Abstract
Transmembrane signaling events that propagate through receptors and transporters, play critical roles in cellular function and regulation. In the Escherichia coli vitamin B12 transporter, BtuB, substrate binding to the extracelluar surface of the protein triggers the unfolding of an energy coupling motif at the periplasmic surface. Here, the molecular interactions mediating this substrate-dependent transmembrane signaling event were investigated in a novel way by combining a two mutant cycle analysis with site-directed spin labeling (SDSL). SDSL was used to monitor the unfolding and conformational equilibrium of the energy-coupling motif, and a thermodynamic two-mutant cycle analysis was used to estimate pair-wise interaction free energies for a pair of charged residues (D316 and R14) within the protein interior. The data indicate that D316 and R14 are critical to this structural transition. Substrate binding is shown to reduce the interaction free energy between these residues, thereby triggering the unfolding of the energy coupling motif of this membrane transporter. The result indicates that SDSL when used in combination with a mutant cycle analysis provides an approach to examine the molecular interactions mediating signaling events in membrane proteins.
Keywords: Membrane proteins, spin labeling, EPR spectroscopy, ion pair, mutant cycle analysis
The uptake of rare nutrients by Gram negative bacteria is facilitated by a series of high-affinity outer-membrane transporters that derive energy for transport by coupling to the inner-membrane protein TonB.1 The N-terminal end of these TonB-dependent transporters contains a highly conserved energy-coupling segment termed the Ton box, which is localized at the periplasmic side of the transporter. In BtuB, substrate binding on the extracellular surface of the protein unfolds the Ton box,2 producing a disordered protein segment that includes the first 15 residues on the N-terminus.3 This order-disorder transition results in the extension of the Ton box 20 to 30 Angstroms into the periplasmic space (Fig. 1a) where it may function as a recognition signal to promote binding of the transporter to TonB.4
Figure 1.
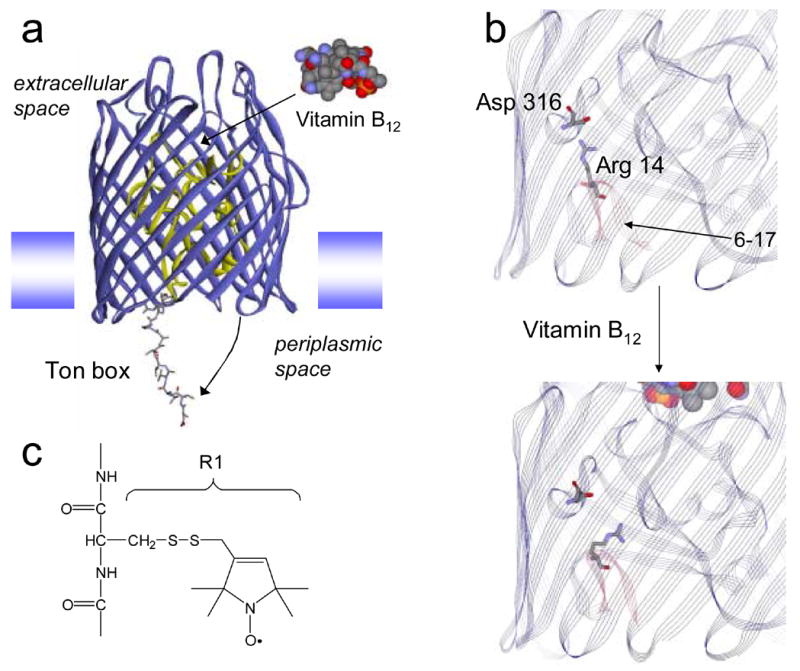
Molecular models of BtuB. (a) Upon the binding of substrate (vitamin B12) the energy coupling segment, or Ton box, of BtuB undergoes an unfolding so that it projects into the periplasm. The model shown for BtuB is based upon x-ray crystallography, except that the structure has been modified to show an unfolded state of the Ton box that is consistent with EPR derived constraints.4 The N-terminal or hatch region of BtuB is shown in yellow and residues 6–17 are shown in a stick representation. A weak ion pair is formed between Arg14 on the C-terminal side of the Ton box and Asp 316 in the β-barrel. (b) a portion of the crystal structure of BtuB in its apo form showing Arg 14 and Asp 316 (PDB ID: 1NQE). In this structure, the two side-chains are separated by approximately 4.2 Angstroms. This ion pair is slightly closer in a structure obtained from meso phase crystallization (3.5 Angstroms) (PDB ID: 2GUF).6 Upon vitamin B12 addition, the ligand bound crystal structure (PDB ID: 1NQH) indicates that the separation increases to approximately 7.5 Angstroms. Residues 6–17 are highlighted in red, the Ton box includes residues 6–12. The Ton box is seen in its folded form in the substrate bound structure, a configuration that results from the high solute concentrations that are present in the crystallization buffer.7 (c) A native residue is replaced by the nitroxide side-chain R1 by derivatizing a single cysteine mutation with the sulfhydryl specific reagent 1-oxyl-2,2,5,5-tetramethyl-3-pyrroline-3-methyl methanethiosulfonate.
At the present time, the molecular interactions leading to this transmembrane signaling event are not understood; however, inspection of the high-resolution models of BtuB,5; 6 suggests that an ion pair within the protein interior might be involved in this structural change. The positively charged side chain of Arg 14 on the C-terminal side of the Ton box is in close proximity to the side chain of Asp 316 on the inner surface of the BtuB barrel (see Fig. 1b). This suggests that these side-chains interact electrostatically, and that a favorable interaction between these residues might prevent the Ton box from unfolding. Addition of substrate increases the distance between this charge pair from 4.3 to 7.5 Angstroms, which may weaken and subsequently break the interaction. A loss of this interaction might then lead to the unfolding event seen in Fig. 1a. This conformational transition is seen by EPR spectroscopy and by chemical derivatization, but is not revealed by crystallography due to the solute conditions used to produce protein crystals.7
Combining a two mutant cycle analysis with SDSL
Ion pairs have been observed to act as conformational switches in membrane proteins,8 and in the present work, the role of the R14-D316 pair in the Ton box unfolding was investigated in a novel way by combining a two mutant cycle analysis 9; 10; 11 with site-directed spin labeling (SDSL).12; 13 SDSL was used to quantitate the populations of folded and unfolded forms of the Ton box, and thereby measure changes in the Ton box equilibrium and free energy differences between the protein in different states with different mutants. X-band EPR spectra undergo motional averaging on the ns time-scale, which is fast compared to the rate of conformational exchange between the two Ton box forms. As a result, the populations of folded and unfolded Ton box yield distinct components in the EPR spectrum and they may be estimated through spectral deconvolution.14
Four mutant variants of BtuB were prepared, expressed, purified and reconstituted into palmitoyloleoylphosphatidylcholine (POPC) bilayers. Each mutant had a single cysteine incorporated into position 10 of the Ton box, to which the spin-label side chain R1 was derivatized (Fig 1c). The EPR spectrum of R1 is highly sensitive to steric contact of the spin labeled side-chain as well as the dynamics of the protein backbone. In the folded conformation of the Ton box, the spectrum of V10R1 is broad, indicating that it is in tertiary contact within the protein interior. The spectrum of V10R1 in the unfolded form of the Ton box is narrow due to motional averaging of the nitroxide magnetic interactions. Because the lineshapes in the two states are dramatically different, the fractions of Ton box in the folded and unfolded configurations may be easily estimated directly from the EPR spectrum of V10R1. Shown in Fig. 2 are four pairs of EPR spectra of the V10R1 label without (Fig. 2a) and with (Fig. 2b) substrate for the wild-type R14-D316 side chains and 3 mutants where the R14 and D316 side chains are separately and then simultaneously replaced with alanine.
Figure 2.
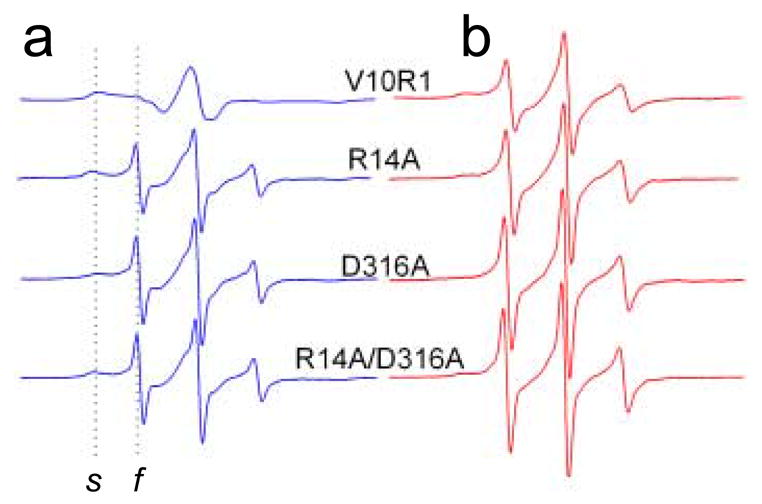
Normalized X-band EPR spectra from the BtuB Ton box. (a) EPR spectra of V10R1 in the wild-type protein, and V10R1 in the presence of R14A, D316A, and R14A/D316A. The dashed lines indicate the spectral components resulting from a population of spin label (V10R1) that is static or slowly diffusing on the ns time-scale (s) and label that is fast or highly mobile (f). (b) Spectra shown in a, except in the presence of the BtuB substrate, vitamin B12. EPR spectra have a sweep width of 100 Gauss and were recorded for samples in quartz capillaries using a loop-gap resonator (Medical Advances, Milwaukee, WI) with an incident microwave power of 2.0 mW and a modulation of 1 Gauss peak-to-peak. All measurements were carried out at room temperature (295K). Point mutations were introduced into BtuB by two-step PCR-based site-directed mutagenesis as described previously,18 and the resulting clones were sequenced for integrity using dye terminator sequencing (GeneWiz, Inc.; New Brunswick, NJ). The BtuB mutants were expressed in minimal media using E. coli strain RK5016 (metE). This strain is a derivative of strain MC4100 [Δ(argF-lac)U169 araD139 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR22 non-9 gyrA219] with additional mutations (metE70 argH btuB recA), so that vitamin B12 is required for methionine biosynthesis. Outer membranes were isolated as described previously.19, and then solubilized by octyl glucoside, spin-labeled, and purified by ion exchange chromatography.3. Purified BtuB was reconstituted into POPC vesicles as described elsewhere.20
Estimating the energy of interaction between D316 and R14
Each of the EPR spectra in Fig. 2 is a composite of a highly immobile population of spins corresponding to the folded state of the Ton box, and a mobile population corresponding to the unfolded state (spectral features resulting from these two components are labeled s and f, respectively). For the V10R1 labeled BtuB containing the wild-type R14 and D316 side chains, the Ton box is almost entirely in the folded form, yielding a broad EPR spectrum having a low amplitude. Addition of substrate results in an increase in the population of mobile spins, seen as an increase in the amplitude of the narrow component in the spectrum. This change is a result of the Ton box unfolding transition, which results in the projection of the Ton box into the periplasm.4 When either the Arg 14 or Asp 316 side chains are switched to alanine, the mobile component in the V10R1 spectrum is dramatically increased, even in the absence of substrate, indicating that these mutations unfold the Ton box. The spectra from the alanine mutants in the absence of substrate, qualitatively resemble the spectra seen in the presence of substrate, although the fractions of mobile and immobile components vary with each mutant.
From the mobile and immobile components in these EPR spectra, the relative populations of folded (ordered) and unfolded (disordered) Ton box were estimated by spectral subtraction. This was accomplished, as described previously, by simulating the isotropic highly mobile (or fast) spectral component and subtracting it from the composite spectrum until an immobile lineshape remained.14 Double intergration of the first derivative EPR spectrum gives a value proportional to spin number, and was used to quantitate the conformational equilibrium (or ratio) of folded to unfolded Ton box. The EPR lineshapes corresponding to the two Ton box states are dramatically different at position 10, making an estimate of the two populations relatively simple. The uncertainty in this ratio was approximately +/− 10% or better.
The conformational equilibria were used to estimate the free energy differences between folded and unfolded Ton box forms for each mutant with and without substrate. A two mutant cycle analysis is shown in Fig. 3 along with the change in these free energy differences (which are labeled as Δ°G) between each mutant. As expected, the sum of the energy changes going around the cycle are roughly zero (+/− 0.1 kcal/mole). The fact that Δ°G2 ≠ Δ°G4 and Δ°G1 ≠ Δ°G3 indicates that there is an energy of interaction between these residues.11 If is assumed that R14 and D316 do not interact in the unfolded Ton box state, the energy of interaction between these residues in the folded state may be estimated from the difference between either Δ°G2 and Δ°G4 or Δ°G1 and Δ°G3. In each case, this difference yields a value of about −1.8 kcal/mole (Fig. 3). In the presence of substrate, the energy of interaction between D316 and R14 drops to approximately −0.1 kcal/mole, indicating that the interaction has been largely eliminated. These energies represent the contributions made by these two residues to the Ton box equilibrium either without or with substrate, and the difference in these energies (1.7 kcal/mole) represents the shift in interaction energy that is produced by substrate binding.
Figure 3.
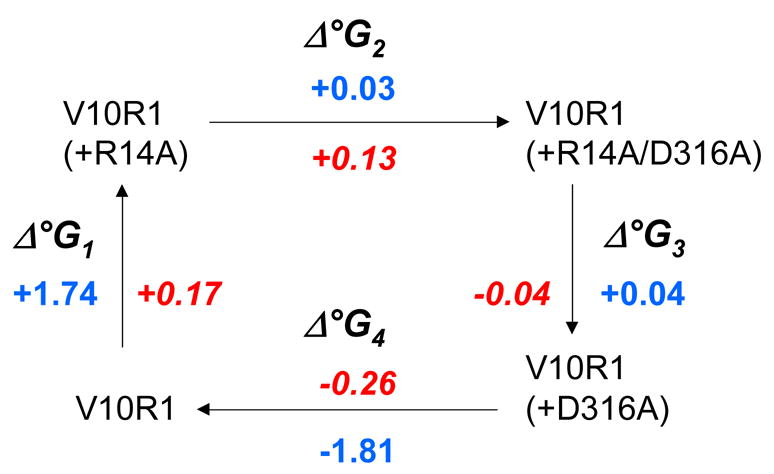
Δ°G changes and mutant cycle analysis for the BtuB Ton box. The values of Δ°G are actually ΔΔ°G changes in the Ton box unfolding equilibrium between the two indicated BtuB mutants. Blue numbers represent free energy changes in kcal/mole for the substrate free BtuB, red numbers indicate the free energy changes found in the presence of substrate. The free energies were estimated from EPR spectra of V10R1. These spectra result from an R1 label that has two motional modes where the narrower spectral component corresponds to the unfolded (undocked) form of the Ton box.14 Spectral subtraction was used to deconvolute the spectrum and determine the populations of docked and undocked Ton box. This resulted in an error in the estimate of the free energy differences of approximately 50 to 100 cal/mole.
The two mutant cycle analysis does not indicate how these residues interact, or how many intervening residues may be involved. However, the proximity between these residues seen in high-resolution models of BtuB suggests that there is a direct ionic interaction between R14 and D316. The energy of interaction estimated from Figure 3, is approximately the electrostatic energy expected from Coulomb’s law for two charges separated by 4 Angstroms in an environment of dielectric constant of 40. While the effective value of the dielectric constant within a protein can be highly variable,15 this high value is not unreasonable given the localization of the Ton box at the periplasmic surface and the presence of water within the protein core.5 In any case, this estimate indicates that a significant electrostatic interaction is likely between these residues.
In the high-resolution models of BtuB, the increase in distance between R14 and D316 that occurs upon substrate binding is due to a movement of portions of the N-terminal (hatch) region relative to the BtuB barrel. In the membrane bilayer, the actual distance change between these two residues upon substrate binding is much larger than that indicated in the crystal structure,4 due to differences between the crystal and membrane associated forms of BtuB.7 In contrast to substrate binding, the binding of colicin E3 is found to stabilize the folded form of the Ton box.14; 16 Interestingly, colicin binding results in a shortening of the distance between R14 and D316, and apparently strengthens this interaction.17
This work demonstrates that an ion pair within the interior of BtuB functions as a switch to mediate a transmembrane signaling event, and to regulate the configuration of the Ton box. It also illustrates how site-directed spin labeling may be used in combination with a mutant cycle analysis to investigate the molecular mechanisms regulating structural changes and transmembrane signaling in membrane proteins.
Acknowledgments
LabView software used in the EPR data analysis was provided by Drs. Wayne Hubbell, and Christian Altenbach. This work was supported by a grant from the National Institutes of Health, NIGMS, GM035215.
Abbreviations
- EPR
electron paramagnetic resonance spectroscopy
- POPC
palmitoyloleoylphos-phatidylcholine
- R1
spin-labeled side chain produced by derivatization of a cysteine with a methanethiosulfonate spin label
- SDSL
site-directed spin labeling
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Postle K, Kadner R. Touch and go: tying TonB to transport. Mol Microbiol. 2003;49:869–882. doi: 10.1046/j.1365-2958.2003.03629.x. [DOI] [PubMed] [Google Scholar]
- 2.Merianos HJ, Cadieux N, Lin CH, Kadner R, Cafiso DS. Substrate-induced exposure of an energy-coupling motif of a membrane transporter. Nat Struct Biol. 2000;7:205–209. doi: 10.1038/73309. [DOI] [PubMed] [Google Scholar]
- 3.Fanucci GE, Coggshall KA, Cadieux N, Kim M, Kadner RJ, Cafiso DS. Substrate-Induced conformational changes of the perplasmic N-terminus of an outer-membrane transporter by site-directed spin labeling. Biochemistry. 2003;42:1391–1400. doi: 10.1021/bi027120z. [DOI] [PubMed] [Google Scholar]
- 4.Xu Q, Ellena JF, Kim M, Cafiso DS. Substrate-dependent unfolding of the energy coupling motif of a membrane transport protein determined by double electron-electron resonance. Biochemistry. 2006;45:10847–10854. doi: 10.1021/bi061051x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chimento DP, Mohanty AK, Kadner RJ, Wiener MC. Substrate-induced transmembrane signaling in the cobalamin transporter BtuB. Nat Struct Biol. 2003;10:394–401. doi: 10.1038/nsb914. [DOI] [PubMed] [Google Scholar]
- 6.Cherezov V, Yamashita E, Liu W, Zhalnina M, Cramer WA, Caffrey M. In meso structure of the cobalamin transporter, BtuB, at 1.95 A resolution. J Mol Biol. 2006;364:716–34. doi: 10.1016/j.jmb.2006.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fanucci GE, Lee JY, Cafiso DS. Spectroscopic evidence that osmolytes used in crystallization buffers Inhibit a conformation change in a membrane protein. Biochemistry. 2003;42:13106–13112. doi: 10.1021/bi035439t. [DOI] [PubMed] [Google Scholar]
- 8.Rao VR, Oprian DD. Activating mutations of rhodopsin and other G protein-coupled receptors. Annu Rev Biophys Biomol Struct. 1996;25:287–314. doi: 10.1146/annurev.bb.25.060196.001443. [DOI] [PubMed] [Google Scholar]
- 9.Carter PJ, Winter G, Wilkinson AJ, Fersht AR. The use of double mutants to detect structural changes in the active site of the tyrosyl-tRNA synthetase (Bacillus stearothermophilus) Cell. 1984;38:835–40. doi: 10.1016/0092-8674(84)90278-2. [DOI] [PubMed] [Google Scholar]
- 10.Ackers GK, Smith FR. Effects of site-specific amino acid modification on protein interactions and biological function. Annu Rev Biochem. 1985;54:597–629. doi: 10.1146/annurev.bi.54.070185.003121. [DOI] [PubMed] [Google Scholar]
- 11.Horovitz A, Fersht AR. Strategy for analysing the co-operativity of intramolecular interactions in peptides and proteins. J Mol Biol. 1990;214:613–7. doi: 10.1016/0022-2836(90)90275-Q. [DOI] [PubMed] [Google Scholar]
- 12.Hubbell WL, Gross A, Langen R, Lietzow MA. Recent advances in site-directed spin labeling of proteins. Curr Op Struct Biol. 1998;8:649–656. doi: 10.1016/s0959-440x(98)80158-9. [DOI] [PubMed] [Google Scholar]
- 13.Fanucci GE, Cafiso DS. Recent advances and applications of site-directed spin labeling. Curr Opin Struct Biol. 2006;16:644–653. doi: 10.1016/j.sbi.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 14.Fanucci GE, Cadieux N, Kadner R, Cafiso DS. Competing ligands stabilize alternate conformations of the energy coupling motif of a TonB-dependent outer membrane transporter. Proc Natl Acad Sci USA. 2003;100:11382–11387. doi: 10.1073/pnas.1932486100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Warshel A, Sharma PK, Kato M, Parson WW. Modeling electrostatic effects in proteins. Biochim Biophys Acta. 2006;1764:1647–76. doi: 10.1016/j.bbapap.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 16.Cadieux N, Phan PG, Cafiso DS, Kadner RJ. Differential substrate-induced signaling through the TonB-dependent transporter BtuB. Proc Natl Acad Sci USA. 2003;100:10688–10693. doi: 10.1073/pnas.1932538100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurisu G, Zakharov SD, Zhalnina MV, Bano S, Eroukova VY, Rokitskaya TI, Antonenko YN, Wiener MC, Cramer WA. The structure of BtuB with bound colicin E3 R-domain implies a translocon. Nat Struct Biol. 2003;10:948–54. doi: 10.1038/nsb997. [DOI] [PubMed] [Google Scholar]
- 18.Cadieux N, Kadner RJ. Site-directed disulfide bonding reveals an interaction site between energy coupling protein TonB and BtuB, the outer membrane cobalamin transporter. Proc Natl Acad Sci USA. 1999;96:10673–10678. doi: 10.1073/pnas.96.19.10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coggshall KA, Cadieux N, Piedmont C, Kadner R, Cafiso DS. Transport-defective mutations alter the conformation of the energy-coupling motif of an outer membrane transporter. Biochemistry. 2001;40:13946–13971. doi: 10.1021/bi015602p. [DOI] [PubMed] [Google Scholar]
- 20.Fanucci GE, Cadieux N, Piedmont CA, Kadner RJ, Cafiso DS. Structure and dynamics of the β-barrel of the membrane transporter BtuB by site-directed spin labeling. Biochemistry. 2002;41:11543–11551. doi: 10.1021/bi0259397. [DOI] [PubMed] [Google Scholar]


