Summary paragraph:
bicoid mRNA localises to the anterior of the Drosophila egg, where it is translated to form a morphogen gradient of Bicoid protein that patterns the head and thorax of the embryo. Although bicoid was the first identified localised cytoplasmic determinant1-4, little is known about how the mRNA is coupled to the microtubule-dependent transport pathway that targets it to the anterior, and it has been proposed that it is recognised by a complex of many redundant proteins, each of which binds to the localisation element in its 3'UTR with little or no specificity5. Indeed, the only known RNA-binding protein that co-localises with bicoid mRNA is Staufen, which binds non-specifically to dsRNA in vitro6, 7. Here we show that mutants in all subunits of the ESCRT-II complex (Vps22, Vps25 and Vps36) abolish the final Staufen-dependent step in bcd RNA localisation. ESCRT-II is a highly conserved component of the pathway that sorts ubiquitinated endosomal proteins into internal vesicles8, 9, and functions as a tumour-suppressor by removing activated receptors from the cytoplasm10, 11. However, the role of ESCRT-II in bicoid localisation appears to be independent of endosomal sorting, because mutations in ESCRT-I and III components have no effect of the targeting of bicoid mRNA. Instead, Vps36 functions by binding directly and specifically to stem-loop V of the bicoid 3'UTR through its N-terminal GLUE domain12, making it the first example of a sequence specific RNA-binding protein that recognises the bicoid localisation signal. Furthermore, Vps36 localises to the anterior of the oocyte in a bicoid mRNA-dependent manner, and is required for the subsequent recruitment of Staufen to the bicoid complex. This novel function of ESCRT-II as an RNA-binding complex is conserved in vertebrates, and may explain some of its roles that are independent of endosomal sorting.
Genetic screens for mutations that disrupt anterior-posterior patterning of the Drosophila embryo have identified a few genes that are required at different stages for the anterior localisation of bicoid mRNA, but most of these appear to play an indirect role in the process. Mutations in exuperantia (exu) abolish all stages of bicoid mRNA localisation13, 14, but its function is unclear, since Exu protein is a component of a translational repression complex that co-purifies with oskar mRNA, but not with bicoid itself15. Swallow, γ-Tubulin37C, Dgrip75, Dgrip128, and Minispindles are necessary for the localisation of bicoid mRNA from stage 10B of oogenesis onwards, and function to nucleate anterior microtubules that direct localisation at this stage16-18. Finally, Staufen is required for bicoid mRNA localisation at the end of oogenesis and in the early embryo19, 20. Unlike the other trans-acting factors, Staufen is a dsRNA-binding protein, and associates with bicoid mRNA at the oocyte anterior from stage 10B onwards6.
bicoid mRNA is localised by distinct and partially redundant mechanisms at different stages of oogenesis, which may explain why genetic screens have missed many of the essential trans-acting factors. In mutants that only disrupt early localisation, the localisation of the mRNA in late oocytes can rescue anterior development, whereas mutants that only disrupt late localisation result in a gradient of mRNA that induces some anterior patterning, unless bicoid translation is also impaired21-23. To circumvent this problem, we performed a direct visual screen in germline clones for mutants that alter the localisation of bicoid mRNA in living oocytes using GFP-Staufen as a marker24. This screen identified one complementation group, called larsen, that is required for the anterior localisation of bicoid mRNA. GFP-Staufen fails to localise to the anterior cortex of the oocyte in homozygous germline clones of both larsen alleles (Fig.1 a, b, d, e), whereas its posterior localisation with oskar mRNA is unaffected. The stronger allele, lsn5F3-8, is homozygous lethal, and females with a homozygous mutant germline do not lay eggs. However, the weaker allele, lsn2B63, is only semi-lethal, and mutant germline clones result in eggs in which bicoid mRNA forms a gradient across the anterior half of the embryo (Fig.1 f), a phenotype very similar to that seen staufen mutants6.
Figure1. Mutations in lsn and vps36 disrupt the anterior localisation of GFP-Stau and bcd mRNA.

a and b GFP-Staufen (green) localisation to the anterior cortex of wildtype stage 11 (a) and stage 13 (b) oocytes. The actin cortex is labelled with Rhodamine-Phalloidin (red).
c bcd mRNA localisation in a freshly-laid wildtype egg.
d and e Homozygous vps22/lsn germline clones, showing the absence of GFP-Stau at the anterior pole of the oocyte at stage 10B (d) and at stage 13 (e).
f bcd mRNA localisation in an embryo derived from a vps22/lsn germline clone.
g and h Homozygous vps25Pb2931 germline clones, showing the absence of GFP-Stau at the anterior pole of the oocyte at stage 10B (g) and at stage 13 (h).
i and j Homozygous vps36 (l(3)L5212) germline clones, showing the absence of GFP-Stau at the anterior pole of the oocyte at stage 11 (i) and at stage 13 (j).
Both larsen alleles correspond to mutations in the highly conserved protein, Vps22 (supplementary Fig.1). Vps22p, together with Vps25p and Vps36p, forms the ESCRT-II (for endosomal sorting complex required for transport), one of four complexes (Vps27/Hrs complex, ESCRT-I, -II and -III) that act in a linear pathway to sort mono-ubiquitinated transmembrane proteins within the endosomal compartment into internal vesicles, leading to the formation of multivesicular bodies (MVBs)9, 25. Mutants in Drosophila ESCRT-I and ESCRT-II components have been shown to cause a tumorous phenotype, because activated receptors accumulate on endosomes, rather than being removed from the cytoplasm and degraded10, 11, 26. Mammalian ESCRT-II was independently identified as a complex that binds the RNA polymerase II elongation factor, ELL27, 28, while the fission yeast Vps 22/Larsen homologue regulates the expression of centriolar proteins during meiosis29. However, the relationship between these functions and endosomal protein sorting remains unclear.
To test whether the role in bcd mRNA localisation is specific for Larsen/Vps22 or whether the whole ESCRT-II is involved in the process, we examined mutants in the other two components of this complex, vps25 and vps36. Homozygous germline clones of vps25 and vps36 show an identical phenotype to lsn5F3-8: GFP-Stau does not localise to the anterior of the oocyte (Fig.1g-j), and the flies do not lay eggs. We therefore tested whether other MVB sorting mutants also disrupt bcd mRNA localisation. However, bicoid mRNA localises normally in germ line clones mutant for hrs (vps27), the ESCRT-I components, vps28 and ept, and the ESCRT-III component, vps32 (Fig.2c-f). Thus, the ESCRT-pathway appears to be dispensable for bcd mRNA localisation, indicating that ESCRT-II has another function in addition to the sorting of ubiquitinated proteins into MVBs.
Figure 2. Localisation of GFP-Stau in oocytes mutant for different ESCRT components.
a The anterior localisation of GFP-Stau in a wildtype stage 11 oocyte.
b-f GFP-Staufen localisation in lsn2B6-3 (b), hrsD28 (c), vps28 (l(2)k16503) (d), eptP26 (e) and vps32 (KG01481) (f) germline clones.
Given the role of ESCRT-II in bicoid mRNA localisation, we tested whether any of the proteins of the complex interact with the bicoid localisation signal, using a yeast 3-hybrid assay30. Both Staufen and Vps36, but not Vps22 or Vps25, interact with the bicoid 3′UTR in this assay, resulting in growth of the yeast cells on plates lacking histidine and in the expression of α-galactosidase (Fig.3a). To determine whether Vps36 protein binds directly to bcd RNA, we performed a UV-cross linking assay with purified recombinant Vps36 and in vitro transcribed bicoid 3'UTR. Vps36 cross-links efficiently to the bcd 3′UTR in this assay, indicating that it contacts single-stranded regions of the RNA directly (Fig.3b). Furthermore, this interaction is specific, since the binding is competed by excess unlabelled bcd 3′UTR, but not by control RNAs (Fig.3b, d, e and data not shown). Given the high degree of conservation of ESCRT-II, we tested whether Xenopus laevis Vps36 also interacts with RNA, and found that its N-terminal GLUE domain12 binds specifically to the bcd 3'UTR, indicating that the RNA-binding activity of Vps36 is conserved in vertebrates (Fig.3c).
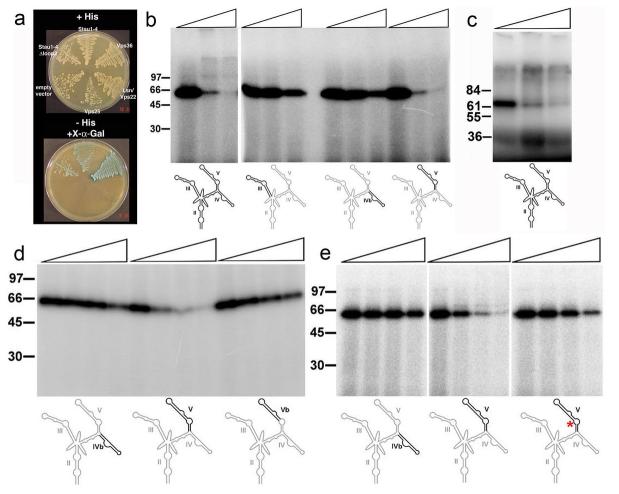
Figure 3. Vps36 binds specifically to stem-loop V of the bicoid 3'UTR.
a 3-Hybrid assay using the full-length bcd 3′UTR as an RNA bait. Growth on media lacking histidine and α-galactosidase activity (blue staining) indicates an interaction between the protein and the RNA. Vps36 interacts with bcd RNA in this assay, as does the region of Staufen containing the first four dsRNA-binding domains with or without the insert in dsRBD2 (Δloop2). In contrast, Lsn/Vps22 and Vps25 show no interaction.
b-e UV cross-linking assay using purified recombinant Drosophila Vps36 protein or purified GST-GLUE domain from Xenopus (c) and full-length 32P-labelled bcd 3′UTR as a probe. b Vps36 binding to the full-length 3'UTR in the presence of increasing amounts of competitor RNA (first lane: no competitor, second lane: 10-fold excess, third lane: 100-fold excess). The binding of Vps36 is inhibited by an excess of the full-length 3′UTR (lanes 1-3) and stem-loop V (lanes 10-12), but not by stem-loops III or IVb (lanes 4-9).
c The Xenopus GLUE domain can be cross-linked to the bcd 3'UTR (first lane: no competitor, second lane: 10-fold excess, third lane: 100-fold excess).
d The distal region of Stem-loop V does not compete for to Vps36 binding, indicating that the binding site must include part of the proximal region (first lane: no competitor, second lane: equal amount, third lane: 5-fold excess, fourth lane: 25-fold excess).
e Stem-loop V carrying three base changes in the central loop region shows an impaired ability to compete for Vps 36 binding. The probe is stem-loop V, and the competitors are Stem-loop IVb, Stem-loop V, and Stem-loop V with the following three mutations: 419 A->G, 427 A->C, 428 C->A (numbering starts with the first nucleotide after the Stop codon, see supplementary figure 2).
To map the binding site within the RNA more precisely, we tested the ability of single stem-loops of the bicoid 3'UTR to compete for binding to Vps36. Stem-loop V competed as well as the full-length 3'UTR, whereas stem-loops III, IVb and the distal part of stem-loop V did not compete (Fig.3b, d). We then performed a random mutagenesis to map the Vps36 binding site within stem-loop V, and found that none of the distal mutations affect the interaction, whereas three base changes in the proximal stem and the central loop-region strongly reduce Vps36 binding (Fig.3e and supplementary Fig.2). Thus, Vps36 binds specifically to the proximal part of stem-loop V, and recognises specific bases, most probably in the context of the RNA structure of this region.
We next examined whether Vps36 associates with bicoid mRNA in vivo by generating transgenic flies expressing Vps36-YFP under the control of its endogenous promoter. Vps36-YFP is ubiquitously expressed in the ovary, and shows a general cytoplasmic localisation with some clouds in the nurse cells (Fig.4a and a′). From stage 10B/11 of oogenesis, the protein localises to the anterior cortex of the oocyte in the same region as bicoid mRNA (Fig.4b, b' and c). This anterior accumulation is abolished in exu mutants, which block bicoid mRNA localisation at an earlier stage (Fig.4d), and in sry δ mutants, in which bcd RNA is not transcribed (Fig.4e). Thus, Vps36 co-localises with bicoid mRNA at the anterior, and this localisation is bicoid mRNA-dependent, indicating that the protein binds directly to the RNA in vivo, as it does in vitro.
Figure 4. Vps36 is recruited to the anterior of the oocyte by bicoid mRNA.
a-f Expression of a YFP -Vps36 fusion protein (green) under the control of the endogenous vps36 promoter. a Vps36 is expressed in both the germ line and the somatic follicle cells of the ovary. The actin cytoskeleton has been counter-stained in red with Rhodamine–Phalloidin (a' shows the YFP channel alone). The protein shows a uniform distribution in the cytoplasm of the oocyte and the follicle cells, with a patchy distribution in the nurse cells, which becomes more disperse as oogenesis progresses. At stage 10B, Vps36 begins to accumulate at the anterior pole of the oocyte. b and b' Close-up of the anterior of a stage 11 egg chamber, showing the strong enrichment of Vps36 at the anterior cortex. The green spheres in the oocyte cytoplasm in this panel and panels D and E are not due to YFP-Vps36, but the background autofluorescence of yolk granules at late stages. c –f Localisation of YFPV-ps36 at stage 11 in wildtype (c), exuVL/exuSC (d), sry-δ14/Df(3R)X3F,P{ry+,sryDB56} (e), and homozygous stauD3 (f) oocytes. The fusion protein does not localise to the anterior cortex in exu mutants, which disrupt all stages of bicoid mRNA localisation, or in sry mutants, which prevent bicoid transcription, but localises normally in staufen mutants.
Both ESCRT-II and Staufen are recruited to bicoid mRNA at stage 10B, and are then required for its localisation during the final stages of oogenesis. staufen mutants have no effect on the anterior recruitment of Vps36 (Fig.4f), however, whereas mutants in larsen/vps22 and vps36 abolish the anterior recruitment of Staufen protein at this stage (Figs.1 and 2). ESCRT-II therefore binds to bicoid mRNA independently of Staufen, and is required for the subsequent recruitment of the latter to form a functional bicoid mRNA localisation complex.
Our results demonstrate that ESCRT-II is required for the anterior localisation of bicoid mRNA, and is the first identified sequence-specific RNA binding complex that recognises the bicoid localisation signal. This novel activity of ESCRT-II seems to be independent of its well-characterised role in endosomal protein sorting, and is conserved, since Xenopus Vps36 also interacts specifically with the bcd 3'UTR, suggesting that ESCRT-II may play a role in mRNA localisation in vertebrates. In future, it will be important to determine whether this conserved RNA-binding activity of ESCRT-II plays any role in endosomal trafficking, or in the other proposed functions of the complex in transcription elongation and the regulation of centriole assembly.
Methods
A more detailed description of the materials and methods used can be found under supplementary information.
Drosophila Genetics
Flies were grown under standard conditions on corn meal medium. Germline clones were induced using the autosomal DFS technique. For detailed information and the genotypes of the flies used see supplementary information.
Yeast 3-Hybrid assay
For the yeast 3-Hybrid assay we used the system based on the transcription factor GAL 4 and the RNA binding protein RevM10 from HIV-1, as well as its binding element (Rev responsive element, RRE)30.
UV-cross linking
The purified proteins were pre-incubated with the competitor RNAs for 20 min at room temperature in X-link buffer (PBS + 5% Glycerol, 2 mM DTT, 0.2% NP-40, 100 μg/ml heparin and 100 μ/ml tRNA). After addition of the probe and a further 20 min of incubation the cross-linking was performed using a ‘Stratalinker 1800’ at maximum power for 5 min. The samples were treated with RNaseA (0.5 μg/ml) for 1 h at 37°C and after SDS-PAGE the radioactivity was visualised using a phosphoimager.
Stainings and microscopy
Stainings were performed as described. Pictures were taken using a BioRad 1024 Confocal Microscope and processed with Adobe Photoshop.
Supplementary Material
Supplementary Figure 1
Comparison of ESCRT-II proteins
a shows a schematic comparison of Vps25, Vps22/Lsn and Vps36 from yeast (Sc, Saccharomyces cerevisiae), humans (Hs, Homo sapiens) and flies (Dm, Drosophila melanogaster). The winged-helix domains A and B are represented as grey boxes (WHA and WHB), the GLUE domain of Vps36 as a red box. Amino acid similarities between the proteins are given as percentages. b sequence alignment between Vps22/Lsn from flies, humans and yeast. The mutations in lsn2B6-3 (blue triangle, R64->C) and lsn5F3-8 (red triangle, R2->Stop) are indicated.
Supplementary Figure 2
Random mutagenesis of stem-loopV
a shows the four pairs of oligonucleotides that were used for the mutagenesis. For each experiment, we replaced one or two of the oligonucleotides with “degenerate” oligonucleotides in which each base had a 10% probability of being mutant. The oligonucleotides were annealed, the internal pairs were phosphorylated, and they were then ligated together and cloned into pSP73, which had been modified to contain MluI and EagI sites. In vitro transcription then results in stem-loopV RNA. In b the mutations in stem-loop V that decrease its affinity for Vps36 are indicated. They correspond to positions 419, 427 and 428 of the full-length 3'UTR.
Acknowledgments
We are very grateful to Roger Williams, Olga Perisec and Hsiangling Teo for giving us the purified recombinant Xenopus laevis Vps36 GLUE domain, and for their helpful advice on ESCRT-II. We would like to thank John Oberdick and Zulung Zhang for sharing their unpublished results on rat Vps36. This work was supported by a Wellcome Trust Principal Research Fellowship to D. St J. and by the Max-Planck-Gesellschaft (U.I.).
References
- 1.Frohnhöfer HG, Nüsslein-Volhard C. Organization of anterior pattern in the Drosophila embryo by the maternal gene bicoid. Nature. 1986;324:120–125. [Google Scholar]
- 2.Berleth T, et al. The role of localization of bicoid RNA in organizing the anterior pattern of the Drosophila embryo. EMBO J. 1988;7:1749–1756. doi: 10.1002/j.1460-2075.1988.tb03004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Driever W, Nüsslein-Volhard C. A gradient of Bicoid protein in Drosophila embryos. Cell. 1988;54:83–93. doi: 10.1016/0092-8674(88)90182-1. [DOI] [PubMed] [Google Scholar]
- 4.Driever W, Nüsslein-Volhard C. The Bicoid protein determines position in the Drosophila embryo in a concentration-dependent manner. Cell. 1988;54:95–104. doi: 10.1016/0092-8674(88)90183-3. [DOI] [PubMed] [Google Scholar]
- 5.Arn EA, Cha BJ, Theurkauf WE, Macdonald PM. Recognition of a bicoid mRNA localization signal by a protein complex containing Swallow, Nod, and RNA binding proteins. Dev Cell. 2003;4:41–51. doi: 10.1016/s1534-5807(02)00397-0. [DOI] [PubMed] [Google Scholar]
- 6.Johnston D, Beuchle D, Nüsslein-Volhard C. staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell. 1991;66:51–63. doi: 10.1016/0092-8674(91)90138-o. [DOI] [PubMed] [Google Scholar]
- 7.Ramos A, et al. RNA recognition by a Staufen double-stranded RNA-binding domain. EMBO J. 2000;19:997–1009. doi: 10.1093/emboj/19.5.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Babst M. A protein's final ESCRT. Traffic. 2005;6:2–9. doi: 10.1111/j.1600-0854.2004.00246.x. [DOI] [PubMed] [Google Scholar]
- 9.Babst M, Katzmann DJ, Snyder WB, Wendland B, Emr SD. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev Cell. 2002;3:283–289. doi: 10.1016/s1534-5807(02)00219-8. [DOI] [PubMed] [Google Scholar]
- 10.Vaccari T, Bilder D. The Drosophila tumor suppressor vps25 prevents nonautonomous overproliferation by regulating Notch trafficking. Dev Cell. 2005;9:687–698. doi: 10.1016/j.devcel.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 11.Thompson BJ, et al. Tumor suppressor properties of the ESCRT-II complex component Vps25 in Drosophila. Dev Cell. 2005;9:711–720. doi: 10.1016/j.devcel.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 12.Teo H, et al. ESCRT-I core and ESCRT-II GLUE domain structures reveal role for GLUE in linking to ESCRT-I and membranes. Cell. 2006;125:99–111. doi: 10.1016/j.cell.2006.01.047. [DOI] [PubMed] [Google Scholar]
- 13.Frohnhöfer HG, Nüsslein-Volhard C. Maternal genes required for the anterior localization of bicoid activity in the embryo of Drosophila. Genes Dev. 1987;1:880–890. [Google Scholar]
- 14.Macdonald PM, Luk SK, Kilpatrick M. Protein encoded by the exuperantia gene is concentrated at sites of bicoid mRNA accumulation in Drosophila nurse cells but not in oocytes or embryos. Genes Dev. 1991;5:2455–2466. doi: 10.1101/gad.5.12b.2455. [DOI] [PubMed] [Google Scholar]
- 15.Wilhelm JE, et al. Isolation of a ribonucleoprotein complex involved in mRNA localization in Drosophila oocytes. J Cell Biol. 2000;148:427–440. doi: 10.1083/jcb.148.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Theurkauf WE, Hazelrigg TI. In vivo analyses of cytoplasmic transport and cytoskeletal organization during Drosophila oogenesis: characterization of a multi-step anterior localization pathway. Development. 1998;125:3655–3666. doi: 10.1242/dev.125.18.3655. [DOI] [PubMed] [Google Scholar]
- 17.Moon W, Hazelrigg T. The Drosophila microtubule-associated protein Mini spindles is required for cytoplasmic microtubules in oogenesis. Curr Biol. 2004;14:1957–1961. doi: 10.1016/j.cub.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 18.Vogt N, Koch I, Schwarz H, Schnorrer F, Nüsslein-Volhard C. The γTuRC components Grip75 and Grip128 have an essential microtubule-anchoring function in the Drosophila germline. Development. 2006;133:3963–3972. doi: 10.1242/dev.02570. [DOI] [PubMed] [Google Scholar]
- 19.Ferrandon D, Elphick L, Nüsslein-Volhard C, Johnston D. Staufen protein associates with the 3′UTR of bicoid mRNA to form particles that move in a microtubule-dependent manner. Cell. 1994;79:1221–1232. doi: 10.1016/0092-8674(94)90013-2. [DOI] [PubMed] [Google Scholar]
- 20.Weil TT, Forrest KM, Gavis ER. Localization of bicoid mRNA in late oocytes is maintained by continual active transport. Dev Cell. 2006;11:251–262. doi: 10.1016/j.devcel.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 21.Johnston D, Driever W, Berleth T, Richstein S, Nüsslein-Volhard C. Multiple steps in the localization of bicoid RNA to the anterior pole of the Drosophila oocyte. Development. 1989;107(Suppl):13–19. doi: 10.1242/dev.107.Supplement.13. [DOI] [PubMed] [Google Scholar]
- 22.Macdonald PM, Kerr K, Smith JL, Leask A. RNA regulatory element BLE1 directs the early steps of bicoid mRNA localization. Development. 1993;118:1233–1243. doi: 10.1242/dev.118.4.1233. [DOI] [PubMed] [Google Scholar]
- 23.Micklem DR, Adams J, Grunert S, Johnston D. Distinct roles of two conserved Staufen domains in oskar mRNA localization and translation. EMBO J. 2000;19:1366–1377. doi: 10.1093/emboj/19.6.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin SG, Leclerc V, Smith-Litière K, Johnston D. The identification of novel genes required for Drosophila anteroposterior axis formation in a germline clone screen using GFP-Staufen. Development. 2003;130:4201–4215. doi: 10.1242/dev.00630. [DOI] [PubMed] [Google Scholar]
- 25.Teo H, Perisic O, Gonzalez B, Williams RL. ESCRT-II, an endosome-associated complex required for protein sorting: crystal structure and interactions with ESCRT-III and membranes. Dev Cell. 2004;7:559–569. doi: 10.1016/j.devcel.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 26.Moberg KH, Schelble S, Burdick SK, Hariharan IK. Mutations in erupted, the Drosophila ortholog of mammalian Tumor Susceptibility Gene 101, elicit non-cell-autonomous overgrowth. Dev Cell. 2005;9:699–710. doi: 10.1016/j.devcel.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 27.Kamura T, et al. Cloning and characterization of ELL-associated proteins EAP45 and EAP20. a role for yeast EAP-like proteins in regulation of gene expression by glucose. J Biol Chem. 2001;276:16528–16533. doi: 10.1074/jbc.M010142200. [DOI] [PubMed] [Google Scholar]
- 28.Schmidt AE, Miller T, Schmidt SL, Shiekhattar R, Shilatifard A. Cloning and characterization of the EAP30 subunit of the ELL complex that confers derepression of transcription by RNA polymerase II. J Biol Chem. 1999;274:21981–21985. doi: 10.1074/jbc.274.31.21981. [DOI] [PubMed] [Google Scholar]
- 29.Jin Y, Mancuso JJ, Uzawa S, Cronembold D, Cande WZ. The fission yeast homolog of the human transcription factor EAP30 blocks meiotic spindle pole body amplification. Dev Cell. 2005;9:63–73. doi: 10.1016/j.devcel.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 30.Putz U, Skehel P, Kuhl D. A tri-hybrid system for the analysis and detection of RNA-protein interactions. Nucleic Acids Res. 1996;24:4838–4840. doi: 10.1093/nar/24.23.4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Comparison of ESCRT-II proteins
a shows a schematic comparison of Vps25, Vps22/Lsn and Vps36 from yeast (Sc, Saccharomyces cerevisiae), humans (Hs, Homo sapiens) and flies (Dm, Drosophila melanogaster). The winged-helix domains A and B are represented as grey boxes (WHA and WHB), the GLUE domain of Vps36 as a red box. Amino acid similarities between the proteins are given as percentages. b sequence alignment between Vps22/Lsn from flies, humans and yeast. The mutations in lsn2B6-3 (blue triangle, R64->C) and lsn5F3-8 (red triangle, R2->Stop) are indicated.
Supplementary Figure 2
Random mutagenesis of stem-loopV
a shows the four pairs of oligonucleotides that were used for the mutagenesis. For each experiment, we replaced one or two of the oligonucleotides with “degenerate” oligonucleotides in which each base had a 10% probability of being mutant. The oligonucleotides were annealed, the internal pairs were phosphorylated, and they were then ligated together and cloned into pSP73, which had been modified to contain MluI and EagI sites. In vitro transcription then results in stem-loopV RNA. In b the mutations in stem-loop V that decrease its affinity for Vps36 are indicated. They correspond to positions 419, 427 and 428 of the full-length 3'UTR.