity for LANA. Fluorescein isothiocyanate and Texas red conjugated to the appropriate secondary antibody were used for visualization of the proteins. The results showed that LANA and Sel10 protein colocalized to the same nuclear compartments in DG75 and 293T cells. (Fig. 1D, Top and Middle). To determine whether endogenous LANA and Sel10 colocalized in similar compartments in KSHV latently infected BCBL1 cells, cells were fixed on slides and probed with rabbit polyclonal antibody against Sel10 and human antiserum specific for LANA. The antibody–protein complexes were visualized with Texas red and FITC-conjugated secondary antibodies, respectively, by confocal microscopy. The results clearly demonstrated that endogenous LANA and Sel10 are colocalized in similar nuclear compartments in BCBL1 cells (Fig. 1D Bottom).
LANA is a large viral protein having 1,162 aa with distinct domains (27). To identify the region of LANA binding to Sel10, Escherichia coli-expressed Sel10 GST fusion protein was prepared and incubated with in vitro-translated truncated polypeptides of LANA. Sel10-GST but not GST alone control, efficiently bound to the C terminus of LANA (762–1,162aa) (Fig. 1E). Further truncation of the carboxyl-terminal domain of LANA identified the extreme carboxyl-terminal 170 aa (992–1,162) as the interacting domain of Sel10. This region strongly bound to Sel10 in our in vitro binding assay (Fig. 1E). Under similar conditions, two amino-terminal truncations (1–435 aa and 1–751 aa) failed to bind to the Sel10-GST fusion protein (Fig. 1E). To ensure that the extreme carboxyl terminus of LANA also interacts with Sel10 in vivo, we performed immunoprecipitation assays, which showed that GFP-tagged Sel10 can be coprecipitated with Myc-tagged C-LANA (SI Fig. 7).
LANA Can Rescue ICN from the Degradation Mediated by Sel10 and Inhibit Ubiquitination of ICN.
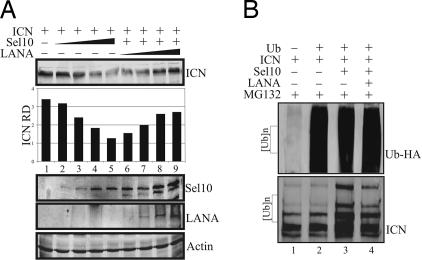
To explore the functional consequences of the interaction between LANA and Sel10, we cotransfected HEK 293T cells with a fixed amount of ICN expression vector with increasing amounts of Sel10 (Fig. 2A, lanes 1–5). Forty-eight hours after transfection, the cells were harvested, and Western blot analysis was performed for determination of ICN levels. Expectedly, the expression levels of ICN gradually decreased with increasing amounts of Sel10 in a dose-dependent manner (Fig. 2A, lanes 1–5). Interestingly, the ICN expression levels were rescued from Sel10-mediated degradation with increasing amounts of LANA (Fig. 2A, lanes 6–9). This suggested that LANA can antagonize Sel10 function as an E3 ligase for ICN ubiquitination by directly competing for interaction.
Fig. 2.
LANA inhibits degradation of ICN. (A) LANA rescues ICN degradation mediated by Sel10. In lanes 1–5, 5 μg of ICN expression vectors along with 0 μg, 2.5 μg, 5 μg, 10 μg, and 15 μg of Sel10 were transfected into 293T cells. In lanes 6-, 5 μg of ICN and 15 μg of Sel10 along with 2.5 μg, 5 μg, 10 μg, and 15 μg of LANA were transfected into 293 cells. Forty-eight hours after transfection, cells were harvested for preparation of lysates and examined by Western blotting. Each protein was checked by specific antibody individually, and actin was used for loading control. (B) LANA inhibits ubiquitination of ICN by Sel10. Transfection of 293T cells with the ICN (5 μg) together with HA-tagged ubiquitin (1.5 μg), Sel10 (10 μg), and 10 μg of LANA by using different combinations indicated by +/−. Forty-eight hours after transfection, cells were harvested for preparation of lysates and IP experiments. MG132 was added to the cell culture 3 h before harvesting. The cell lysates were immunoprecipitated by rabbit anti-ICN antibody and Western blotting for HA-tagged ubiquitin (Upper) as well as ICN (Lower).
ICN degradation occurs through the ubiquitin–proteasome pathway, with Sel10 functioning as the F-box component of the E3 ubiquitin ligase (24–26). To determine whether expression of LANA can suppress ubiquitination of ICN by Sel10, an ICN expression vector was transfected together with hemagglutinin-tagged ubiquitin expression vector (Fig. 2B). Total cell lysates were prepared, and ICN was immunoprecipitated from cellular protein extracts by using a purified polyclonal antibody against ICN. Immunoprecipitated ICN was visualized by Western blots using the specific anti-ICN antibody (28). The ubiquitinated form of ICN degrades rapidly but can be detected in the presence of proteosome inhibitor MG132 (24–26). In our assay, ubiquitination of ICN was greatly enhanced in the presence of Sel10 when compared with samples lacking Sel10 expression (Fig. 2B Lower, lane 3). Surprisingly, in the presence of LANA, the ubiquitination of ICN by Sel10 was greatly reduced to ≈50% based on the quantification (Fig. 2B Lower, lane 4). These results suggest that LANA can enhance ICN stability through interference of Sel10 activities in functioning as an E3 ligase in the context of the ubiquitin–proteasome system. In addition, we also checked the total amount of the ubiquitinated proteins associated with ICN by probing with anti ubiquitin-HA. No specific bands were detected, suggesting that a number of proteins are associated with ICN and can be pulled down by using anti-ICN antibody (Fig. 2B Upper).
LANA Binds to the WD40 Domain of Sel10 and Competes with ICN for Sel10 Binding.
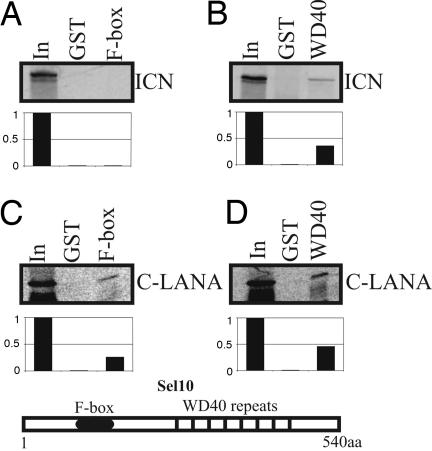
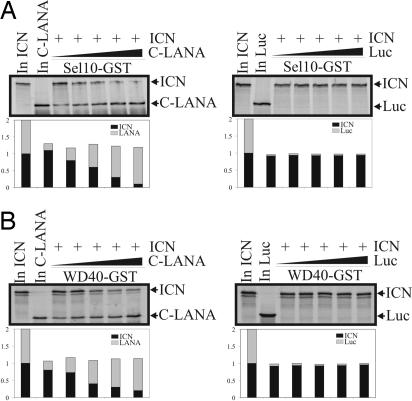
LANA interacts with Sel10 and inhibits Sel10-mediated ubiquitination. To determine the molecular mechanism by which LANA can perform this function, we identified the interacting domain. Sel10 binds to ICN through its WD40 domain, and its F-box domain recruits other components of the E3 ligase (24–26). To determine whether LANA can associate with the WD40 domain and compete with ICN for Sel10 binding, the F-box and WD40 domain of Sel10 were fused to GST and incubated with the carboxyl terminus of LANA, which we showed above was able to bind to the full-length Sel10. The carboxyl terminus of LANA strongly bound to the WD40 domain of Sel10 and was also seen to weakly bind to the F-box domain (Fig. 3 C and D). As expected, ICN bound to the WD40 domain used as a control in this assay (Fig. 3 A and B). This result indicates that LANA can compete with ICN for binding to the WD40 domain of Sel10 in KSHV-infected cells and so prevent or reduce Sel10 association with ICN. This leads to accumulation of ICN in these cells. To further explore the question of competition between LANA and ICN, the carboxyl terminus of LANA known to bind to Sel10 as well as ICN were in vitro-translated with [35S]Met labeling. The in vitro-translated products were incubated with either GST-Sel10 or GST-WD40 to determine the relative binding affinity of LANA and ICN to Sel10 and its WD40 domain. In this binding assay, the amount of ICN was constant, and increasing amounts of LANA were added to determine the impact of LANA levels on the interaction of ICN with Sel10 in vitro. The results of this experiment showed that LANA can directly compete and disrupt ICN from binding to either Sel10 or its WD40 domain with an ≈50% loss of binding activity (Fig. 4 A and B). However, in control experiments, increasing amounts of luciferase control protein did not have any effect on ICN binding to Sel10 or the Sel10 WD40 domain (Fig. 4 A and B), indicating that LANA can specifically compete with ICN for binding to Sel10. To further ensure that the full-length LANA also competes with ICN for Sel10 binding, we performed a similar assay in which the full-length translated LANA protein was used. Clearly, full-length LANA can inhibit ICN binding to Sel10 in a dose-dependent manner (SI Fig. 8).
Fig. 3.
C terminus LANA interacts with both F-box and WD40 domain. ICN was in vitro-transcribed and translated. (A and B) The 35S-labeled products were incubated with GST as well as GST-F-box (A) or GST-WD40 fusion protein (B). (C and D) C terminus LANA was in vitro-transcribed and translated. The 35S-labeled products were incubated with GST as well as GST-F-box (C) or GST-WD40 fusion protein (D). The pulldown products were electrophoresed on 10% SDS/PAGE gel, dried, and exposed to a PhosphorImager. Input control is 10% of total translation product. A schematic diagram of Sel10 is shown below the blots. Sel10 is a 540-aa protein, and the F-box motif and WD40 repeated are indicated.
Fig. 4.
LANA/ICN competition for Sel10 binding. (A) Seven hundred sixty-two to 1,162-aa truncations of LANA and ICN were in vitro-transcribed and translated. The 35S-labeled products were incubated with GST-Sel10. A fixed amount of ICN and increasing amounts of LANA were used. Pulldown products were electrophoresed on 10% SDS/PAGE gels, dried, and exposed to a PhosphorImager. Input controls of 10% for LANA and for ICN were run as well. (Right) A fixed amount of ICN and increasing amounts of luciferase were used. Pulldown products were electrophoresed on 10% SDS/PAGE gels, dried, and exposed to a PhosphorImager. The quantification of each band is shown at the bottom. (B) LANA/ICN competition for Sel10 WD40 domain binding. Seven hundred sixty-two to 1,162-aa truncations of LANA and ICN were in vitro-transcribed and translated. The 35S-labeled products were incubated with GST-WD40. The procedure is the same as that mentioned in A.
ICN Is Stabilized by LANA in Vivo Through Targeting Sel10 and Is Associated with the Proliferation of KSHV-Infected Cells.
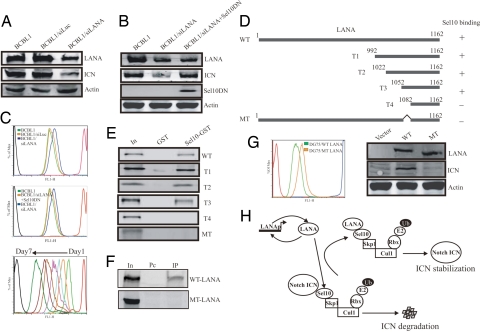
The results above suggest that LANA regulates ICN stability by directly targeting Sel10, the F-box component of the E3 ligase. LANA achieves this effect by directly competing with ICN for Sel10 binding. Thus, the binding of ICN to Sel10 is decreased in KSHV-infected cells, leading to a reduction in ubiquitination of ICN. We further wanted to evaluate the role of LANA on ICN stability using a more physiologically relevant approach. Previously, we showed that ICN accumulation in KSHV-positive cell lines is critical for the proliferation of these cells (17). Additionally, a γ-secretase inhibitor that blocks the production of ICN dramatically slows down the proliferation of KSHV-infected cells (17). In the context of these results, we wanted to know whether there were any functional consequences due to the interactions of LANA, Sel10, and ICN in regulation of cell proliferation. BCBL1 cells infected with KSHV in which LANA expression is at a relatively high level were transfected with an siRNA expression vector that specifically knocks down LANA expression in these cells. Selection of transfected cells resulted in stable cell lines expressing LANA-specific siRNA. A fraction of these stable cells were further transfected with another vector expressing a Sel10 dominant-negative construct. The ICN expression levels of these cells along with mock-transfected BCBL1 were then compared by Western blot analysis (Fig. 5A). The LANA expression level was knocked down in the siRNA-expressing cells (Fig. 5A). Accordingly, the ICN levels were also reduced comparable with the drop in LANA levels (Fig. 5A). This can be explained as a result of decreased binding of LANA to Sel10, resulting in increased ubiquitination of ICN. Interestingly, the ICN level was, in part, rescued by expression of the Sel10 dominant negative (Fig. 5B). This result strongly supports the hypothesis that ICN degradation in LANA siRNA-transfected BCBL1 cells is linked specifically through the Sel10 E3 ligase pathway and suggests that LANA can regulate this activity in a similar manner to the Sel10 dominant-negative molecule in the context of KSHV-infected cells.
Fig. 5.
Stabilization of ICN by LANA is associated with proliferation of KSHV-infected cells. (A) Western blotting showing expression of LANA and ICN in BCBL1, LANA siRNA-transfected BCBL1, and luciferase siRNA-transfected BCBL1 cells. Action level is as the loading control. (B) Western blotting to show expression level of LANA and ICN in BCBL1 cells, LANA siRNA-transfected BCBL1 cells, and LANA siRNA/Sel10 dominant-negative cotransfected BCBL1 cells. (C) CFSE staining showing the mitosis of cells. (Top) BCBL1 was transfected with LANA siRNA or Luciferase siRNA expression vectors and selected for stable clones. (Middle) BCBL1, LANA siRNA-transfected BCBL1, and LANA siRNA/Sel10 dominant-negative cotransfected BCBL1 cells were compared for mitosis. (Bottom) The standard proliferation curve of BCBL1 cells from 1st day to 7th day after CFSE staining. The negative control cell is CFSE nonstaining cells and the positive control cell is CFSE-staining but immediately fixed cell. (D) Mapping of the precise region of LANA responsible for Sel10 binding. A series of truncated LANA T1 (992–1,162), T2 (1,022–1,162), T3 (1,052–1,162), and T4 (1,082–1,162) were constructed, and in vitro binding assays were performed by using translated individual LANA- and GST-fused Sel10. (E) LANA lacking 1,052–1,082 aa were generated and used for in vitro binding assay with GST-Sel10. (F) Immunoprecipitation assays performed with mouse anti-Myc antibody and rabbit anti-GFP antibody showed that WT LANA but not the mutant type LANA (1,052–1,082 deletion) was directly immunoprecipitated with Sel10-GFP in 293T cells. Fifteen million cells were cotransfected with 10 μg of Sel10-GFP and 10 μg of WT LANA expression vector or mutant-type (MT) LANA expression vector. Twenty-four hours after transfection, cell lysates were used for IP analysis with GFP antibody. In, 5% total cell lysate input; Pc, preclear; IP, immunoprecipitate. (G) DG75 cells were -transfected with WT LANA and MT LANA and selected for stable clones. The individual stable cells expressing WT LANA or MT LANA were labeled with CFSE for proliferation assay. This figure shows that the proliferation of WT LANA-expressing cells is faster than that of MT-expressing cells. (H) Proposed model of ICN stability enhanced by KSHV LANA. ICN degradation is a tightly controlled ubiquitin–proteasome pathway mediated by Sel10 F-box protein. In KSHV-infected cells, LANA is expressed at a constant high level because LANA autoactivates its own promoter. In these cells, LANA competes with ICN for Sel10 binding, thus reducing ubiquitination and degradation of ICN. ICN is therefore stabilized in KSHV latently infected tumor cells.
To test the effect on cell proliferation caused by variation of these gene expression in these cells, we used carboxy-fluorescein diacetate succinimidyl ester (CFSE) staining to examine the mitotic activity of these cells by FACS (29). Expectedly, in LANA-specific siRNA-transfected BCBL1 cells, the mitotic activity was dramatically decreased compared with the mock-transfected cells (Fig. 5C). However, there is relatively no change in proliferation comparing the control luciferase siRNA-transfected cells with mock-transfected cells (Fig. 5C). Interestingly, coexpression of the Sel10 dominant-negative molecule in LANA siRNA-transfected BCBL1 cells did increase the proliferative activity when compared with cells expressing LANA siRNA alone, which showed a reduction in proliferation (Fig. 5C). A similar effect was also observed in JSC1 cells that are dually infected with KSHV and EBV but KSHV gene expression is predominant (SI Fig. 9).
The experiments above strongly suggest that the interaction between LANA and Sel10, which results in ICN accumulation, plays a role in KSHV-mediated cell proliferation. To determine the domain of LANA responsible for interacting with Sel10, a series of truncated versions of carboxyl-terminal LANA (Fig. 5D) were made, and we performed an in vitro binding assay. We found that the region between 1,052 and 1,082 aa is responsible for Sel10 binding (Fig. 5E). This was further confirmed as an in vitro-translated 1,052–1,082 deletion mutant of LANA showed loss in binding ability to Sel10 (Fig. 5E). We also performed the immunoprecipitation assay by using the 293T cells, which were transiently cotransfected with Sel10 and WT LANA or 1,052–1,082 deletion mutants, respectively. Interestingly, the mutant LANA was not pulled down with Sel10 antibody (Fig. 5F). This suggested that this domain of LANA is also responsible for Sel10 interaction. Finally, we transfected WT LANA and mutant type LANA into DG75 cells, a KSHV-negative Burkitt lymphoma cell line and obtained the cell clones in which LANA was stably expressed. We further compared the proliferation rate between these two stable cells in RPMI medium 1640 with 1% growth serum. The proliferation of WT LANA-expressing cells is clearly faster than that of the mutant LANA-expressing cells (Fig. 5G Left). In addition, the increased proliferation of WT LANA-expressing cells is, at least in part, due to the increased expression of ICN caused by the interaction between LANA and Sel10 (Fig. 5G Right).
Discussion
KSHV possesses a complex series of molecular strategies to regulate cell proliferation, induce cell transformation, and prevent cell apoptosis. Indeed, a number of single-gene studies have shown that different KSHV genes do possess these functional properties (11, 12, 30, 31). We demonstrated that ICN is accumulated in KSHV-infected cells, and KSHV LANA can enhance the stability of ICN (17). However, the detailed mechanism by which LANA stabilizes ICN is still elusive.
In this article, we further explored the mechanism by which LANA can inhibit degradation of ICN. Interestingly, we found that LANA directly interacts with F-box protein Sel10, thus enhancing the stability of ICN in KSHV-infected cell. In summary, the results obtained above are consistent with a model in which KSHV LANA inhibits ICN degradation by targeting the F-box protein Sel10, an essential component of the ubiquitin E3 ligase (Fig. 5H). In KSHV-infected tumors, LANA is a predominant latent protein and exhibits multiple functions related to viral genome replication, transcription regulation, and cell proliferation (10, 14, 32). It has been shown that LANA autoactivates its own promoter; thus, the expression of LANA is maintained consistently at a high level by the regulation of this positive-feedback loop (33). High expression of LANA in KSHV-infected cells is one strategy for the virus to establish and maintain latent infection because down regulation of LANA expression results in disruption of viral latency (34–36).
Establishment of viral latency is an essential step in pathogenesis induced by KSHV. So far, understanding the mechanism by which the KSHV encoded LANA drives oncogenesis is still incomplete. LANA plays a critical role in inhibiting the degradation of ICN in KSHV-infected tumor cells (17). This reveals a strategy by which KSHV induces abnormal proliferation of the tumor cells. In this study, we further addressed the related mechanism.
Sel10 was first identified in Caenorhabditis elegans as a negative regulator of Notch (LIN-12) (37). Biochemical evidence has suggested that both Notch1 and Notch4 (24–26) as well as presenilin (38) and cyclin E (39–41) are targets for ubiquitylation mediated by mammalian Sel10. Furthermore, the products of two protooncogenes, c-Jun and c-Myc, have recently been added to the list of proteins that are ubiquitylated via the Sel10-dependent ubiquitin pathway (42, 43). Accumulating evidence suggests that Sel10 functions as a tumor-suppressor gene. Indeed, the prevalence of Sel10 mutations in human cancers has recently turned out to be higher than was expected a few years ago and is now shown to be similar to that of p53 (39–41). Whereas such mutations were originally identified in a few breast and ovarian cancer cell lines, they have subsequently been also found in endometrial and colorectal cancers (44, 45). Most recently, it has been shown that inactivation of Sel10 can cause chromosome instability and that Sel10 is a p53-dependent tumor-suppressor gene in knockout mouse experiments in vivo (46). Sel10 plays a critical role in maintenance of cell homeostasis. Our study not only provides an explanation for the stabilization of ICN in KSHV-infected cells but establishes a link between the KSHV major viral antigen LANA and the Sel10 tumor suppressor. LANA directly interacts with Sel10 and inhibits its ability to ubiquitinate ICN, implying that LANA may also use this mechanism to stabilize other protooncogenes. Further exploration of the functional consequences of LANA–Sel10 interaction would provide insights elucidating KSHV-mediated oncogenesis.
Materials and Methods
Antibodies, Cell Lines, and Plasmids.
KSHV LANA rabbit polyclonal antibody was a kind gift from Bala Chandran (Rosalind Franklin University of Medicine and Science, North Chicago, IL). Human polyclonal serum that recognizes LANA is designated as HS and was provided by Gary Nabel (Vaccine Institute, National Institutes of Health, Bethesda, MD). Notch rabbit antiserum was provided by Jon C. Aster and Elliott Kieff (Brigham and Women's Hospital, Boston, MA). Val-1744 antibody was purchased from Cell Signaling Technology, Beverly, MA. Sel10 (Fbw7) rabbit polyclonal antibody was purchased from Abcam (Cambridge, MA). GFP rabbit polyclonal antibody was purchased from BD Biosciences (San Diego, CA). Myc- or HA-tagged proteins were detected by supernatants from 9E10 or 12CA5 hybridoma, respectively.
HEK293T cells were obtained from Jon Aster (Brigham and Women's Hospital). DG75 is a KSHV-negative B cell line provided by Elliott Kieff (Harvard Medical School, Boston, MA). BCBL1 and BC3 are KSHV-positive body cavity-based lymphoma-derived cell lines obtained from Dr. Don Ganem (University of California School of Medicine, San Francisco, CA) and the American Type Culture Collection (Manassas, VA), respectively. JSC1 was a kind gift from Richard F. Ambinder (Johns Hopkins University School of Medicine, Baltimore, MD). A human osteosarcoma cell line U2OS was obtained from the American Type Culture Collection.
The 293T cells were grown in high-glucose DMEM supplemented with 5% bovine growth serum (BGS; HyClone, Logan, UT), 2 mM l-glutamine, 25 units/ml penicillin, and 25 μg/ml streptomycin. DG75, BCBL1, BC3, and JSC1 were grown in RPMI medium 1640 supplemented with 10% BGS, 2 mM l-glutamine, 25 units/ml penicillin, and 25 μg/ml streptomycin.
HA-tagged Notch ICN, pflu-ICN, and untagged pCDNA3.1-ICN expression vector were described (28). Myc-tagged full-length LANA and different truncations were described (36, 47, 48). Sel10-GFP and Sel10-myc expression vectors (25) were provided by Dr. Urban Lendahl (Department of Cell and Molecular Biology, Medical Nobel Institute, Karolinska Institute, Stockholm, Sweden). Based on these Sel10 vectors, F-box (amino acids 1–207), WD40 (amino acids 184–540), and Sel10 gene sequences were cloned into pGEX-2T to generate GST fusion proteins. To construct the pCDNA-Sel10 expression vector, a Sel10 fragment was obtained by digestion of Sel10-myc plasmid by using EcoRI and inserted into the EcoRI site of pCDNA3.1+. HA-tagged ubiquitin expression vector was described (49, 50). siRNAs complementary to the carboxyl-terminal (GCTAGGCCACAACACATCT) fragment of LANA as described (14) were cloned into the pSIREN vector according to the instructions of the manufacturer (Clontech, Palo Alto, CA) to generate the si-LANA construct. A pSIREN vector with luciferase target sequence was used as control.
Transfection.
HEK293T, DG75, or BCBL1 cells were transfected by electroporation using a Gene Pulser II electroporator (Bio-Rad, Hercules, CA). Ten to 15 million cells harvested in exponential phase were collected and washed in PBS and then resuspended in 400 μl of RPMI medium 1640 or DMEM with DNA for transfection. Resuspended cells were transferred to a 0.4-cm cuvette and electroporated at 975 μF and 220 V (for 293T and DG75) or 250 V (for BCBL1) and incubated at 37°C and 5% CO2. Transfections were harvested after 24 or 48 h and assayed for activity.
Immunoprecipitation and Western Blotting.
The transfected cells were lysed in RIPA buffer [50 mM Tris (pH7.6), 150 mM NaCl, 2 mM EDTA, 1% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride, aprotinin (1 μg/ml), and pepstatin (1 μg/ml)] for 1 h on ice with brief vortexing every 15 min. A portion of the lysate was removed for use as the control. The lysates were precleared by 1-h incubation with control antibody conjugated to protein A beads. Specific antibodies were incubated with the lysates overnight at 4°C. Immunoprecipitates were collected by rotating with protein A and G-Sepharose beads for 1 h and washed four times in RIPA buffer. The protein was then heated in SDS/2-mercaptoethanol lysis buffer and analyzed by SDS/PAGE. Western blot analyses were done by using antibodies specific for the detection. For BC3 or BCBL1 cells, a total of 3 × 107 cells were used following the same protocol.
Preparation of GST Fusion Proteins and in Vitro Binding Assays.
BL21 cells were transformed with the plasmid constructs for each fusion protein, and single colonies were picked and grown overnight in 2 ml of LB medium. Five hundred milliliters of LB medium was inoculated and allowed to shake at 37°C until midexponential growth phase, induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), harvested and sonicated and the proteins solubilized in the presence of protease inhibitors (36). Solubilized proteins were incubated with GST-Sepharose beads overnight at 4°C with rotation and then collected by centrifugation and washed three times in NETN buffer and stored with protease inhibitors at 4°C. To determine binding of GST-Sel10, GST-F-box or GST-WD40 fusion to full-length and different truncations of LANA, proteins were translated in vitro by using the in vitro transcription and translation (TNT) system (Promega, Madison, WI). Labeled LANA protein was incubated with equivalent amounts of GST fusion proteins bound to beads and rotated at 4°C and washed four times in NETN with protease inhibitors. Bound proteins were eluted from the bead lysis buffer by heating at 95°C for 10 min and fractionated by 10% SDS/PAGE. Dried gels were analyzed with a PhosphorImager (Molecular Dynamics, Sunnyvale, CA), and signals were quantified with ImageQuant software.
Immunofluorescence.
Immunofluorescent assays were performed essentially as described (36, 47, 48). Slides were visualized with an Olympus XI70 inverted fluorescence microscope (Olympus, Melville, NY) and photographed by using a digital PixelFly camera and software (Cooke, Warren, MI).
Flow Cytometry and Proliferation Assay.
Flow cytometric analysis was conducted on a FACSCalibur cytometer (Becton Dickinson, San Jose, CA) and analyzed with FlowJo software (Tree Star, Ashland, OR). In CFSE labeling experiments, briefly, 5 million cells were washed and incubated with 2.5 μM CFSE (Molecular Probes, Eugene, OR) in PBS for 10 min in the dark at room temperature. Unbound CFSE was quenched by the addition of bovine growth serum (BGS). The labeled cells were washed twice with 5% BGS in PBS and replated at the concentration of 300,000 cells per milliliter in RPMI medium 1640 containing 10% BGS. After 48 or 72 h in culture, cells were collected, fixed, and applied to FACS analysis.
Supplementary Material
Acknowledgments
We thank Dr. Urban Lendahl for providing Sel10-GFP and Sel10-myc expression vectors. This work was supported by grants from the Leukemia and Lymphoma Society of America, Public Health Service Grants CA072510 and CA091792 from the National Cancer Institute, National Institute of Dental and Craniofacial Research Grants DE01436-04 and DE017338, and National Institute of Allergy and Infectious Diseases Grant A1067037 (to E.S.R.). K.L. is a Special Fellow and E.S.R. is a Scholar of the Leukemia and Lymphoma Society of America.
Abbreviations
- CFSE
carboxy-fluorescein diacetate succinimidyl ester
- ICN
intracellular activated Notch
- KSHV
Kaposi's sarcoma-associated herpesvirus
- LANA
latency-associated nuclear antigen.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.U.J. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0703508104/DC1.
References
- 1.Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 2.Russo JJ, Bohenzky RA, Chien MC, Chen J, Yan M, Maddalena D, Parry JP, Peruzzi D, Edelman IS, Chang Y, Moore PS. Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boshoff C, Weiss RA. Philos Trans R Soc London Ser B. 2001;356:517–534. doi: 10.1098/rstb.2000.0778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boshoff C, Chang Y. Annu Rev Med. 2001;52:453–470. doi: 10.1146/annurev.med.52.1.453. [DOI] [PubMed] [Google Scholar]
- 5.Brander C, Raje N, O'Connor PG, Davies F, Davis J, Chauhan D, Hideshima T, Martin J, Osmond D, Kedes DH, et al. Blood. 2002;100:698–700. doi: 10.1182/blood.v100.2.698. [DOI] [PubMed] [Google Scholar]
- 6.Brander C, Suscovich T, Lee Y, Nguyen PT, O'Connor P, Seebach J, Jones NG, van Gorder M, Walker BD, Scadden DT. J Immunol. 2000;165:2077–2083. doi: 10.4049/jimmunol.165.4.2077. [DOI] [PubMed] [Google Scholar]
- 7.Sarid R, Flore O, Bohenzky RA, Chang Y, Moore PS. J Virol. 1998;72:1005–1012. doi: 10.1128/jvi.72.2.1005-1012.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cotter MA, II, Subramanian C, Robertson ES. Virology. 2001;291:241–259. doi: 10.1006/viro.2001.1202. [DOI] [PubMed] [Google Scholar]
- 9.Ballestas ME, Kaye KM. J Virol. 2001;75:3250–3258. doi: 10.1128/JVI.75.7.3250-3258.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ballestas ME, Chatis PA, Kaye KM. Science. 1999;284:641–644. doi: 10.1126/science.284.5414.641. [DOI] [PubMed] [Google Scholar]
- 11.Friborg J, Jr, Kong W, Hottiger MO, Nabel GJ. Nature. 1999;402:889–894. doi: 10.1038/47266. [DOI] [PubMed] [Google Scholar]
- 12.Radkov SA, Kellam P, Boshoff C. Nat Med. 2000;6:1121–1127. doi: 10.1038/80459. [DOI] [PubMed] [Google Scholar]
- 13.Fujimuro M, Hayward SD. J Virol. 2003;77:8019–8030. doi: 10.1128/JVI.77.14.8019-8030.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujimuro M, Wu FY, ApRhys C, Kajumbula H, Young DB, Hayward GS, Hayward SD. Nat Med. 2003;9:300–306. doi: 10.1038/nm829. [DOI] [PubMed] [Google Scholar]
- 15.Knight JS, Cotter MA, II, Robertson ES. J Biol Chem. 2001;276:22971–22978. doi: 10.1074/jbc.M101890200. [DOI] [PubMed] [Google Scholar]
- 16.Si H, Robertson ES. J Virol. 2006;80:697–709. doi: 10.1128/JVI.80.2.697-709.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lan K, Choudhuri T, Murakami M, Kuppers DA, Robertson ES. J Virol. 2006;80:6411–6419. doi: 10.1128/JVI.00239-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lan K, Murakami M, Choudhuri T, Kuppers DA, Robertson ES. Virology. 2006;351:393–403. doi: 10.1016/j.virol.2006.03.047. [DOI] [PubMed] [Google Scholar]
- 19.Greenwald I. Genes Dev. 1998;12:1751–1762. doi: 10.1101/gad.12.12.1751. [DOI] [PubMed] [Google Scholar]
- 20.Artavanis-Tsakonas S, Rand MD, Lake RJ. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 21.Ellisen LW, Bird J, West DC, Soreng AL, Reynolds TC, Smith SD, Sklar J. Cell. 1991;66:649–661. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- 22.Jhappan C, Gallahan D, Stahle C, Chu E, Smith GH, Merlino G, Callahan R. Genes Dev. 1992;6:345–355. doi: 10.1101/gad.6.3.345. [DOI] [PubMed] [Google Scholar]
- 23.Girard L, Hanna Z, Beaulieu N, Hoemann CD, Simard C, Kozak CA, Jolicoeur P. Genes Dev. 1996;10:1930–1944. doi: 10.1101/gad.10.15.1930. [DOI] [PubMed] [Google Scholar]
- 24.Wu G, Lyapina S, Das I, Li J, Gurney M, Pauley A, Chui I, Deshaies RJ, Kitajewski J. Mol Cell Biol. 2001;21:7403–7415. doi: 10.1128/MCB.21.21.7403-7415.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oberg C, Li J, Pauley A, Wolf E, Gurney M, Lendahl U. J Biol Chem. 2001;276:35847–35853. doi: 10.1074/jbc.M103992200. [DOI] [PubMed] [Google Scholar]
- 26.Gupta-Rossi N, Le Bail O, Gonen H, Brou C, Logeat F, Six E, Ciechanover A, Israel A. J Biol Chem. 2001;276:34371–34378. doi: 10.1074/jbc.M101343200. [DOI] [PubMed] [Google Scholar]
- 27.Dourmishev LA, Dourmishev AL, Palmeri D, Schwartz RA, Lukac DM. Microbiol Mol Biol Rev. 2003;67:175–212. doi: 10.1128/MMBR.67.2.175-212.2003. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aster JC, Robertson ES, Hasserjian RP, Turner JR, Kieff E, Sklar J. J Biol Chem. 1997;272:11336–11343. doi: 10.1074/jbc.272.17.11336. [DOI] [PubMed] [Google Scholar]
- 29.Wells AD, Gudmundsdottir H, Turka LA. J Clin Invest. 1997;100:3173–3183. doi: 10.1172/JCI119873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng EH, Nicholas J, Bellows DS, Hayward GS, Guo HG, Reitz MS, Hardwick JM. Proc Natl Acad Sci USA. 1997;94:690–694. doi: 10.1073/pnas.94.2.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cesarman E, Mesri EA, Gershengorn MC. J Exp Med. 2000;191:417–422. doi: 10.1084/jem.191.3.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Renne R, Barry C, Dittmer D, Compitello N, Brown PO, Ganem D. J Virol. 2001;75:458–468. doi: 10.1128/JVI.75.1.458-468.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeong JH, Orvis J, Kim JW, McMurtrey CP, Renne R, Dittmer DP. J Biol Chem. 2004;279:16822–16831. doi: 10.1074/jbc.M312801200. [DOI] [PubMed] [Google Scholar]
- 34.Godfrey A, Anderson J, Papanastasiou A, Takeuchi Y, Boshoff C. Blood. 2005;105:2510–2518. doi: 10.1182/blood-2004-08-3052. [DOI] [PubMed] [Google Scholar]
- 35.Ye FC, Zhou FC, Yoo SM, Xie JP, Browning PJ, Gao SJ. J Virol. 2004;78:11121–11129. doi: 10.1128/JVI.78.20.11121-11129.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lan K, Kuppers DA, Verma SC, Robertson ES. J Virol. 2004;78:6585–6594. doi: 10.1128/JVI.78.12.6585-6594.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hubbard EJ, Wu G, Kitajewski J, Greenwald I. Genes Dev. 1997;11:3182–3193. doi: 10.1101/gad.11.23.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu G, Hubbard EJ, Kitajewski JK, Greenwald I. Proc Natl Acad Sci USA. 1998;95:15787–15791. doi: 10.1073/pnas.95.26.15787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moberg KH, Bell DW, Wahrer DC, Haber DA, Hariharan IK. Nature. 2001;413:311–316. doi: 10.1038/35095068. [DOI] [PubMed] [Google Scholar]
- 40.Strohmaier H, Spruck CH, Kaiser P, Won KA, Sangfelt O, Reed SI. Nature. 2001;413:316–322. doi: 10.1038/35095076. [DOI] [PubMed] [Google Scholar]
- 41.Koepp DM, Schaefer LK, Ye X, Keyomarsi K, Chu C, Harper JW, Elledge SJ. Science. 2001;294:173–177. doi: 10.1126/science.1065203. [DOI] [PubMed] [Google Scholar]
- 42.Nateri AS, Riera-Sans L, Da Costa C, Behrens A. Science. 2004;303:1374–1378. doi: 10.1126/science.1092880. [DOI] [PubMed] [Google Scholar]
- 43.Welcker M, Orian A, Jin J, Grim JE, Harper JW, Eisenman RN, Clurman BE. Proc Natl Acad Sci USA. 2004;101:9085–9090. doi: 10.1073/pnas.0402770101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rajagopalan H, Jallepalli PV, Rago C, Velculescu VE, Kinzler KW, Vogelstein B, Lengauer C. Nature. 2004;428:77–81. doi: 10.1038/nature02313. [DOI] [PubMed] [Google Scholar]
- 45.Spruck CH, Strohmaier H, Sangfelt O, Muller HM, Hubalek M, Muller-Holzner E, Marth C, Widschwendter M, Reed SI. Cancer Res. 2002;62:4535–4539. [PubMed] [Google Scholar]
- 46.Mao JH, Perez-Losada J, Wu D, Delrosario R, Tsunematsu R, Nakayama KI, Brown K, Bryson S, Balmain A. Nature. 2004;432:775–779. doi: 10.1038/nature03155. [DOI] [PubMed] [Google Scholar]
- 47.Lan K, Kuppers DA, Verma SC, Sharma N, Murakami M, Robertson ES. J Virol. 2005;79:7453–7465. doi: 10.1128/JVI.79.12.7453-7465.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lan K, Kuppers DA, Robertson ES. J Virol. 2005;79:3468–3478. doi: 10.1128/JVI.79.6.3468-3478.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Knight JS, Sharma N, Robertson ES. Proc Natl Acad Sci USA. 2005;102:18562–18566. doi: 10.1073/pnas.0503886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Knight JS, Sharma N, Robertson ES. Mol Cell Biol. 2005;25:1749–1763. doi: 10.1128/MCB.25.5.1749-1763.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.