Abstract
A new type of high avidity binding molecule, termed “peptabody” was created by harnessing the effect of multivalent interaction. A short peptide ligand was fused via a semi-rigid hinge region with the coiled-coil assembly domain of the cartilage oligomeric matrix protein, resulting in a pentameric multivalent binding molecule. In the first peptabody (Pab-S) described here, a peptide (S) specific for the mouse B-cell lymphoma BCL1 surface Ig idiotype, was selected from a phage display library. A fusion gene was constructed encoding peptide S, followed by the 24 aa hinge region from camel IgG and a modified 55 aa cartilage oligomeric matrix protein pentamerization domain. The Pab-S fusion protein was expressed in Escherichia coli in a soluble form at high levels and purified in a single step by metal-affinity chromatography. Pab-S specifically bound the BCL1 surface idiotype with an avidity of about 1 nM, which corresponds to a 2 × 105-fold increase compared with the affinity of the synthetic peptide S itself. Biochemical characterization showed that Pab-S is a stable homopentamer of about 85 kDa, with interchain disulfide bonds. Pab-S can be dissociated under denaturing and reducing conditions and reassociated as a pentamer with full-binding activity. This intrinsic feature provides an easy way to combine Pab molecules with two different peptide specificities, thus producing heteropentamers with bispecific and/or chelating properties.
Keywords: peptide ligand, multivalent binding, idiotype specific, phage display
In recent years, the understanding of molecular interactions in the realm of biomolecules, such as proteins and nucleic acids, greatly benefited from the isolation of artificial polypeptide “ligands” with de novo binding activities for various “receptors.” A powerful means of developing artificial ligands is offered by the screening of large phage libraries, displaying billions of different polypeptide sequences fused with coat proteins on the surface of filamentous bacteriophage (1, 2). For example, isolation of new peptide ligands allowed the mapping of antibody binding sites, the characterization of important residues in HLA-DR molecules, and the identification of protease substrates or inhibitors (for review see ref. 3). However, apart from some exceptions (4, 5), only low-affinity (micromolar range) ligands have been isolated from peptide libraries (6–8). This can be readily explained by the high degree of conformational freedom and small number of contact residues within a short peptide molecule.
Interestingly, nature provides us with numerous examples of molecules with low-affinity binding sites, yet capable of high avidity interactions with their targets due to multivalent binding. For instance, the low affinity of IgM produced during the primary immune response is compensated by its pentameric structure resulting in a high avidity toward repetitive antigenic determinants present on the surface of bacteria or viruses (9). Similarly, the complement factor C1q binds with low affinity (100 μM) to individual IgG molecules present in serum, whereas when the same IgG are clustered in immune complexes the avidity of C1q is drastically increased (to about 1 μM and 3 nM for IgG dimers and tetramers, respectively) leading to activation of the complement cascade (10).
We have brought together the advantage of sequence diversity, provided by phage-displayed random peptide libraries, and the benefits of multivalency, provided by the cartilage oligomeric matrix protein (COMP) assembly domain (11, 12), to create a new type of binding molecule, which we termed “peptabody.” In this newly designed recombinant molecule, a short peptide ligand is fused via a semi-rigid hinge at the N terminus of the COMP pentamerization domain. Here we describe the first peptabody (Pab-S), specific for the surface Ig idiotype of the BCL1 mouse lymphoma (13). In vitro studies of Pab-S revealed several unique features of the Pab molecule, suggesting a spectrum of potential scientific and industrial applications.
MATERIALS AND METHODS
Bacterial Strains. E.
coli TG1 (14) was used for propagation of plasmids and phage and E. coli SG13009 (Qiagen, Chatsworth, CA) was used for production of fusion proteins.
Cells and Antibodies.
The BALB/c-derived B cell lymphoma BCL1 (13) and the mouse hybridoma B1, secreting an anti-idiotype mAb B1 of IgG1 isotype (15) were kindly provided by Kris Thielemans (Medical School, VUB, Brussels). BCL1 cells were propagated in BALB/c mice by i.p. injection of 106 cells. The BCL1 soluble IgM idiotype was purified from the serum of a mouse with large BCL1 tumors. B1 IgG was purified by protein G-Sepharose (Pharmacia). Fab′ fragments were obtained by limited digestion with pepsin followed by reduction and alkylation, as described (16).
Peptide Selection.
Two filamentous bacteriophage libraries of about 107 independent members displaying random hexapeptides, called Smith (6) and Doorbar (7), as well as a combinatorial library of about 1012 independent members displaying a tandem of random decapeptides, called Fisch (5), were used.
The screening of the phage display libraries was performed essentially as described (5). Specific inhibition of phage binding to BCL1 IgM was performed by addition of mAb B1 at 100 μg/ml. The DNA fragments encoding selected peptides were amplified by PCR and sequenced as described (5).
Peptide Synthesis.
Peptides were synthesized using standard fluorenylmethoxycarbonyl solid phase chemistry on a peptide synthesizer (Applied Biosystems). For competition studies, peptides were dissolved in PBS (pH 7.4), and peptide concentrations were determined by the method of Waddell based on absorption difference at 215 and 225 nm (17).
Plasmid Construction.
The plasmid p3bCOMP encoding a 64 aa COMP assembly domain (11) was a kind gift of V. Efimov from J. Engel’s laboratory (Biozentrum, Basel). All further constructs were made using standard methods of DNA manipulation (18). The DNA fragment encoding 55 aa of the COMP domain (residues 26–80, ref. 11) was amplified by PCR from the p3bCOMP template using COMP-specific primers. The resulting PCR product contained the 2 aa substitutions Lys-29 → Cys and Ala-30 → Cys, as well as XhoI and SpeI restriction sites at the 5′ and 3′ ends, respectively.
The DNA duplexes encoding the peptides S, F, and D, were prepared by oligonucleotide annealing. All three duplexes contained BamHI and XhoI cohesive ends on the 5′ and 3′ ends, respectively. By means of three-part ligation, the duplexes encoding peptides and the PCR-amplified COMP domain, restricted with XhoI and SpeI enzymes, were joined together in the modified pDS78 vector (19), linearized with BamHI and SpeI enzymes, in front of a six-histidine tail present in the vector. This generated pSC6H, pDC6H, and pFC6H plasmids, which encode S, D, and F peptides, respectively.
A DNA fragment encoding the 24 aa hinge region derived from camel IgG [(PQ)2PK(PQ)4PKPQPK(PE)2], called PX, was prepared by annealing of the oligonucleotides encoding the plus and minus strands of the duplex and containing XhoI and SalI cohesive ends on the 5′ and 3′ ends, respectively. The PX duplex was ligated into linearized with XhoI enzyme and dephosphorylated pSC6H, pDC6H, and pFC6H plasmids, to generate the expression plasmids pSPXC6H, pDPXC6H, and pFPXC6H, respectively. The final constructs were verified by dideoxynucleotide sequencing (20) using Sequenase 2.0 (United States Biochemical).
Expression and Purification of Pabs.
Pab fusion proteins were expressed in E. coli SG13009. The cultures were grown in a shaker at 37°C to OD600; ≈0.5, then 1 mM isopropyl β-d-thiogalactoside was added to induce protein synthesis, followed by a further 4-h incubation at 30°C. Bacteria were harvested by centrifugation (8000 × g for 15 min at 4°C) and frozen at −70°C. Bacterial pellets were resuspended in PBS (pH 7.4), 1 mM EDTA/1 mg/ml lysozyme, incubated for 30 min at room temperature, and subjected to three rounds of freezing/thawing (liquid nitrogen/37°C). The lysates were incubated for 15 min at 25°C with 0.1 mg/ml of DNase I and the supernatants collected, after centrifugation (23,000 × g for 15 min at 4°C). Imidazol was added to a final concentration of 5 mM and the recombinant protein was absorbed on 2 ml of Ni-NTA resin (Qiagen), equilibrated in 5 mM imidazol/PBS (pH 7.4). After extensive washing with PBS containing 5 mM and 20 mM imidazol retained proteins were eluted with PBS containing 250 mM imidazol. Imidazol was removed by dialysis against PBS (pH 7.4)/1 mM EDTA; proteins were concentrated 5-fold and stored at −20°C.
Labeling.
Typically, Pab-S (0.2 nmol), mAb B1 IgG ( 0.5 nmol), or B1 Fab′ fragments (1 nmol) were labeled in PBS with 100 μCi (1 Ci = 37 GBq) of 125I in Iodo-Gen (Bio-Rad, 10 μg) coated tubes for 20 min (2 h for Pab-S) at 4°C. Uncoupled iodine was removed by gel filtration on a PD-10 column (Pharmacia). About 40% of the radioactivity for Pab-S and 70% for B1 IgG and B1 Fab′ was recovered. The specific activity ranged from 70 to 200 μCi/nmol.
Cell Binding.
To determine maximal immunoreactivity and nonimmunoreactive fraction (NIF), 125I-labeled Pab-S, mAb B1 IgG, or Fab′ (20 nCi) were incubated with serial dilutions of freshly harvested BCL1 cells (0.3–100 × 106 cells per ml). At the binding plateau (reached with 25 × 106 cells per ml) the maximum immunoreactivity from a representative experiment was 52 ± 5.4%, 66.1 ± 0.7%, and 81.3 ± 1.2% for Pab-S, mAb B1 IgG, or Fab′, respectively. NIF was obtained by subtracting the maximum immunoreactivity from 100%. For competition assays, the labeled compounds were incubated with serial dilutions of unlabeled competitors and 3 × 106 BCL1 cells per ml. Nonspecific binding (NSB) was determined with an irrelevant mAb IgG labeled with 125I, or by measuring the binding of 125I-labeled Pab-S, mAb B1 IgG, or Fab′ in excess of competitor. NSB was always below 1%. Comparative experiments were performed in triplicate, the same day with the same batch of cells, in V-shaped 96-well plates (final volume 300 μl of PBS supplemented with 1 mg/ml of BSA at 4°C under agitation for 2.5 h). After centrifugation, the amounts of both free and bound 125I were measured as above from an aliquot of the supernatant and of the pellet washed once with ice-cold PBS.
Equilibrium Binding Constants.
Cell dilution experiments and homogenous competition curves (i.e., competition for cell binding between the labeled and unlabeled forms of the same ligand) were analyzed by a linear equilibrium model. The free (F) and bound (B) radioactivity were corrected by subtracting the NIF and NSB, respectively, and expressed in moles per liter, taking into account the specific activity of each isotopic dilution. The parameters Kd (dissociation equilibrium constant) and R (molarity of binding sites) were fitted to the experimental data by regression of the corrected Scatchard equation (B − NSB)/(F − NIF) = R/Kd − (B − NSB)/Kd, using excell solver (Microsoft).
Biochemical Characterization of Pab-S.
The concentration of purified Pab-S was determined by the method of Waddell (17), as well as by Bradford protein assay (21) (Bio-Rad). As previously shown (11), the COMP assembly domain can be completely reduced under native conditions without denaturation. Thus, a completely reduced form of Pab-S was obtained by incubation with 100 mM DTT at 37°C for 30 min (as in ref. 11), followed by extensive dialysis against PBS (1 mM EDTA/1 mM 2-mercaptoethanol). Pab-S was dissociated by boiling for 20 min in 4 M urea in the presence of 100 mM DTT and renatured by extensive dialysis against PBS (1 mM EDTA). Circular dichroism (CD) spectra between 180 and 250 nm were recorded on an Aviv 62 DS CD spectrometer at a protein concentration of 40 μg/ml in water at 25°C. Gel filtration was performed using fast protein liquid chromatography (FPLC) (Pharmacia) on a Superdex G-200 column, equilibrated in PBS (± 1 mM EDTA/± 1 mM 2-mercaptoethanol). The elution was monitored at 280 nm. The proteins were analyzed by 10–15% gradient SDS/PAGE (Pharmacia) under nonreducing conditions and stained with Coomassie blue R250.
Intracellular Ca2+ Release and Protein Tyrosine Phosphorylation.
To measure Ca2+ release, BCL1 cells were washed once with buffer P2 (PBS supplemented with 2% FCS) and incubated at 2 × 106 cells per ml in P2 buffer supplemented with 2 μM Indo-1-AM (Sigma) for 1 h at 37°C. Cells were washed three times with P2 buffer and kept at 37°C until addition of activators (control ionophore A23187, Pab-S, mAb B1 IgG, or control Pab-F). The emission ratio (405/530 nm versus time) was analyzed by FACS (Becton Dickinson) and the maximal values were plotted against concentration for each activator. For the phosphotyrosine assay, 2 × 105 BCL1 cells were incubated with 100 nM of Pab-S, mAb B1 IgG, or control Pab-F, for 1 min and the cell lysates were electrotransferred from a reducing SDS/10% PAGE to nitrocellulose and probed with the monoclonal anti-phosphotyrosine antibody 4G10 (Upstate Biotechnology, Lake Placid, NY). The same membrane was striped and stained with a goat anti-mouse μ chain peroxidase conjugate (Sigma).
Molecular Modeling.
The Pab-S molecule was modeled using the computer graphics program tourbo-frodo (22). The structure was further refined by using x-plor, Version 3.1 (23) using the following procedure: a 5-ps molecular dynamics simulation at 300 K followed by 1000 steps of conjugate gradient minimization, carried out with a 1/r dependent dielectric constant.
RESULTS
Selection of Peptides Specific for the BCL1 Surface Ig Idiotype.
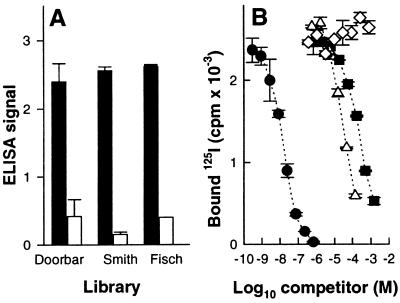
Peptide ligands specific for the mouse B-cell lymphoma BCL1 idiotype were selected from three different peptide libraries displayed on phage, namely Smith (6), Doorbar (7), and Fisch (5). Specific phages were selected on purified BCL1 IgM, individual phage clones were isolated, and DNA fragments encoding peptides were amplified by PCR and sequenced (Table 1). The idiotype specificity of selected peptides was demonstrated by inhibition of phage binding with the anti-idiotype antibody, mAb B1 (Fig. 1A). Synthetic peptides corresponding to the selected sequences were prepared as outlined in Table 1.
Table 1.
Amino acid sequences of selected peptide ligands specific for BCL1 Ig idiotype
Names of the phage-displayed peptide libraries.
Peptides selected from random sequences are in bold-face type, framework residues included in the synthesis are in normal type.
An unusual N to Y substitution in the constant part of Doorbar library was observed.
Figure 1.
Specificity of selected phage and synthetic peptides for the BCL1 idiotype. (A) Binding of selected phage clones to BCL1 IgM coated on plastic in the absence (solid bars) or presence (open bars) of 100 μg/ml of anti-Id B1 IgG (mean of triplicates ± SD). (B) The binding of 125I-labeled B1 IgG to BCL1 IgM coated on plastic (3 μg/ml in PBS) was competed with increasing amounts of B1 IgG (•), peptide S (▪), peptide D (Δ), and peptide F (⋄). Data shown are means of duplicates ± SD.
Interestingly, two distal cysteine residues, potentially capable of forming a disulfide bonded loop, were found in the hexapeptide, called peptide S, selected from the Smith library. Oxidation of peptide S resulted in the loss of two hydrogen atoms, as shown by mass spectroscopy, which is consistent with cyclization of the peptide. All synthetic peptides were tested for inhibition of 125I-labeled mAb B1 binding to BCL1 IgM coated on plates (Fig. 1B). The D and S peptides gave 50% inhibition (IC50) at about 60 μM and 200 μM, respectively. No inhibition was seen with peptide F up to 2 mM, which was used, thereafter, as a negative control. Substitution of cysteine residues by serines in peptide S resulted in a 10-fold increase of IC50, indicating that the disulfide bonded turn conformation of the peptide is favorable for binding to BCL1 IgM idiotype.
Pab: Molecular Design and Gene Construct.
The Pab fusion protein consisted of four distinct parts: (i) A selected peptide ligand represented the N-terminal binding domain. (ii) A 24 aa sequence derived from a long camel Ig hinge region (24) provided the space necessary for multivalent binding. (iii) The 55 aa pentamerization domain, a modification of the coiled-coil COMP assembly domain (11), formed a five-stranded α-helical bundle. The modification concerns the introduction of additional disulfide bonds. The wild-type COMP is known to have interchain disulfide bonds at the C terminus of the assembly domain (11). Based on the structural model of COMP assembly domain (25), two substitutions, Lys-29 → Cys and Ala-30 → Cys, were introduced to allow the formation of additional interchain disulfide bonds near the N-terminal portion of the assembly domain, where the hinge and peptide sequence have been fused. (iv) Six histidine residues were placed at the C terminus of the fusion molecule to facilitate protein purification via metal-chelating affinity chromatography.
Fusion genes encoding three different Pab molecules were constructed, as illustrated in Fig. 2 and described in Materials and Methods. Essentially, the DNA sequences encoding the BCL1 idiotype-specific peptides, as well as the hinge region, were assembled from oligonucleotide duplexes. The COMP pentamerization domain was amplified by PCR, simultaneously introducing point mutations, and the fusion genes were assembled into the pDS-78 expression vector (19).
Figure 2.
Schematic representation of the Pab fusion gene. P/O stands for the E. coli phage T5 promoter and two lac operator sequences. Restriction sites used for cloning are indicated.
Expression and Purification of Pab Fusion Proteins.
The fusion genes were expressed in E. coli and the amounts of soluble full-length proteins were determined after purification on a Ni-NTA column. The Pab-S and Pab-F fusion proteins were produced at high levels (>30 mg/liter), allowing efficient one-step metal-chelating affinity purification under native conditions. The amounts of soluble Pab-D protein were much lower, preventing an efficient purification and further characterization.
Affinity-purified Pab-S and Pab-F molecules were analyzed by FPLC on a Superdex G-200 column. A single elution peak corresponding to a protein of about 85 kDa was observed for Pab-S, whereas two major elution peaks corresponding to proteins of about 90 and 180 kDa were observed for Pab-F. The high molecular weight fraction presumably resulted from dimer formation by unsaturated cysteine present in peptide F. In both cases, fractions of the elution peak corresponding to a 85–90-kDa protein were collected, pooled, and used for binding studies.
Equilibrium Binding Studies.
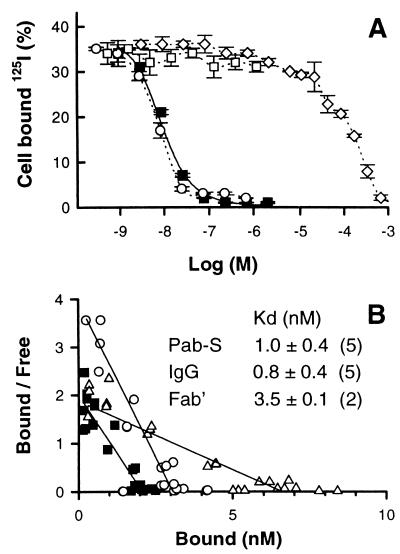
In preliminary solid-phase competition assay, Pab-S was found to compete for binding of 125I-labeled mAb B1 IgG to BCL1 IgM idiotype coated on plates (data not shown). However, since spatial arrangement of target molecules on the surface may influence multivalent binding, the equilibrium binding parameters of Pab-S were determined directly on live BCL1 cells, which represent a more relevant biological surface. Purified Pab-S was labeled with 125I and was shown to bind to BCL1 idiotype on the cell surface. The binding can be competed by unlabeled Pab-S, mAb B1 IgG, and by a much higher concentration of peptide S, but not by the control Pab-F pentamer (Fig. 3A). The competition of 125I-labeled Pab-S binding by unlabeled Pab-S at three different cell concentrations, as well as a cell dilution experiment, were used to calculate the equilibrium binding parameters (see Materials and Methods).
Figure 3.
Equilibrium binding of Pab-S to the BCL1 cell-surface idiotype. (A) BCL1 cells were incubated with trace amounts of 125I-labeled Pab-S and increasing concentrations of Pab-S (▪), S-peptide (⋄), mAb B1 IgG (○), or Pab-F as control (□). After 2.5 h at 4°C cell bound 125I was counted (mean ± SD of triplicates). Broken lines are guidelines for the eyes. Solid lines are calculated from the fitted parameters (see text). (B) Scatchard plot of a representative equilibrium binding experiment. BCL1 cells were incubated with trace amounts of 125I-labeled Pab-S (▪), mAb B1 Fab′ (Δ), or IgG (○) and increasing concentrations of the same unlabeled proteins. Parameters (Kd, binding sites per cell, NSB) were fitted according to a linear model. Kd values are the mean of three independent experiments ± SD. The number of apparent binding sites per cell were 1.17, 0.54, and 0.38 × 106 for mAb B1 Fab, IgG, and Pab-S, respectively.
The Scatchard representation and equilibrium binding constants of Pab-S, B1 IgG, and B1 Fab′ are shown in Fig. 3B. The apparent equilibrium binding constant of Pab-S (≈1 nM) was found to be similar to that of B1 IgG, which represents a 2 × 105-fold increase in avidity compared with the peptide S itself (IC50, ≈200 μM). Because both the Kd of the 125I-labeled ligand Pab-S (1 nM) and its concentration in the inhibition test (≈0.1 nM), as well as the concentration of binding sites (2 nM), are negligible compared with the peptide IC50 (2 × 105 nM), we can consider that IC50 = Kd. The same binding experiments were performed with 125I-labeled B1 IgG and B1 Fab′ fragment. The binding constants obtained for mAb B1 IgG and B1 Fab′ fragment tested in parallel were consistent with recently reported data (C. Manetti, personal communication and ref. 16). Scatchard analysis also showed a lower molarity of binding sites for Pab-S compared with mAb B1 IgG and B1 Fab′ fragment (Fig. 3B), providing evidence for the multivalent nature of Pab-S binding.
In another series of experiments the capacity of soluble BCL1 IgM idiotype to compete for binding of either Pab-S or mAb B1 to BCL1 cells was compared. The results showed that a much higher concentration of soluble BCL1 IgM was needed to compete out Pab-S than mAb B1 IgG (100 nM and 4 nM of soluble BCL1 IgM, respectively, for 80% displacement). These data indicate a selective avidity of Pab-S for the cell-surface-immobilized BCL1 IgM, as opposed to soluble BCL1 IgM. Interestingly, no inhibition of Pab-S binding to BCL1 IgM idiotype was observed in the presence of up to 30% (vol/vol) of mouse or human serum (data not shown).
Biochemical Characterization of Pab-S Protein.
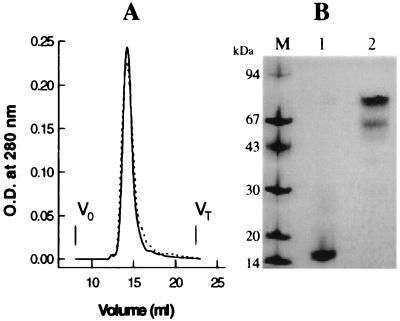
The Pab-S molecule was characterized using FPLC gel filtration, SDS/PAGE, and CD spectrometry. Affinity purification, followed by gel filtration, yielded the oxidized form of Pab-S with interchain disulfide bonds, whereas the reduced form was prepared as described. Under nondenaturing conditions, gel filtration on a Superdex G-200 column gave a single elution peak, corresponding to a protein of about 85 kDa for both reduced and nonreduced forms of Pab-S (Fig. 4A). Under denaturing conditions in SDS/PAGE, a major protein band with an apparent Mr of about 85 kDa was observed for the nonreduced form of Pab-S, whereas a single protein band with an apparent Mr of about 17 kDa was observed for the reduced form, in agreement with a covalent pentameric structure of Pab-S (Fig. 4B). The minor protein band with an apparent molecular weight of about 68 kDa observed for the nonreduced Pab-S (Fig. 4B, lane 2) suggests the presence of a small percentage of incompletely oxidized molecules, in which only four chains out of five are covalently linked by disulfide bonds.
Figure 4.
Biochemical characterization of Pab-S chimeric protein. (A) FPLC gel filtration on a Superdex G-200 column of reduced (broken line) and oxidized (solid line) forms of the Pab-S molecule. V0, exclusion volume; VT, total bed volume. (B) Coomassie blue staining of reduced (lane 1) and nonreduced (lane 2) forms of Pab-S analyzed by 10–15% gradient SDS/PAGE.
Analysis of CD spectra of Pab-S pentamer allowed for the determination of an α-helical content of about 54% by fitting the experimental data with a set of reference proteins, as described elsewhere (26) (data not shown). This is in good agreement with the length of the α-helical coiled-coil domain within the entire molecule. Taken together, these data indicate that the Pab-S molecule is a stable homopentamer of about 85 kDa with monomer subunits of about 17 kDa held together by an α-helical coiled-coil bundle, and covalently linked by disulfide bonds.
It has been shown that coiled-coil structures can undergo reversible denaturation (27). Indeed, the Pab-S molecule was denatured in urea at 95°C under reducing conditions and refolded into the pentamer by simple dialysis against PBS, as demonstrated by FPLC gel filtration of renatured 125I-labeled Pab-S. Importantly, renaturation of Pab-S restored full-binding activity, as demonstrated by binding competition assays on BCL1 cells (data not shown).
Pab-S-Induced Signal Transduction in BCL1 Cells.
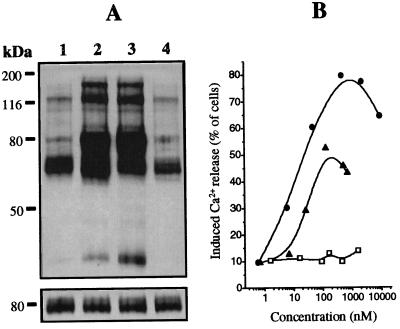
Pab-S was found to induce a strong Ca2+ release upon contact with BCL1 cells in a dose-dependent manner. Pab-S induced Ca2+ release at lower concentrations and in a larger proportion of cells than mAb B1 IgG at similar concentrations (Fig. 5B). No Ca2+ release was observed after treatment of BCL1 cells with control Pab-F. A slight decrease seen at high concentrations of Pab-S or mAb B1 IgG can be explained by surface saturation resulting in an elevated proportion of monovalent binding. In addition, we have demonstrated that Pab-S induces a specific phosphorylation of intracellular protein tyrosine, similarly to anti-idiotypic mAb B1 IgG (Fig. 5A Upper). Only background phosphorylation was seen with control Pab-F. The equal amount of material loaded on each lane was verified using anti-mouse μ chain antibody, detecting the BCL1 surface IgM (Fig. 5A Lower).
Figure 5.
Effect of Pab-S on signal transduction in BCL1 cells. (A) (Upper) Western blot showing intracellular protein tyrosine phosphorylation in BCL1 cells after 1 min incubation with medium (lane 1), mAb B1 IgG (lane 2), Pab-S (lane 3), or control Pab-F (lane 4). (Lower) To show equal loading in all lanes the same membrane was striped and stained with anti-mouse μ chain antibody. (B) Induction of intracellular Ca2+ release in BCL1 cells after addition of Pab-S (•), mAb B1 IgG (▴), or control Pab-F (□).
Computer Modeling of the Pab-S Molecule.
To visualize the spatial arrangement of Pab-S and to ensure that its geometry allows multivalent binding to the BCL1 Ig idiotype, a model of the Pab-S three-dimensional structure was developed as described in Materials and Methods. For the pentamerization domain, a previous model of five-stranded α-helical coiled-coil structure (25) was taken with minor modifications at the junction with the hinge region. Concerning the hinge region, a molecular dynamic simulation with subsequent energy minimization resulted in a set of possible conformations. To assess the dimension of Pab-S potential pentavalent interaction, we selected one of these conformations, which gave maximal spreading of peptide heads.
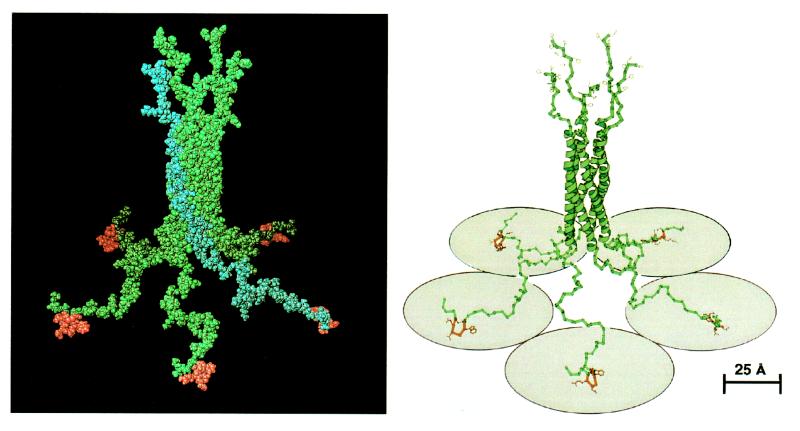
For peptide S, stereochemical analysis showed that the type I conformation of β-turn allowed the formation of a disulfide bond between the two cysteines of peptide S. This conformation was chosen as the most likely for the peptide ligand. Fig. 6 shows a 5-fold symmetrical structure of Pab-S, which was obtained by 72° × n (n = 0 to 4) rotation of a monomer molecule around the coiled-coil axes. The analysis of the modeled Pab structure showed that such a molecule should be capable of simultaneous binding to five surface receptors, provided they are located up to 80 Å apart (Fig. 6, ribbon presentation). This criterion is largely satisfied in the case of the surface Ig receptor, whose variable domains can be brought as close as 50 Å, as predicted from their van der Waals contours.
Figure 6.
Space-filling (Left) and ribbon (Right) representations of a model of the three-dimensional structure of Pab-S. Binding peptides are in red. The upper part of the structure shows six histidine residues at each C terminus. One chain within the pentameric molecule is highlighted in blue in the space-filling representation. Five shaded circles (radius of 40 Å) under the ribbon structure schematically denote receptor molecules. The ribbon representation was generated with the program molscript (28).
DISCUSSION
We have described here a novel pentameric protein allowing a multivalent high avidity binding of selected peptide ligands to “receptors.” A fusion gene was engineered, encoding sequentially a selected peptide ligand, the hinge region derived from camel IgG, and the assembly domain derived from COMP. Transfected into bacteria, the fusion gene was translated into a 17-kDa protein chain, which spontaneously assembled into a stable homopentamer of 85 kDa, termed Pab. We characterized the first Pab, Pab-S, where the N-terminal peptide ligand was selected from a phage display library for specific binding to the idiotype of the mouse BCL1 lymphoma.
Equilibrium binding studies show that Pab-S has an avidity for BCL1 cells, which is nearly 2 × 105 times higher than the affinity of the free peptide and similar to that of the anti-idiotype mAb B1. Comparison of parameters derived from Scatchard analysis shows a lower molarity of binding sites for Pab-S as compared with mAb B1 IgG and B1 Fab′ fragment, consistent with the multivalent nature of Pab binding. The high avidity of Pab-S molecules toward BCL1 cells is likely to be the result of multiple simultaneous low affinity interactions of peptide heads with several BCL1 surface idiotypes, representing a cooperative binding to the surface-immobilized target molecules. In contrast, in solution, Pab-S should bind independently to different individual IgM molecules, resulting in lower avidity. We can speculate that the preferential binding of Pab-S to cell-surface-immobilized BCL1 IgM, as compared with soluble IgM, will be a valuable advantage in vivo, allowing targeting of the B-cell lymphoma in the presence of relatively high levels of circulating idiotype.
The functional activity of the Pab-S molecule is still under investigation. We have already demonstrated here that binding of Pab-S to BCL1 cells induces a strong intracellular Ca2+ influx as well as significant tyrosine phosphorylation, known to be hallmarks of cell activation upon crosslinking of surface Ig receptors (29, 30).
The Pab architecture is based on three major structural units, each contributing to the unique properties of this newly designed molecule. First, the specificity of the Pab is provided by a short peptide ligand, representing a “minimal” binding domain, where the primary structure information is sufficient for recognition. Second, the semi-rigid proline-rich sequence of the hinge region should favor the cooperative binding of peptide heads. Third, we took advantage of the COMP assembly domain, which spontaneously forms a five-stranded α-helical bundle, the highest oligomerization state known for a compact coiled-coil structure. As shown recently (31), various forms of this domain can be readily produced in E. coli and easily purified to near homogeneity under nondenaturing conditions. These properties, taken together with a remarkable solubility in salt-free water (up to 20 mg/ml) and thermostability, make the COMP assembly domain an ideal pentamerization tool for protein engineering. Peptide heads and spacers do not require any particular folding to assemble into a pentamer. Thus, the display of short peptides in a pentameric form on Pab molecules bypasses the folding problems and the difficulties previously encountered during the expression of oligomeric forms of relatively complex proteins, such as single-chain Fv fragments (32).
Moreover, we have shown that Pab-S can undergo reversible denaturation, without any reduction in binding activity. Thus, heteropentamers could be readily obtained by mixing Pabs with different specificities (i.e., with different peptide ligands) under reducing denaturing conditions followed by dialysis against physiologic buffer. This intrinsic property of Pab opens an interesting approach to the production of a heteropentameric Pab with chelating properties, as recently revealed for a chelating bispecific single chain Fv fragment (33).
As shown here for the Pab-S molecule, a Pab with relatively high avidity can be obtained starting from a peptide ligand of very low intrinsic affinity. However, if we consider peptide ligands with higher affinity, either selected from phage display libraries (4, 5) or derived from naturally occurring peptide hormones, such as the somatostatin analogue (34), an exceptionally high avidity may be reached by expressing these peptides in a Pab format. Furthermore, a Pab expressing the somatostatin analogue with different tailor-made hinge regions may have new diagnostic and therapeutic properties.
As an important step for future development, we have recently observed that a Pab molecule (e.g., Pab-S) can be expressed as a fusion with the filamentous phage g3p coat protein and displayed in a pentameric form on the phage (unpublished observation). Thus, it should be possible to display random peptide library directly in the Pab format on the surface of phages. The screening of such a phage display Pab library should allow a more rapid isolation of new Pab molecules, benefiting from the power of selection and experience accumulated in the field of the phage display technology.
Finally, the C-terminal end of Pab remains free to generate additional fusion domains. Indeed, in the wild-type full-length COMP molecule, the assembly domain used here is followed at the C terminus by other functional domains (35). Thus, the fusion to Pab of different relevant polypeptides, such as an Fc receptor-binding domain, would provide new functional properties to this molecule, in addition to the multivalent high avidity binding capacity described here.
Acknowledgments
We especially thank Drs. J. Engel, V. Efimov, and G. Winter for valuable help at the initial stage of this work. We are grateful to Dr. C. Servis for peptide synthesis, Drs. H. Vogel and M. Eisenhauer for help in CD spectra measurements, Dr. D. Stüber for the supply of expression vectors, and Dr. C. Manetti for providing data prior to publication. We thank Dr. S. Betz-Corradin for editorial assistance.
ABBREVIATIONS
- Pab
peptabody
- Pab-S
peptabody S
- COMP
cartilage oligomeric matrix protein
- FPLC
fast protein liquid chromatography
- CD
circular dichroism
- NSB
nonspecific binding
- NIF
nonimmunoreactive fraction
References
- 1.Smith G P. Science. 1985;228:1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 2.Winter G, Griffiths A D, Hawkins R E, Hoogenboom H R. Annu Rev Immunol. 1994;12:433–455. doi: 10.1146/annurev.iy.12.040194.002245. [DOI] [PubMed] [Google Scholar]
- 3.Scott J K, Craig L. Curr Opin Biotechnol. 1994;5:40–48. doi: 10.1016/s0958-1669(05)80068-0. [DOI] [PubMed] [Google Scholar]
- 4.Goodson R J, Doyle M V, Kaufman S E, Rosenberg S. Proc Natl Acad Sci USA. 1994;91:7129–7133. doi: 10.1073/pnas.91.15.7129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fisch I, Kontermann R E, Finnern R, Hartley O, Soler-Gonzales A S, Griffiths A D, Winter G. Proc Natl Acad Sci USA. 1996;93:7761–7766. doi: 10.1073/pnas.93.15.7761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scott J K, Smith G P. Science. 1990;249:386–390. doi: 10.1126/science.1696028. [DOI] [PubMed] [Google Scholar]
- 7.Doorbar J, Winter G. J Mol Biol. 1994;244:361–369. doi: 10.1006/jmbi.1994.1736. [DOI] [PubMed] [Google Scholar]
- 8.Renschler M F, Bhatt R R, Dower W J, Levy R. Proc Natl Acad Sci USA. 1994;91:3623–3627. doi: 10.1073/pnas.91.9.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roitt I M. Essential Immunology. 7th Ed. London: Oxford/Blackwell; 1991. pp. 65–84. [Google Scholar]
- 10.Male D, Champion B, Cooke A. In: Advanced Immunology. Male D, Champion B, Cooke A, editors. London: Gower Medical; 1987. pp. 2.1–2.13. [Google Scholar]
- 11.Efimov V P, Lustig A, Engel J. FEBS Lett. 1994;341:54–58. doi: 10.1016/0014-5793(94)80239-4. [DOI] [PubMed] [Google Scholar]
- 12.Tomschy A, Fauser C, Landwehr R, Engel J. EMBO J. 1996;15:3507–3514. [PMC free article] [PubMed] [Google Scholar]
- 13.Slavin S, Strober S. Nature (London) 1978;272:624–626. doi: 10.1038/272624a0. [DOI] [PubMed] [Google Scholar]
- 14.Wertman K F, Wyman A R, Botstein D. Gene. 1986;49:253–262. doi: 10.1016/0378-1119(86)90286-6. [DOI] [PubMed] [Google Scholar]
- 15.Brissinck J, Demanet C, Moser M, Leo O, Thielemans K. J Immunol. 1991;147:4019–4026. [PubMed] [Google Scholar]
- 16.Manetti, C., Rouvier, E., Gautherot, E., Loucif, E., Barbet, J. & Le Doussal, J.-M. (1997) Int. J. Cancer, in press. [DOI] [PubMed]
- 17.Wolf P. Anal Biochem. 1983;129:145–155. doi: 10.1016/0003-2697(83)90062-3. [DOI] [PubMed] [Google Scholar]
- 18.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1990. [Google Scholar]
- 19.Stüber D, Matile A, Garotta G. In: Immunological Methods. Lefkovits I, Perris B, editors. New York: Academic; 1990. pp. 121–152. [Google Scholar]
- 20.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bradford M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 22.Roussel A, Cambillan C Silicon Graphics, editors. Silicon Graphics Geometry Partner Directory (Fall 1989) Mountain View, CA: Silicon Graphics; 1989. pp. 77–78. [Google Scholar]
- 23.Brunger A T. x-plorVersion 3.1: A System for X-Ray Crystallography and NMR. New Haven, CT: Yale Univ. Press; 1992. [Google Scholar]
- 24.Hamers-Casterman C, Atarhouch T, Muyldermans S, Robinson G, Hamers C, Bajyana-Songa E, Bendahman N, Hamers R. Nature (London) 1993;363:446–448. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- 25.Kajava A V. Proteins. 1996;24:218–226. doi: 10.1002/(SICI)1097-0134(199602)24:2<218::AID-PROT8>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 26.Vogel H. Biochemistry. 1987;26:4562–4572. doi: 10.1021/bi00388a060. [DOI] [PubMed] [Google Scholar]
- 27.Potekhin S A, Medvedkin V N, Kashparov I A, Venyaminov S Yu. Protein Eng. 1994;7:1097–1101. doi: 10.1093/protein/7.9.1097. [DOI] [PubMed] [Google Scholar]
- 28.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 29.Pleiman C M, D’Ambrosio D, Cambier J C. Immunol Today. 1994;15:393–399. doi: 10.1016/0167-5699(94)90267-4. [DOI] [PubMed] [Google Scholar]
- 30.Flaswinkel H, Barner M, Reth M. Semin Immunol. 1995;7:21–27. doi: 10.1016/1044-5323(95)90004-7. [DOI] [PubMed] [Google Scholar]
- 31.Efimov V P, Engel J, Malashkevich V N. Proteins. 1996;24:259–262. doi: 10.1002/(SICI)1097-0134(199602)24:2<259::AID-PROT13>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 32.Pack P, Müller K, Zahn R, Plückthun A. J Mol Biol. 1995;246:28–34. doi: 10.1006/jmbi.1994.0062. [DOI] [PubMed] [Google Scholar]
- 33.Neri D, Momo M, Prospero T, Winter G. J Mol Biol. 1995;246:367–373. doi: 10.1006/jmbi.1994.0091. [DOI] [PubMed] [Google Scholar]
- 34.Reubi J C, Waser B, Horisberger U, Krenning E, Lambers S W, Gebbers J O, Gerbach P, Laissue J A. Blood. 1993;82:2143–2151. [PubMed] [Google Scholar]
- 35.Lawler J, Duquette M, Urry L, McHenry K, Smith T F. J Mol Evol. 1993;36:509–516. doi: 10.1007/BF00556355. [DOI] [PubMed] [Google Scholar]