Abstract
RNA polymerase core enzyme of Escherichia coli is composed of two α subunits and one each of the β and β′ subunits. The C-terminal domain of the RNA polymerase α subunit plays a key role in molecular communications with class I transcription factors and upstream (UP) elements of promoter DNA, using the same protein surface. To identify possible differences in the functional roles of the two α subunits, we have developed a reconstitution method for hybrid RNA polymerases containing two distinct α subunit derivatives in a defined orientation (“oriented α-heterodimer”). The binding sites of two α C-terminal domains on the UP element DNA were determined by hydroxyl radical-based DNA cleavage mediated by (p-bromoacetamidobenzyl)-EDTA·Fe, which was bound at Cys-269 on the UP recognition surface of one or both α subunits. The results clearly indicated that the two α subunits bind in tandem to two helix turns of the rrnBP1 UP element, and that the β′-associated α subunit is bound to the promoter–distal region.
Keywords: transcription activation, molecular assembly, protein–DNA contact
Escherichia coli RNA polymerase holoenzyme is composed of core enzyme with the subunit structure α2ββ′ forming the catalytic unit for RNA polymerization and one of the multiple species of σ subunit, which provide promoter recognition activity. The core enzyme is assembled in vivo and in vitro in the sequence 2α → α2 → α2β → α2ββ′ (reviewed in ref. 1). The α subunit, consisting of 329 amino acid residues, plays a key role in RNA polymerase assembly. The N-terminal region from residue 20 to 235 is necessary and sufficient for enzyme assembly in vitro (2–5) and in vivo (6, 7). On the other hand, the C-terminal region, 94 residues long, plays a key role in molecular communications with class I transcription factors (reviewed in refs. 8–10), including cAMP receptor protein (CRP) (11–13), MarA (14), OxyR (15, 16), OmpR (12), Rob (17), SoxS (18), TyrR (19), and GalR (20) [transcription factors that require the C-terminal domain of RNA polymerase α subunit, directly or indirectly, for action are designated as class I factors (9)]. The C-terminal domain of α is also involved in interactions with the promoter upstream (UP) element, which has transcription enhancer activity (21–24).
The N-terminal region of α subunit for RNA polymerase assembly and the C-terminal region for transcription regulation form structurally independent domains, which are connected by a protease-sensitive flexible linker (25, 26). The tertiary structure of the C-terminal domain from residue 235 to the terminus (residue 329), as revealed by NMR (27), implied that the same protein surface is involved in contact with both a set of class I factors and the rrnBP1 UP element DNA. Systematic mutagenesis of the C-terminal regulatory domain showed that, even though these protein factors and UP DNA sequences interact with the same protein surface, the individual amino acid residues forming this surface differ subtly in importance for activation as between CRP and the UP element (28). Nevertheless, among the amino acid residues constituting the contact surface, Arg-265 was found to play a major role in response not only to the protein factor but also to the DNA element.
In the assembly of RNA polymerase, the two molecules of α subunit are considered to play different roles, with one adjacent to the β subunit and the other to the β′ (1, 7). In terms of subsequent function, however, the role of each α subunit remains to be elucidated. For instance, it is not yet known whether one or both of the α subunits are required for molecular contacts with protein factors and UP DNA sequences, and if both are required, whether each α subunit plays a different role in the process leading to transcription activation or repression. To investigate the functional role of each α subunit in transcription regulation, we have developed a method to reconstitute hybrid RNA polymerases containing two different α subunits in a defined orientation with respect to contact with the two large subunits, β and β′. Using the hybrid RNA polymerases containing “oriented α-heterodimers,” we studied the role of each α subunit in making contact with the rrnBP1 UP element by analysis of DNA cleavage caused by free radicals, which were generated by (p-bromoacetamidobenzyl)-EDTA·Fe (Fe·BABE), attached to the UP contact surface of one or both α subunits.
MATERIALS AND METHODS
Plasmids.
A list of plasmids used in this work is presented in Table 1. Plasmid pGEMAX185 (11), encoding α under the control of T7 promoter, was modified to make pGEMAX190 by site-directed mutagenesis in two points: (i) two BamHI sites were introduced into pGEMAX185 before and after the rplQ gene, and a segment between the two BamHI sites was then deleted to remove rplQ; and (ii) a single EcoT22I site was introduced, overlapping codons 1 and 2 of rpoA. For production of α proteins with hexahistidine (His6) tags, a sequence including six histidine codons was inserted at this EcoT22I site. Starting from pGEMAX190, a number of expression plasmids for α derivatives were constructed. pGEMA(45A)-H6 was constructed by inserting into pGEMAX190 a PCR product generated using pETMA-R45A (5) as the template and a 5′ primer including the EcoT22I and His6-tag sequences, whereas pGEMACD235(45A)-H6 was also constructed by inserting a PCR product generated using pETMA-R45A as a template and the same 5′ primer, but with a 3′ primer including a TAA stop codon at position 236.
Table 1.
Plasmids used
| Plasmid | Protein | Source |
|---|---|---|
| pGEMAX185 | Wild-type α | Ref. 11 |
| pGEMAX190 | Wild-type α | This work |
| pGEMACD235 | α-235 | Ref. 11 |
| pETMA-R45A | [45A]α | Ref. 5 |
| pGEMA(45A)-H6 | [45A]α with His6 tag | This work |
| pGEMACD235(45A)-H6 | [45A]α235 with His6 tag | This work |
| pET-α-HA | α with HA tag | This work |
| pGEMA(45A)-H6-HA | [45A]α with His6 and HA tags | This work |
| pGEMA269C | [269C]α | Unpublished work* |
| pGEMA(45A)269C | [45A269C]α | This work |
| pLAW2 | Wild-type α | Ref. 13 |
| pLAW2-H6 | Wild-type α with His6 tag | Ref. 7 |
| pLA-R45A-H6 | [45A]α with His6 tag | This work |
HA, hemagglutinin.
O. Ozoline, K.M., T. Negishi, and A.I.
For construction of the plasmid pET-α-HA for production of α-WT with the influenza hemagglutinin (HA) nonapeptide tag at its C terminus, a DNA duplex formed from two synthetic oligonucleotides (XBHA-UP with sequence TCGAGTATCCGTATGATGTCCCCCAGTATGCGTGAG and XBHA-LOW with sequence GATCCTCACGCATAGTCGGGCACATCATACGGATAC), including a sequence for the HA tag and having XhoI and BamHI termini at the 5′ and 3′ ends, respectively, was inserted into pLAW2-H6 (7), which had previously been treated with XhoI and BamHI. The XbaI–BamHI fragment, including the rpoA-HA sequence, was isolated and inserted into pET-21a (Novagen) between its corresponding sites. Plasmid pGEMA(45A)-H6-HA encoding [45A]α with an His6 tag at the N terminus and an HA tag at the C terminus was constructed by replacing the EcoRI–BamHI fragment of pET-α-HA with the corresponding fragment from pGEMA(45A)-H6. On the other hand, the expression plasmid, pLA-R45A-H6, of C-terminal His6-tagged [45A]α under the control of lpp-lac promoter was constructed from pETMA-R45A after substitution of its XbaI–HindIII fragment by the corresponding fragment of pLAW2-H6. All plasmid constructions were checked by DNA sequencing.
Isolation of RNA Polymerase Complexes Containing His6-Tagged α Subunits.
The expression plasmids for His6-tagged α derivatives were transformed into E. coli HN198 (rpoA+). Expression of the plasmid-encoded α subunits and preparation of the cell lysates were carried out essentially as described by Kimura and Ishihama (7). To isolate protein complexes containing His6-tagged α subunits, the cell lysates were directly subjected to affinity chromatography on a Ni2+-charged nitrilotriacetic acid (NTA)-Sepharose (His·Bind resin, Novagen) column. The elution buffer contained 300 mM of imidazole.
Reconstitution and Purification of RNA Polymerases with Oriented α Heterodimers.
α-WT, β, β′, σ70, and all α derivatives were expressed and purified as described (5, 11, 28). Core enzymes containing various combinations of α heterodimers were reconstituted by mixing 5- to 10-fold molar excess of His6-tagged [45A]α with α, β, and β′ at a molar ratio of 1:1:1 in the dissociation buffer [50 mM Tris·HCl, pH 8.0 at 4°C/1 mM EDTA/10 mM DTT/10 mM MgCl2/0.2 M KCl/20% (vol/vol) glycerol] containing 6 M deionized urea. The mixtures were then dialyzed against the reconstitution buffer [50 mM Tris·HCl, pH 8.0 at 4°C/1 mM EDTA/1 mM DTT/10 mM MgCl2/0.3 M KCl/20% (vol/vol) glycerol] to remove urea and the premature core enzymes thus formed were activated by incubation for 30 min at 30°C. The activated samples were diluted with 2 vol of TGED buffer [10 mM Tris·HCl, pH 8.0 at 4°C/5% (vol/vol) glycerol/0.1 mM EDTA/0.1 mM DTT] and applied to a Protein Pak G-DEAE column (Waters) equilibrated with TG buffer [10 mM Tris·HCl, pH 8.0 at 4°C/5% (vol/vol) glycerol] containing 0.1 M NaCl. Core enzymes thus assembled were separated from unassembled subunits by elution with a linear gradient of 0.1–0.7 M NaCl in the same buffer. Core enzyme fractions were pooled and applied onto a Ni2+-affinity column [HiTrap-chelating column (Pharmacia) or Ni-NTA (Qiagen, Chatsworth, CA)]. The column was washed successively with 10 × bed volumes of washing buffer [50 mM Tris·HCl, pH 8.0 at 4°C/0.2 M NaCl/5 mM imidazole/5% (vol/vol) glycerol]. RNA polymerases were eluted by 6 × bed volumes of elution buffer [50 mM Tris·HCl, pH 8.0 at 4°C/0.2 M NaCl/0.5 M imidazole/5% (vol/vol) glycerol] and concentrated by step-wise elution from a DEAE column in an HPLC system or through centrifugal ultrafiltration [Centricon-100 filter units (Amicon)]. Holoenzymes were prepared by mixing the purified core enzymes and 4-fold molar excess of purified σ70 at 30°C for 20 min.
Protein–Protein Cross-Linking.
Cross-linking of holoenzymes with dimethyl suberimidate (DMS) was carried out in 50 mM Hepes (pH 8.0), 0.3 M KCl, 0.1 mM EDTA, 0.1 mM DTT, 5% (vol/vol) glycerol at a protein concentration of 0.06 mg/ml. DMS was dissolved immediately before use in 50 mM Hepes buffer (30 mg/ml), and the pH was readjusted to 8.5. After addition of the freshly prepared DMS solution (10 μl) to sample solutions (final volume, 60 μl), the mixture was incubated at room temperature for 5 min, and the reaction then quenched by addition of 17 μl of 5 × SDS/gel loading buffer. Proteins were separated by electrophoresis on SDS/8% polyacrylamide gels and then transferred from the gel onto a poly(vinylidene difluoride) membrane using a semi-dry transfer apparatus (Bio-Rad). RNA polymerase subunits on the membrane were detected using antibodies against α, β, β′, σ70, and influenza virus HA nonapeptide, and visualized using an enhanced chemiluminescence Western blotting detection system (Amersham). Anti-HA antibody was purchased from Boehringer Mannheim.
Conjugation of RNA Polymerases with Fe·BABE.
Two α derivatives, [269C]α with a single cysteine at residue 269 (O. Ozoline, K.M., T. Negishi, and A.I., unpublished work) and [45A269C]α (R45A derivative of [269C]α), was used for conjugation with Fe·BABE. Conjugation was initiated by mixing 500 μl of a 90 μM protein solution in 20 mM Mops (pH 8.0 at 37°C), 10 mM MgCl2, 0.2 M KCl, 0.1 mM EDTA, 6 M urea with 25 μl of an 18 mM Fe·BABE solution in dimethyl sulfoxide. The reaction was carried out at 37°C for 1 h and then terminated by adding 500 μl of 1 M Tris·HCl. Unconjugated Fe·BABE was removed by overnight dialysis at 4°C against 500 ml of the dissociation buffer without DTT. The conjugated α derivatives were stored at −80°C until needed. The hybrid RNA polymerase carrying Fe·BABE on the β′-associated α was reconstituted by mixing αWT, [45A269C]α·Fe, β, and β′ in a molar ratio of 1:3:1:1.
Fe·EDTA-Mediated Cleavage of DNA.
Mixtures of 32P-end-labeled DNA fragments and Fe·BABE-conjugated RNA polymerase (20 nM) were incubated at 37°C for 10 min in 50 μl of 10 mM Tris·HCl (pH 7.8 at 37°C), 3 mM magnesium acetate, 1 mM EDTA, 50 mM NaCl, 25 μg/ml BSA, 10% (vol/vol) glycerol. DNA cleavage was initiated by the addition of sodium ascorbate (final 2 mM), followed by incubation at 37°C for 20 min. The DNA was extracted with phenol/chloroform, precipitated with ethanol, and analyzed by electrophoresis on 6% polyacrylamide gel containing 8 M urea. Bands were visualized with Bioimage Analyzer BAS2000 (Fujix, Tokyo). The template used was the XhoI(−160)–HindIII(+50) fragment of pSL9 carrying the rrnBP1 promoter region from −88 to +50 (29), which was 32P-labeled on the upper strand at its 5′ XhoI end.
RESULTS
Theoretical Background and Experimental Approach.
The α subunit of E. coli RNA polymerase is composed of two structural domains, each carrying distinct functions (reviewed in refs. 8 and 9). The N-terminal domain plays a key role in RNA polymerase assembly by providing the contact surface for α dimerization and the binding of β and β′ subunits, whereas the C-terminal domain plays a regulatory role in transcription by providing the contact surface for transcription regulatory factors including many trans-acting protein factors and cis-acting DNA elements.
The RNA polymerase core enzyme consists of two molecules of the α subunit and one molecule each of the β and β′ subunits. The two α subunits are considered to contact the other subunits differentially and thus to play different roles in RNA polymerase assembly and plausibly also in transcription regulation (1, 6). The elucidation of the role of each α subunit requires a strategy to distinguish between the two α subunits within the assembled RNA polymerase. For this purpose, we have developed a system to reconstitute RNA polymerase containing two different α derivatives in a defined arrangement. The method is based on the following findings: On the N-terminal domain of the α subunit, the β and β′ subunit assembly sites are located in two different places. The β binding sites are located at two positions, one at residues 45–48 and the other around residue 80, whereas the β′ binding sites are also located at two positions, one around residue 80 and the other at residues 173–200 (3–5, 7, 30). An α mutant having the Arg-45-Ala substitution, hereafter designated as [45A]α, retains the ability to dimerize but cannot form α2β complex (5).
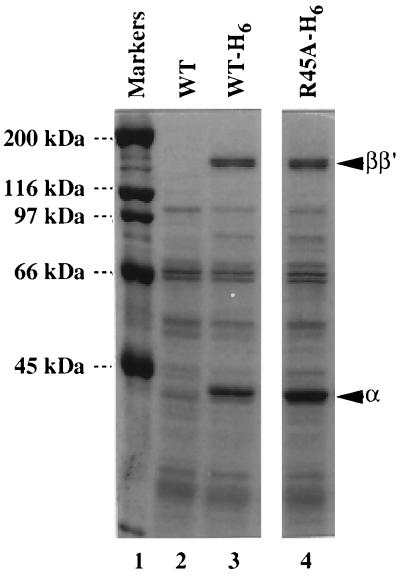
This mutant α can, however, be assembled in vivo into RNA polymerase. Fig. 1 shows the Ni2+-affinity column chromatogram of extracts of cells harboring plasmid pLAW2, pLAW2-H6, or pLA-R45A-H6 for expression of wild-tpe α, His6-tagged wild-type α or His6-tagged [45A]α subunit, respectively. Both wild-type (lane 3) and mutant (lane 4) His6-tagged α subunits were retained on the column and eluted with a buffer containing 300 mM imidazole, whereas wild-type α without His6 tag was recovered in the flow-through fraction (lane 2). Two large subunits, β and β′, were recovered in the column-bound fractions for not only His6-tagged wild-type (lane 3) but also His6-tagged [45A]α (lane 4), indicating that the assembly-defective mutant α was assembled in vivo into RNA polymerase by forming heterodimers with the chromosome-coded wild-type α. In fact, the [45A]α subunit was assembled in vitro into core enzyme by mixing with wild-type α subunit (see below). In the core enzyme containing the α heterodimer, it is expected that the α-WT associates with the β subunit but the mutant [45A]α, being defective in β contact, binds to the β′ subunit.
Figure 1.
Assembly of mutant [45A]α into RNA polymerase. Extracts of E. coli cells expressing wild-type α (lane 2), His6-tagged α (lane 3), or His6-tagged mutant [45A]α (lane 4) were fractionated by chromatography on Ni2+-chelating columns. Bound proteins were eluted with a buffer containing 300 mM imidazole and fractionated by SDS/9% PAGE. The gel was stained with Coomassie brilliant blue.
To obtain pure preparations of such hybrid RNA polymerases, free from α-WT homodimers and containing different species of the α subunit in defined orientations (oriented α-heterodimer), we carried out the mixed reconstitution using the [45A]α mutant with an His6 tag at its N terminus and α-WT, and isolated the hybrid RNA polymerase by Ni2+-affinity column chromatography. An exposed hexa-histidine stretch in a protein molecule immobilizes it on a Ni2+-chelating resin (31, 32). This strategy has already been successfully applied for the isolation of RNA polymerase having the His6 tag on α at either N or C terminus (7, 33).
Reconstitution of Hybrid RNA Polymerases.
For construction of the hybrid RNA polymerases containing one wild-type and one C-terminally truncated α in both of the possible orientations, we carried out mixed reconstitution using two combinations of α subunits: α-WT plus a mutant α carrying both C-terminal deletion of 94 residues and Arg-to-Ala substitution at residue 45 ([45A]α235) and α-235 plus full-length α with R45A mutation. In both cases, the α derivatives carrying the R45A mutation should become associated with the β′ subunit. The first reconstitution mixture included one α-WT and one His6-tagged C-terminally truncated α-235 with [45A]α mutation, and should yield a hybrid enzyme in which α-WT is associated with the β subunit and a C-terminally truncated α is associated with β′ (hereafter designated as αβ-α235β′). The second mixture included α-235 and [45A]α-His6, and should yield the hybrid enzyme α235β-αβ′. None of the α derivatives carrying the [45A] mutation used in this study were able to form RNA polymerase in the absence of intact helper α subunit (data not shown), but all α heterodimers were assembled into complete enzyme as efficiently as α-WT homodimer (data not shown).
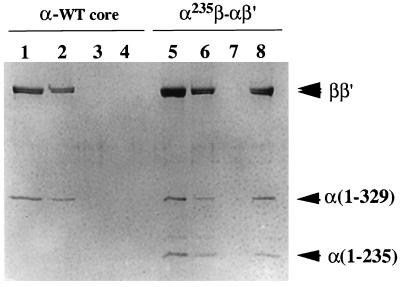
The RNA polymerase preparations produced in this way contained, after purification by DEAE column chromatography, two different species of core enzyme, one containing α homodimers and the other containing α heterodimers with an His6 tag. By Ni2+ column chromatography we could purify the RNA polymerases containing α heterodimers. To prevent their possible contamination by molecules containing α homodimers due to aggregation of the core enzyme, which is favored at low salt concentrations, the enzyme purification was carried out at 0.2 M NaCl concentration. Elution profiles of the Ni2+ column chromatography are shown in Fig. 2. All RNA polymerase without the His6 tag was recovered in the flow-through and wash fractions (Fig. 2, lanes 2 and 3), and none in the eluted fraction (Fig. 2, lane 4). On the other hand, the α235β-αβ′ hybrid RNA polymerase carrying the His6 tag at the N terminus of [45A]α bound to the Ni2+ column, and was recovered in the eluted fraction (Fig. 2, lane 8). The αβ-α235β′ hybrid RNA polymerase with His6 tag at the N terminus of [45A] α235 was also isolated by the same procedure (data not shown). The binding ability of the hybrid enzymes to the Ni2+ column was, however, not high under the conditions employed, and approximately one-half of the His-tagged enzymes were eluted in the flow-through and wash fractions (Fig. 2, lanes 6 and 7). Possibly, the His6 tag associated with the N terminus of the α subunit is partially buried in the assembled core RNA polymerase. We also subjected the purified core enzymes to Ni2+ column chromatography after the addition of σ70 subunit. The holoenzymes carrying the His6 tag at one α N terminus were recovered in the eluted fraction, but the holoenzyme without the His6 tag did not bind to the Ni2+ column (data not shown).
Figure 2.
Subunit composition of the reconstituted RNA polymerases. A hybrid RNA polymerase with the subunit composition α235β-αβ′ was reconstituted from isolated β, β′, [45A]α with the His6 tag, and α-235 subunits. Reconstituted wild-type core enzyme (lanes 1–4) and the hybrid enzyme (lanes 5–8) were subjected to Ni2+ column chromatography as described. An aliquot of each fraction was analyzed by SDS/10% PAGE and Coomassie brilliant blue staining. Lanes: 1 and 5, the samples loaded; 2 and 6, flow-through fractions; 3 and 7, wash fractions; 4 and 8, fractions eluted with imidazole.
Subunit Composition of the Hybrid RNA Polymerases.
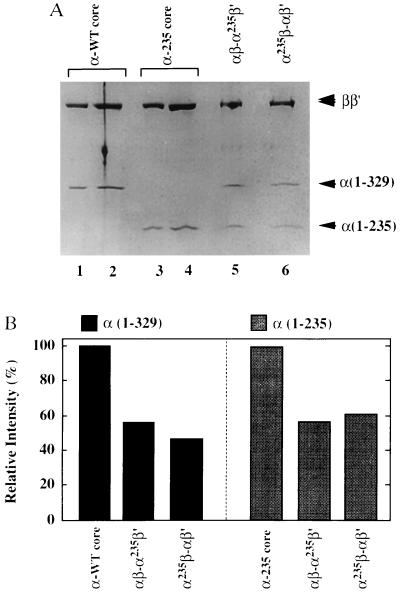
As one way to assess the purity of the hybrid RNA polymerases, we have determined the stoichiometry of the two types of α subunit in the final RNA polymerase preparations. As standards, we used both native core enzyme that had been purified from E. coli cells by repeated chromatography on phosphocellulose columns, and reconstituted core enzyme containing α-235 that has been purified by repeated chromatography on DEAE columns in a HPLC system. The test samples and standards were subjected in parallel to SDS/12% PAGE and stained with Coomassie brilliant blue (Fig. 3A). The intensity of each stained band was quantified by using a PDI-imaging analyzer (PDI Imaging Systems, Huntington Station, NY) and the amounts of the α subunits were calculated after normalizing their intensities against those of the ββ′ subunits (Fig. 3B). Because the relative intensities of intact α and α-235 were almost the same between αβ-α235β′ and α235β-αβ′, we conclude that both of the hybrid enzymes contain the two different α subunits in a stoichiometry of 1:1.
Figure 3.
Stoichiometry of the two α subunits in the reconstituted RNA polymerases. (A) Reconstituted and purified RNA polymerase core enzymes with various combinations of α subunits were analyzed by electrophoresis on an SDS/10% polyacrylamide gel and stained with Coomassie brilliant blue. Lanes: 1 and 2, wild-type core enzyme isolated from E. coli cells; 3 and 4, the reconstituted core enzyme carrying α-235 homodimers; 5 and 6, the hybrid core enzymes containing α heterodimers in the defined orientations indicated above the lanes. The amounts of core enzyme analyzed were: lanes 1, 3, 5, and 6, 1 μg; lanes 2 and 4, 2 μg. (B) Band intensities of the full-length α [α(1–329)] or α-235 [α(1–235)] subunits were quantified and normalized against those of ββ′ subunit bands. The value of 100% represents 2 mol of α subunits per mol each of β and β′ subunits.
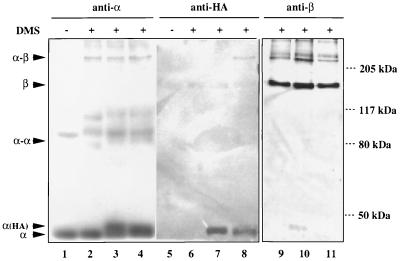
Although [45A]α by itself is completely defective in β subunit binding (5), it has not yet been concluded that [45A]α might regain the ability to associate with β subunit after forming a heterodimer with α-WT. To examine the orientation of the two α subunits within the reconstituted hybrid RNA polymerases directly, we performed protein–protein cross-linking experiments using the reagent DMS. To discriminate between the α subunits in the hybrid enzymes, we added a tag of influenza virus HA nonapeptide sequence (HA tag) at the C terminus of wild-type or [45A] α subunit, and then reconstituted two types of hybrid core enzyme, αHAβ-[45A]αβ′ and αβ-[45A]αHAβ′. The core enzymes were converted into holoenzymes by adding excess σ70 subunit and treated for cross-linking with DMS. After SDS/8% PAGE, Western blotting using specific antibodies against α, β, β′, and σ70 was carried out to identify the cross-linked products. Using anti-α antibody, at least two novel protein bands were detected at the molecular mass of about 80 kDa and above 205 kDa (Fig. 4, lanes 2–4). From the results of Western blotting with antibodies specific for β (Fig. 4, lanes 9–11), β′, and σ70 (data not shown), and taking the molecular mass of each subunit into account, we concluded that the complexes, labeled α-α and α-β in Fig. 4, are products of cross-linking between two α monomers (α dimer) and between α monomer and β subunit (α-β complex). However, no detectable α-β′ cross-link was observed. The fate of the HA-tagged α subunit was then analyzed using anti-HA antibody. The cross-linked α-β complex band from DMS-treated αHAβ-[45A]αβ′ (Fig. 4, lane 8), but not that from αβ-[45A]αHAβ′ (Fig. 4, lane 7), was found to cross-react with anti-HA antibody. Thus, it was confirmed that the [45A]α mutant does not associate with the β subunit.
Figure 4.
Western blotting of the reconstituted RNA polymerases cross-linked with DMS. The reconstituted RNA polymerase holoenzymes were treated with DMS and fractionated by SDS/gel electrophoresis. Gels were treated for immunostaining using anti-α (lanes 1–4), anti-HA (lanes 5–8), and anti-β (lanes 9–11) antibodies. Lanes 1, 2, 5, 6, and 9, holoenzyme with α-WT; lanes 3, 7, and 10, hybrid holoenzyme having β′ subunit-associated HA-tagged α (αβ-[45A]αHAβ′); lanes 4, 8, and 11, hybrid holoenzymes with β subunit-associated HA-tagged α (αHAβ-[45A]αβ′). Lanes 1 and 5 include the control unmodified samples. Subunit monomers (α and β) and cross-linked subunit complexes (α-α and α-β) are shown on the left, whereas the migration positions of molecular weight marker proteins are shown on the right.
Identification of Contact Sites on the UP Element by the Hybrid RNA Polymerases.
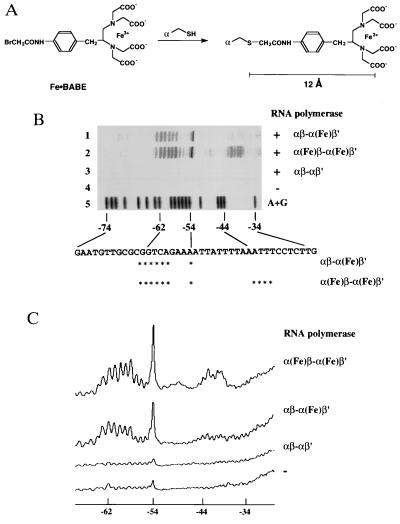
The UP element, an AT-rich stretch of about 20 bases in length located upstream of certain promoters, has transcription-enhancing activity and is recognized by the C-terminal domain of the α subunit (21). To determine the binding sites of the C-terminal domains of the two α subunits on the rrnBP1 UP element DNA, we employed DNA affinity cleavage by a reagent attached to a specific site on proteins (34–36). In this study, a hydroxyl radical-based DNA cleavage moiety, Fe·BABE (refs. 37 and 38; Fig. 5A) was linked to Cys-269 on the α subunit.
Figure 5.
DNA cleavage by RNA polymerase-bound Fe·BABE. (A) Structure of Fe·BABE and its conjugation to a cysteine residue in RNA polymerase α subunit. The length of the reagent is about 12 Å. (B) DNA cleavage by the RNA polymerase having Fe·BABE at Cys-269 of the α subunit. A DNA fragment, 32P-end labeled on the top strand of rrnBP1, was incubated with RNA polymerase reconstituted from Fe·BABE-conjugated α subunits and then subjected to DNA cleavage by adding sodium ascorbate. Lanes: 1, DNA cleavage by the hybrid RNA polymerase modified by Fe·BABE only on the β′-associated α subunits [αβ-α(Fe)β′]; 2, DNA cleavage by the RNA polymerase having both α subunit Fe·BABE modified [α(Fe)β-α(Fe)β′]; 3, control reaction using wild-type RNA polymerase; 4, control reaction without RNA polymerase; 5, A+G-specific Maxam–Gilbert sequence. The nucleotide sequence shown at the bottom represents the top strand of rrnBP1. Asterisks show the main cleavage sites. (C) Cleavage patterns in B were scanned with a Bioimage Analyzer BAS2000 (Fujix).
The wild-type α subunit has four cysteine residues, three in the N-terminal assembly domain (residues 54, 131, and 176) and one in the C-terminal regulatory domain (residue 269). The three cysteine residues in the N-terminal domain could be removed without interfering with the RNA polymerase assembly or transcription activities (O. Ozoline, K.M., T. Negishi, and A.I., unpublished work). Both NMR and site-directed mutagenesis studies indicate that Cys-269 is located on the UP element recognition helix of the α C-terminal domain, the side chain of Cys-269 being partly involved in interaction with the UP element (22, 27, 28). The α derivative [269C]α, having only one cysteine residue (Cys-269) remaining, with alanine substituting for all the other three cysteine residues in the N-terminal domain, was used for site-specific conjugation of Fe·BABE to the α subunit. Isolated mutant α subunit was first alkylated at Cys-269 with Fe·BABE, and the modified α subunit was then used for reconstitution of the RNA polymerase. Since the EDTA linker arm of Fe·BABE is about 12 Å long (Fig. 5A) and the hydroxyl radicals produced diffuse within an effective range of about 10 Å (39), the DNA cleavage reaction should be confined to a region located within a distance of about 22 Å from Cys-269 on the α C-terminal domain. Moreover we performed the DNA cleavage reaction in the presence of 10% glycerol, a known hydroxyl radical scavenger, which improved the resolution of cleavage (34).
We prepared two forms of modified RNA polymerase, one carrying Fe·BABE on a single α subunit and the other carrying Fe·BABE on both α subunits. To introduce Fe·BABE on to just one of the two α subunits within RNA polymerase, we alkylated the α derivative, [45A269C]α (an R45A derivative carrying a single cysteine only at position 269), with Fe·BABE and then reconstituted hybrid RNA polymerase from a mixture of purified α-WT, [45A269C]α-Fe·BABE, β, and β′. This combination yielded the hybrid enzyme carrying Fe·BABE attached at Cys-269 of the β′-associated α subunit. As shown in Fig. 5B, the RNA polymerase modified by Fe·BABE on the C-terminal domains of both α subunits cleaved two separate regions; region I from −43 to −40 and region II from −63 to −54, of the rrnBP1 promoter (Fig. 5B, lane 2). On the other hand, the hybrid RNA polymerase having Fe·BABE only on the β′-associated α subunit cleaved only region II, and not region I (Fig. 5B, lane 1). These results indicate that the two α subunits of the RNA polymerase bind in tandem to the UP element of rrnBP1, and the β′-associated α appears to bind to the promoter–distal region of the UP element.
DISCUSSION
We have developed a new method for reconstitution of hybrid RNA polymerases containing α heterodimers in defined orientations with respect to the two large subunits, β and β′. Our success was based on a combination of the use of a mutant α subunit defective in β binding with the attachment of an His tag to this mutant α, which still retains the ability to bind to the β′ subunit. The hybrid RNA polymerases thus constructed are considered to retain their subunit arrangement during storage and transcription cycles because: (i) the assembly process in vitro is virtually irreversible at the steps of α dimerization and α2β complex formation (1); (ii) the [45A]α mutant is completely inactive in β binding even if it is dissociated from the assembled RNA polymerase (5); (iii) a known temperature-sensitive assembly-defective α mutant carries the rpoA112 mutation (40, 41), which leads to Arg-to-Cys substitution at the same residue (42), supporting the notion that Arg-45 plays a critical role in β binding; (iv) the exchange of α subunits between different forms of reconstituted RNA polymerase does not take place in vitro (43); and (v) protein–protein cross-linking experiments using DMS detected no rearrangement of assembled subunits (this paper).
Extensive studies have been made to analyze the molecular interactions between the α subunit and class I transcription factors and between α and DNA UP elements (for examples see refs. 8, 9, 21, 25, and 28). Several lines of evidence including mutant studies, DNA footprinting, cross-linking experiments, and biophysical measurements indicate direct contacts between the α subunit and transcription factors. The tertiary structure of the α C-terminal domain supports most, if not all, of these previous genetic and biochemical observations (27). One major problem that remained unsolved was the role each of the two individual α subunits plays in RNA polymerase assembly and transcription regulation. The new reconstitution method developed in this study provided us with a valuable tool for identification of the specific functions associated with the individual α subunits in RNA polymerase. In this study we have used a hybrid RNA polymerase containing oriented α heterodimers, in conjunction with the DNA cleavage mediated by the small protein-bound cleaving agent Fe·BABE, to map the two α subunits on the rrnBP1 UP element.
By DNase I footprinting experiments, the α subunit binding site on the rrnBP1 UP element is known to be located approximately between base pairs −40 and −60 relative to the transcription start site (21, 22, 28). It has been believed that the two α C-terminal domains bind in tandem to two helix–turns of DNA. The results of hydroxyl radical-mediated DNA cleavage produced by Fe·BABE, which had previously been linked to Cys-269 on the UP element contact surface of the α C-terminal domain, now indicate that the two α C-terminal domains are aligned in a defined order on the UP element DNA, the β ′-associated α being positioned at the promoter-distal half site. The observed α-DNA interaction is specific because Fe·BABE-modified RNA polymerase did not cleave DNA when it bound to a mutant rrnBP1 without the UP element (data not shown).
The reconstitution of hybrid RNA polymerases with oriented α heterodimers and the conjugation of specific probes at particular residues on the α subunit could together be useful for the identification of molecular contacts between RNA polymerase and trans-acting protein factors with transcription activation or repression activities. As specific probes, we have so far successfully employed those giving fluorescence (O. Ozoline, N. Fujita, K.M., and A.I., unpublished work) or generating cleavage activities toward target DNA or associated proteins (this paper).
Acknowledgments
We thank R. Miyake for technical advice; W. Ross, R. L. Gourse, S. Busby, S. Garges, O. Ozoline, and N. Fujita for discussion; and R. H. Hayward for critical reading of the manuscript. This work was supported by Grants-in-Aid from the Ministry of Education, Science and Culture of Japan and the Proposal-Based Advanced Industrial Technology R&D Program from the New Energy and Industrial Technology Development Organization (NEDO) of Japan.
ABBREVIATIONS
- UP
upstream
- HA
hemagglutinin
- DMS
dimethyl suberimidate
- Fe·BABE
(p-bromoacetamidobenzyl)-EDTA·Fe
References
- 1.Ishihama A. Adv Biophys. 1981;14:1–5. [PubMed] [Google Scholar]
- 2.Igarashi K, Fujita N, Ishihama A. J Mol Biol. 1991;218:1–6. [PubMed] [Google Scholar]
- 3.Kimura M, Fujita N, Ishihama A. J Mol Biol. 1994;242:107–115. doi: 10.1006/jmbi.1994.1562. [DOI] [PubMed] [Google Scholar]
- 4.Kimura M, Ishihama A. J Mol Biol. 1995;248:756–767. doi: 10.1006/jmbi.1995.0258. [DOI] [PubMed] [Google Scholar]
- 5.Kimura M, Ishihama A. J Mol Biol. 1995;254:342–349. doi: 10.1006/jmbi.1995.0621. [DOI] [PubMed] [Google Scholar]
- 6.Hayward R, Igarashi K, Ishihama A. J Mol Biol. 1991;221:23–29. doi: 10.1016/0022-2836(91)80197-3. [DOI] [PubMed] [Google Scholar]
- 7.Kimura M, Ishihama A. Genes Cells. 1996;1:517–528. doi: 10.1046/j.1365-2443.1996.d01-258.x. [DOI] [PubMed] [Google Scholar]
- 8.Ishihama A. Mol Microbiol. 1992;6:3283–3288. doi: 10.1111/j.1365-2958.1992.tb02196.x. [DOI] [PubMed] [Google Scholar]
- 9.Ishihama A. J Bacteriol. 1993;175:2483–2489. doi: 10.1128/jb.175.9.2483-2489.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ebright R H, Busby S. Curr Opin Genet Dev. 1995;5:197–203. doi: 10.1016/0959-437x(95)80008-5. [DOI] [PubMed] [Google Scholar]
- 11.Igarashi K, Ishihama A. Cell. 1991;32:319–325. [Google Scholar]
- 12.Igarashi K, Hanamura A, Makino K, Aiba H, Aiba H, Mizuno T, Nakata A, Ishihama A. Proc Natl Acad Sci USA. 1991;88:8958–8962. doi: 10.1073/pnas.88.20.8958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zou C, Fujita N, Ishihama A. Mol Microbiol. 1992;6:2599–2605. doi: 10.1111/j.1365-2958.1992.tb01437.x. [DOI] [PubMed] [Google Scholar]
- 14.Jair K-W, Martin R G, Rosner J L, Fujita N, Ishihama A, Wolf R E., Jr J Bacteriol. 1995;177:7100–7104. doi: 10.1128/jb.177.24.7100-7104.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tao K, Fujita N, Ishihama A. Mol Microbiol. 1993;7:859–864. doi: 10.1111/j.1365-2958.1993.tb01176.x. [DOI] [PubMed] [Google Scholar]
- 16.Tao K, Zou C, Fujita N, Ishihama A. J Bacteriol. 1995;177:6740–6744. doi: 10.1128/jb.177.23.6740-6744.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jair K-W, Yu X, Skarstad K, Thony B, Fujita N, Ishihama A, Wolf R E., Jr J Bacteriol. 1996;178:2507–2513. doi: 10.1128/jb.178.9.2507-2513.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jair K-W, Fawcett W P, Fujita N, Ishihama A, Wolf R E., Jr Mol Microbiol. 1996;19:307–317. doi: 10.1046/j.1365-2958.1996.368893.x. [DOI] [PubMed] [Google Scholar]
- 19.Lawley B, Fujita N, Ishihama A, Pittard A J. J Bacteriol. 1995;177:238–241. doi: 10.1128/jb.177.1.238-241.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choy H E, Park S W, Aki T, Parrack P, Fujita N, Ishihama A, Adhya S. EMBO J. 1995;14:4523–4530. doi: 10.1002/j.1460-2075.1995.tb00131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ross W, Gosink K K, Salomon J, Igarashi K, Zou C, Ishihama A, Severinov K, Gourse R L. Science. 1993;262:1407–1413. doi: 10.1126/science.8248780. [DOI] [PubMed] [Google Scholar]
- 22.Gaal T, Ross W, Blatter E E, Tang H, Jia X, Krishnan V V, Assa-Munt N, Ebright R H, Gourse R L. Genes Dev. 1996;10:16–26. doi: 10.1101/gad.10.1.16. [DOI] [PubMed] [Google Scholar]
- 23.Giladi H, Murakami K, Ishihama A, Oppenheim A B. J Mol Biol. 1996;260:484–491. doi: 10.1006/jmbi.1996.0416. [DOI] [PubMed] [Google Scholar]
- 24.Tang Y, Murakami K, Ishihama A, deHaseth P L. J Bacteriol. 1996;178:6945–6951. doi: 10.1128/jb.178.23.6945-6951.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blatter E, Ross W, Tang H, Gourse R L, Ebright R H. Cell. 1994;78:889–896. doi: 10.1016/s0092-8674(94)90682-3. [DOI] [PubMed] [Google Scholar]
- 26.Negishi T, Fujita N, Ishihama A. J Mol Biol. 1995;248:723–728. doi: 10.1006/jmbi.1995.0254. [DOI] [PubMed] [Google Scholar]
- 27.Jeon Y H, Negishi T, Shirakawa M, Yamazaki T, Fujita N, Ishihama A, Kyogoku Y. Science. 1995;270:1495–1497. doi: 10.1126/science.270.5241.1495. [DOI] [PubMed] [Google Scholar]
- 28.Murakami K, Fujita N, Ishihama A. EMBO J. 1996;15:4358–4367. [PMC free article] [PubMed] [Google Scholar]
- 29.Rao L, Ross W, Appleman J A, Gaal T, Leirmo S, Schlax P J, Record T, Jr, Gourse R L. J Mol Biol. 1994;235:1421–1435. doi: 10.1006/jmbi.1994.1098. [DOI] [PubMed] [Google Scholar]
- 30.Heyduk T, Heyduk E, Severinov K, Tang H, Ebright R H. Proc Natl Acad Sci USA. 1996;93:10162–10166. doi: 10.1073/pnas.93.19.10162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Houchuli E, Doebeli H, Schachner A. J Chromatogr. 1987;411:177–184. doi: 10.1016/s0021-9673(00)93969-4. [DOI] [PubMed] [Google Scholar]
- 32.Smith M C, Furman T C, Ingolia T D, Pidgeon C. J Biol Chem. 1988;263:7211–7215. [PubMed] [Google Scholar]
- 33.Tang H, Severinov K, Goldfarb A, Ebright R H. Proc Natl Acad Sci USA. 1995;92:4902–4906. doi: 10.1073/pnas.92.11.4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dumoulin P, Ebright R H, Knegtel R, Kaptein R, Granger-Schnarr M, Schnarr M. Biochemistry. 1996;35:4279–4286. doi: 10.1021/bi9529162. [DOI] [PubMed] [Google Scholar]
- 35.Heilek G M, Noller H F. Science. 1996;272:1659–1662. doi: 10.1126/science.272.5268.1659. [DOI] [PubMed] [Google Scholar]
- 36.Lavoie B D, Shaw G S, Millner A, Chaconas G. Cell. 1996;85:761–771. doi: 10.1016/s0092-8674(00)81241-6. [DOI] [PubMed] [Google Scholar]
- 37.Rana T M, Meares C F. Proc Natl Acad Sci USA. 1991;88:10578–10582. doi: 10.1073/pnas.88.23.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Greiner, D. P., Miyake, R., Moran, J. K., Jones, A. D., Negishi, T., Ishihama, A. & Meares, C. F. (1996) Bioconj. Chem., in press. [DOI] [PubMed]
- 39.Dervan P B. Methods Enzymol. 1991;208:497–515. doi: 10.1016/0076-6879(91)08026-e. [DOI] [PubMed] [Google Scholar]
- 40.Ishihama A, Shimamoto N, Aiba H, Kawakami K, Nashimoto H, Tsugawa A, Uchida H. J Mol Biol. 1980;137:137–150. doi: 10.1016/0022-2836(80)90321-6. [DOI] [PubMed] [Google Scholar]
- 41.Kawakami K, Ishihama A. Biochemistry. 1980;19:3491–3495. doi: 10.1021/bi00556a013. [DOI] [PubMed] [Google Scholar]
- 42.Igarashi K, Fujita N, Ishihama A. Nucleic Acids Res. 1990;18:5945–5948. doi: 10.1093/nar/18.20.5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zou C, Fujita N, Ishihama A. J Mol Biol. 1994;236:1283–1288. doi: 10.1016/0022-2836(94)90057-4. [DOI] [PubMed] [Google Scholar]