Abstract
We have previously identified and characterized a heparin-binding cell surface protein (heparin/heparan sulfate-interacting protein, or HIP) present on epithelial and endothelial cells. A synthetic peptide mimicking a heparin-binding domain of HIP is now shown to bind a small subset of heparin molecules with high affinity and, therefore, presumably recognizes a specific structural motif in the heparin molecule. Further analyses revealed that the heparin molecules exhibiting a high affinity for the HIP peptide also show an extremely high affinity for antithrombin III (AT-III), a cofactor required for heparin’s anticoagulant activity. The HIP peptide was shown to compete with AT-III for binding to heparin and to neutralize the anticoagulant activity of heparin in blood plasma assays. Furthermore, the heparin subfraction that binds to the HIP peptide with high affinity exhibits an extremely high anticoagulant activity. We conclude that although the HIP peptide shows no sequence similarity with AT-III, the two proteins recognize the same or similar structural motifs in heparin.
The glycosaminoglycans heparin (Hep) and heparan sulfate (HS) appear to exert their biological functions through regulatory interactions with specific target proteins. Because of the microheterogeneity of the sulfate substitutions of the polysaccharides, it has been speculated that specific monosaccharide sequences in Hep bind to selective domains in target proteins (1–5). A Hep hexasaccharide can theoretically occur in >105 different structural forms, which allows for sufficient structural variations, expected for an information molecule. On the other hand, the biosynthesis of a structurally heterogeneous glycosaminoglycan seems to result from a series of incomplete enzymatic reactions, raising concern about the genetic control of glycosaminoglycan fine structure (6). At least one protein, antithrombin III (AT-III), binds to a specific sequence present in some Hep molecules but not in others (1–3). Other proteins, such as the FGFs bind to Hep with surprisingly high affinity; however, attempts to identify specific binding sites in Hep for different growth factors have given inconclusive and sometimes conflicting results. Here we report on a synthetic peptide derived from a cell surface Hep/HS-interacting protein (HIP), which binds to a Hep sequence that is similar or identical to that specifically recognized by AT-III.
HIP has been identified in human uterine epithelia and a variety of other human epithelial and endothelial cells (7, 8). Analysis of the predicted amino acid sequence of HIP revealed a putative Hep/HS-binding motif. A synthetic peptide corresponding to this motif (HIP peptide) selectively binds Hep and certain forms of HS expressed by human cell lines and supports attachment of a variety of mammalian adherent cell lines in a HS-dependent fashion (S.L. and D.D.C., unpublished work). In the current study, we used HIP peptide affinity chromatography to further investigate the interactions between HIP peptide and Hep polysaccharide chains. Our data indicate that HIP peptide binds a subset (1–5% of total) of Hep from commercial preparations with much higher affinity than unfractionated Hep. The subset of Hep that binds HIP with high affinity (HA-Hep) is also the subset that binds AT-III with highest affinity. Coagulation assays using diagnostic tests containing normal human plasma demonstrated that HIP peptide can compete with AT-III for Hep binding and neutralize AT-III inhibition of both factor Xa (FXa) and thrombin activity. Furthermore, HA-Hep showed an anticoagulant activity ≈10 times higher than that of unfractionated Hep. Finally, purified intact HIP from HEC cells, a human uterine cell line, also inhibited the anticoagulant activities of the AT-III–Hep complex. Therefore, these results suggest that HIP may play a physiological role as a modulator of blood coagulation.
MATERIALS AND METHODS
Materials.
[3H]Hep (0.44 mCi/mg; 1 Ci = 37 GBq) was purchased from DuPont/NEN. Dulbecco’s PBS was from GIBCO. Imject activated immunogen conjugation kits were purchased from Pierce. COATEST Heparin and COATEST Antithrombin kits were purchased from Helena Laboratories. AT-III-agarose, sodium chloride, potassium chloride, and sodium phosphate were purchased from Sigma. All chemicals used were reagent grade or better.
Peptide Synthesis and HIP Peptide Affinity Matrix.
The synthetic HIP peptide, derived from a segment of the predicted amino acid sequence of HIP (7), CRPKAKAKAKAKDQTK, was synthesized on a Vega 250 peptide synthesizer using fluorenylmethoxycarbonyl (Fmoc) methodology (9). This synthetic peptide was conjugated to maleimide-activated BSA (Pierce) through the sulfhydryl group of cysteine in the peptide following the coupling procedures provided by the manufacturer. HIP peptide affinity matrix was formed by cross-linking the BSA-conjugated HIP peptide to cyanogen bromide-activated Sepharose (Sigma) in the presence of N-acetylated Hep. Inclusion of acetylated Hep was adopted to produce a more stable affinity matrix by shielding the Hep binding sites of the HIP peptide–BSA complex from cross-linking to the Sepharose beads. Acetylation of Hep was performed following the method previously described (10).
HIP Peptide Affinity Chromatography.
The HIP peptide affinity matrix was packed into a 1-ml fast protein liquid chromatography (FPLC) column (Pharmacia) and conditioned by repeated washing in 10 mM phosphate buffer (pH 7.4) with a gradient of 0.15–4.0 M NaCl. Commercial [3H]Hep was resuspended in 0.15 M NaCl/10 mM phosphate, pH 7.4, and loaded into the column. The column was eluted with a linear gradient from 0.15 to 4.0 M NaCl in a 10 mM phosphate buffer (pH 7.4) at a rate of 0.5 ml/min. Fractions of 0.5 ml were collected, and aliquots were analyzed for radioactivity. Large-scale fractions were performed on a 5 ml of HIP peptide affinity matrix. [3H]Hep resuspended in 0.15 M NaCl/10 mM phosphate, pH 7.4, was loaded into the column. The column then was washed sequentially with 0.15 M NaCl/10 mM phosphate, pH 7.4 (run-through Hep, or RT-Hep), and 0.45 M NaCl/10 mM phosphate, pH 7.4 (low-affinity Hep, or LA-Hep), extensively. Finally, the column was eluted with 3.0 M NaCl/10 mM phosphate, pH 7.4 (HA-Hep). Radioactivity in an aliquot (50 μl) from each fraction (1 ml per fraction) was determined by liquid scintillation counting. Each of these affinity classes of Hep was collected, extensively dialyzed against doubly distilled, deionized H2O, lyophilized, and used for further analysis.
Hep oligosaccharides were obtained by partial deaminative cleavage of the polysaccharide with nitrous acid at pH 1.5 (11). The reaction was allowed to proceed for 10 min and interrupted by the addition of ammonium sulfamate in a 4-fold molar excess over nitrite. The resulting mixture was separated by gel permeation chromatography on a 1.5 × 170 cm Sephadex G-50 (superfine) column and eluted with 0.5 M NH4HNO3 at a flow rate of 6.5 ml/hr. Fractions of 2.0 ml were collected and uronic acid concentration of individual fraction was determined by the carbazole method (12). The peaks corresponding to tetra-, hexa-, octa- and decasaccharides, as determined by elution positions relative to an internal [3H]octasaccharide standard, were pooled and reductively labeled with [3H]NaBH4. Each size-fractionated mixture was repurified by a second round of gel permeation chromatography performed under the identical conditions to eliminate possible carryover from adjacent peaks. The eluted fractions were analyzed for radioactivity and peak fractions representing size-homogenous oligosaccharides were collected. Estimated specific activity of the radiolabeled oligosaccharides was 2–20 × 104 dpm/nmol. The [3H]Hep oligosaccharides were loaded onto the HIP affinity matrix, and the column was extensively washed with 1 M NaCl/10 mM phosphate, pH 7.4. Bound Hep oligosaccharides were eluted with 3 M NaCl/10 mM phosphate, pH 7.4. Radioactivity in each fraction was determined by liquid scintillation counting.
Determination of Size and Charge Density of Hep Species.
The size of the Hep fractions was analyzed on a 1 × 30 cm Superose 12 column (Pharmacia) equilibrated and eluted with 2 M guanidine hydrochloride, 0.01% (wt/vol) octyl glucoside, 20 mM Tris-acetate (pH 7.0), and 0.02% (wt/vol) sodium azide at a flow rate of 0.7 ml/min at room temperature, with a back pressure of ≈300 psi (1 psi = 6.89 kPa). Fractions were collected every 0.5 min, and the radioactivity was determined by liquid scintillation counting.
The charge density of Hep fractions was analyzed by anion exchange liquid chromatography on a 0.5 × 5 cm Mono Q column (Pharmacia). Samples were loaded into a column equilibrated with 0.5 M urea, 0.01% (wt/vol) octyl glucoside, 20 mM Tris-acetate (pH 7.0), and 0.02% (wt/vol) sodium azide, and the column was eluted with a gradient of 0–4 M NaCl in the same buffer. The column was pumped at a flow rate of 1 ml/min at room temperature with a back pressure of ≈450 psi. Fractions were collected every 0.5 min, and the radioactivity determined by liquid scintillation counting.
AT-III and Basic Fibroblast Growth Factor (bFGF) Affinity Chromatography.
AT-III-agarose (Sigma) was resuspended in 10 mM phosphate (pH 7.4), packed into a 1-ml FPLC column, and conditioned by repeated gradient washing of 0–2.0 M NaCl/10 mM phosphate, pH 7.4. Unfractionated [3H]Hep or HA-Hep were loaded onto the AT-III-agarose column. Then the column was washed with 10 mM phosphate (pH 7.4) and eluted with a gradient of 0–2.0 M NaCl at a flow rate of 0.5 ml/min. Fractions of 0.5 ml were collected and analyzed for radioactivity. Recombinant bFGF was coupled to CNBr-activated Sepharose 4B in the presence of a 5-fold molar excess of N-acetylated Hep to protect potential Hep-binding sites as described (10). The bFGF affinity chromatography of unfractionated [3H]Hep and HA-Hep was performed using the procedures described above for affinity chromatography on AT-III-agarose.
Coagulation Assays.
COATEST Heparin (for determination of Hep in plasma) and COATEST Antithrombin (for determination of AT-III–Hep cofactor activity), were employed to examine the effect of HIP peptide on the anticoagulatory activity of Hep and AT-III, respectively. Coagulation assays were performed following the procedures provided by manufacturer. The principle for determination of Hep concentration in plasma is as follows: (i) Hep + AT-III (excess) → [AT-III–Hep]; (ii) [AT-III–Hep] + FXa (excess) → [AT-III–Hep–FXa] + FXa (remaining); and (iii) S-2222 + FXa (remaining) → peptide + pNA (yellow). S-2222 (Bz-Ile-Glu-Gly-Arg-pNA) is a chromogenic substrate susceptible to FXa (pNA stands for p-nitroaniline). Briefly, standard samples including normal human plasma, 0.1 unit/ml of AT-III, and varying concentrations of Hep from 0.01 to 0.07 unit/ml (Elkins-Sinn, Cherry Hill, NJ) were incubated at 37°C for 3–4 min, then FXa (0.355 nkat) kept at room temperature was added and mixed well. The mixture was incubated at 37°C for 30 sec and the chromogenic substrate S-2222 (100 μmol, 37°C) was added and mixed well. The mixture was further incubated at 37°C for exactly 3 min. The reaction was stopped by adding 20% (vol/vol) acetic acid. To test effect of HIP peptide, different concentrations of HIP peptide were added to the standard sample containing 0.07 unit/ml of Hep, and the Hep activity was analyzed as described above. The principle for determination of AT-III–Hep cofactor activity in plasma is as follows: (i) AT-III + Hep (excess) → [AT-III–Hep]; (ii) [AT-III–Hep] + thrombin (excess) → [AT-III–Hep–thrombin] + thrombin (residual); and (iii) S-2238 + thrombin (residual) → peptide + pNA (yellow). S-2238 (H-d-Phe-Pip-Arg-pNA) is a chromogenic substrate susceptible to thrombin. Briefly, standard samples containing normal human plasma at different dilutions (creating varying concentrations of plasma AT-III) were incubated, in the presence of Hep, at 37°C for 3–6 min. Then, thrombin was added (1.77 nkat) and mixed well, and the solution further incubated at 37°C for 30 sec. The S-2238 substrate was added and incubated at 37°C for 30 sec. The reaction was stopped with 20% (vol/vol) acetic acid. To test the effect of HIP peptide, different concentrations of HIP peptide were added to the standard sample containing 100% normal AT-III concentration ([Hep] ≈ 0.05 unit/ml), and the assay was carried out as described above.
Determination of Anticoagulant Activities of Fractionated Hep.
Hep (1000 units/ml; Elkins-Sinn) was fractionated in a HIP peptide affinity column using stepwise elution as described above resulting in three affinity classes of Hep: RT-Hep, from HIP peptide affinity chromatography; LA-Hep, low-affinity (0.45 M NaCl eluate) Hep from HIP peptide affinity chromatography; and HA-Hep, high-affinity (3.0 M NaCl eluate) Hep from HIP peptide affinity chromatography. The anticoagulant activity of each Hep fraction was determined by FXa-dependent coagulation assay (COATEST Heparin) as described above. Comparison of specific activities was based upon the uronic acid content determined by carbazole assay (12).
RESULTS
A Subset of Hep Chains Bind to HIP Peptide with High Affinity via a Small Oligosaccharide Motif.
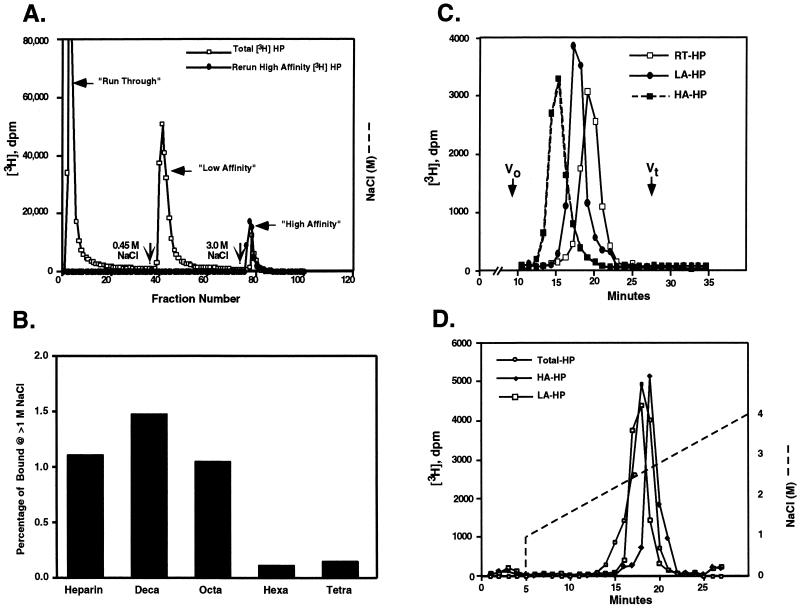
Commercial [3H]Hep was fractionated by HIP peptide affinity chromatography using salt gradient elution. Initial, continuous salt gradient elutions revealed that most [3H]Hep appeared in the run-through fractions (0.15 M NaCl/PBS; RT-Hep) while another portion of [3H]Hep bound to the column and eluted at 0.3–0.4 M NaCl/PBS elution (LA-Hep). A small percentage of [3H]Hep (≈1–5% of total [3H]Hep) bound to HIP peptide affinity matrix tightly and eluted at 2.2 M NaCl (HA-Hep). Large-scale preparations of three affinity classes of Hep for further study were obtained using a stepwise elution procedure with 0.15 M, 0.45 M, and 3.0 M NaCl (Fig. 1A). Rechromatography of each affinity class of [3H]Hep after dialysis resulted in near quantitative elution (>90%) at the salt concentration used for the initial fractionation. These results suggest that HIP peptide binds to a particular structural motif present only in a subset of Hep molecules. To determine the minimum size of the structural motif recognized by HIP, we tested the ability of [3H]Hep oligosaccharides of different sizes to bind to the HIP peptide affinity matrix. As shown in Fig. 1B, a small percentage of both octa- and decasaccharides bound to HIP peptide affinity matrix and eluted at >1.0 M NaCl, whereas no high affinity binding of tetra- and hexasaccharides to HIP peptide affinity matrix was detected. In generating the oligosaccharides, the N-sulfated glucosamine residue originally at the reducing end is effectively destroyed. The next N-sulfated glucosamine required would occur two residues in toward the nonreducing terminus. Therefore, we conclude that the motif recognized by HIP is contained in a penta- or hexasaccharide of intact Hep molecules—i.e., the minimum size required for AT-III binding (1–3).
Figure 1.
HIP peptide affinity chromatography of Hep and Hep oligosaccharides. (A) [3H]Hep dissolved in 0.15 M NaCl/PBS, pH 7.4, was applied to a 1-ml HIP peptide affinity FPLC column equilibrated with the same buffer (□). The column then was eluted first with 0.15 M NaCl then with 0.45 M or 3.0 M NaCl in PBS (indicated by vertical arrows) at a flow rate of 0.5 ml/min to generate RT-, LA-, and HA-Hep fractions, respectively. Rechromatography of the high-affinity fraction (•) demonstrated quantitative rebinding and elution in the 3.0 M NaCl eluate. Each fraction (0.5 ml per fraction) was collected and radioactivity determined by liquid scintillation counting. (B) [3H]Hep oligosaccharide or [3H]Hep was resuspended in 0.15 M NaCl/10 mM phosphate, pH 7.4, and loaded onto a 1-ml HIP peptide affinity FPLC column equilibrated with 0.15 M NaCl/10 mM phosphate, pH 7.4. The column then was washed with 1.0 M NaCl/10 mM phosphate, pH 7.4, extensively. Finally, the column was eluted with 3.0 M NaCl/10 mM phosphate, pH 7.4. The percentage of [3H]Hep or [3H]Hep oligosaccharide binding to the column and eluting at >1.0 M NaCl is shown. (C) Run-through [3H]Hep (RT-Hep, □), low-affinity [3H]Hep (LA-Hep, •), and high-affinity [3H]Hep (HA-Hep, ▪) isolated as described above were analyzed on a Superose 12 column. The elution positions of blue dextran (Vo) and [3H]glucosamine (Vt) standards are indicated. (D) Bulk [3H]Hep (○), LA-Hep (□), and HA-Hep (♦) were analyzed by Mono Q anion exchange liquid chromatography column. The NaCl gradient conditions are indicated by a dashed line.
HA-Hep Has a Larger Molecular Weight and Higher Negative Charge Density than Unfractionated Hep, RT-Hep, and LA-Hep.
To determine the average size and charge density of different HIP-binding Hep fractions, each affinity class of Hep was subjected to gel permeation and ion exchange liquid chromatography. HA-Hep has the largest median size when analyzed on a column of Superose 12, LA-Hep has an intermediate median size, and RT-Hep is enriched in smaller size classes of Hep chains (Fig. 1C). Comparison of elution positions with polysaccharide standards indicate that the median molecular weight of HA-Hep is ≈40,000—i.e., ≈4 times greater than that of unfractionated Hep (median molecular weight of 10,000). Thus, HA-Hep is enriched in larger species of Hep chains. Results of anion exchange chromatography indicate that HA-Hep also has a slightly higher average negative charge density than either LA-Hep or unfractionated Hep (Fig. 1D), indicating that the Hep chains selectively recognized by HIP peptide may have an unusually high sulfate density. Collectively, these results indicate that HA-Hep chains differ, both in terms of size and charge, from unfractionated and other HIP peptide affinity classes of Hep.
HA-Hep Contains Polysaccharide Structures Recognized by AT-III with High Affinity and Is Highly Enriched in Anticoagulant Activity.
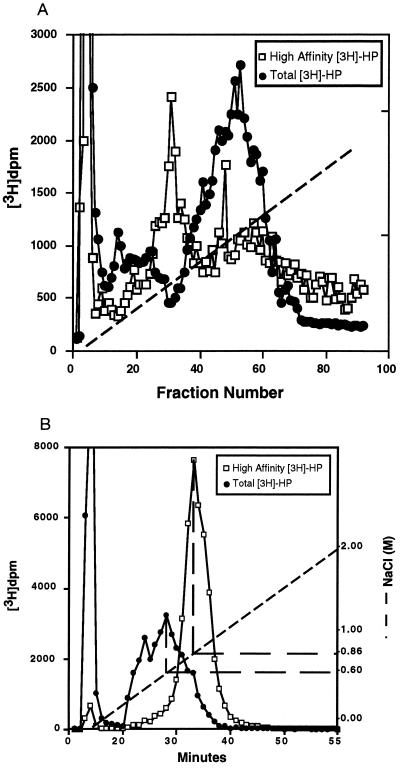
Since HIP peptide recognizes a specific structural motif in Hep, it was of interest to determine if similar structural features were required for Hep binding to other Hep-binding proteins, such as AT-III or bFGF (1, 3, 13–15). Therefore, HA-Hep was subjected to both AT-III and bFGF affinity chromatography. HA-Hep was not enriched in species that specifically bound to bFGF affinity matrix with high affinity (Fig. 2A). In contrast, HA-Hep almost quantitatively bound to AT-III affinity matrix and was eluted as a single homogeneous peak at 0.86 M NaCl. As shown in Fig. 2B, AT-III affinity chromatography of unfractionated [3H]Hep resulted in several distinct fractions—i.e., one eluting at the run-through and another eluting at 0.6 M NaCl with two smaller peaks at 0.4 M NaCl and 0.86 M NaCl. Thus, HA-Hep is highly enriched in Hep species that bind to AT-III with highest affinity. These observations suggest that the same or similar Hep motif(s) are recognized by both HIP peptide and AT-III. Therefore, the structural requirements for high affinity Hep binding to HIP peptide and bFGF appear to be distinct. Hep octasaccharides, which bound to HIP peptide with high affinity, also were greatly enriched for species that bound to AT-III with high affinity. Approximately 50% of these octasaccharides bound to AT-III-agarose and required more than 0.45 M NaCl for elution (data not shown). These observations suggest that HA-Hep contains specific sequence motifs critical for high-affinity binding to both HIP peptide and AT-III.
Figure 2.
bFGF and AT-III affinity chromatography of bulk Hep and HA-Hep. (A) Unfractionated [3H]Hep (•) or HA-Hep (□) were resuspended in PBS (pH 7.4) and then loaded onto a 1-ml bFGF affinity FPLC column equilibrated in PBS (pH 7.4). The column was eluted with a gradient of 0–2 M NaCl/10 mM phosphate, pH 7.4 (dashed line), at a flow rate of 0.5 ml/min. Each fraction (0.5 ml per fraction) was collected and radioactivity determined by liquid scintillation counting. (B) Unfractionated [3H]Hep (•) or HA-Hep (□) resuspended in PBS (pH 7.4) was loaded onto a 1-ml AT-III affinity FPLC column equilibrated in PBS (pH 7.4). The column was eluted with a gradient of 0–2 M NaCl/10 mM phosphate, pH 7.4 (dashed line), at a flow rate of 0.5 ml/min. Each fraction (0.5 ml per fraction) was collected, and radioactivity was determined by liquid scintillation counting.
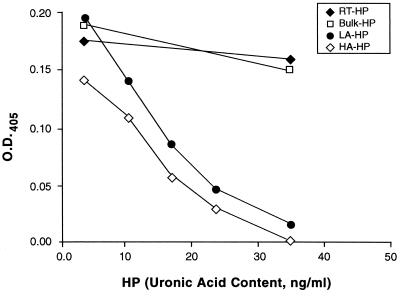
Since the anticoagulant activity of Hep primarily is exerted via binding to and activation of AT-III, it was of interest to determine the specific anticoagulant activity of Hep fractions obtained by HIP peptide affinity chromatography. To this end, we used a FXa-dependent coagulation assay kit. As shown in Fig. 3, HA-Hep displayed the highest anticoagulant activity and was at least 10-fold more potent than unfractionated Hep on a mass basis. Since the median size of HA-Hep is ≈4 times that of unfractionated Hep, anticoagulant activity of HA-Hep is at least 40-fold higher on a molar basis; however, larger Hep chains are more likely to have multiple high-affinity binding sites than smaller chains and so may further increase the apparent specific activity. LA-Hep also had higher anticoagulant activity than that displayed by unfractionated Hep. This activity was consistently lower than that of HA-Hep, but may not be significantly different. RT-Hep had marginal anticoagulant activity. Collectively, these results demonstrate that HA-Hep contains Hep molecules with exceptionally high anticoagulant activity.
Figure 3.
Anticoagulant activity of different affinity classes of Hep selected by HIP peptide affinity chromatography. FXa-dependent chromogenic reaction was performed as described. Bulk (□), RT-Hep (♦), LA-Hep (•), and HA-Hep (⋄) were added to the assay at the indicated concentrations (uronic acid content). The concentration of 36.25 ng/ml (uronic acid content) of unfractionated Hep is equivalent to 0.01 unit/ml of bulk Hep. Results are means of duplicate determinations with SDs <15%.
HIP Peptide and Purified HIP Antagonize AT-III–Hep Actions on Blood Coagulation Activities.
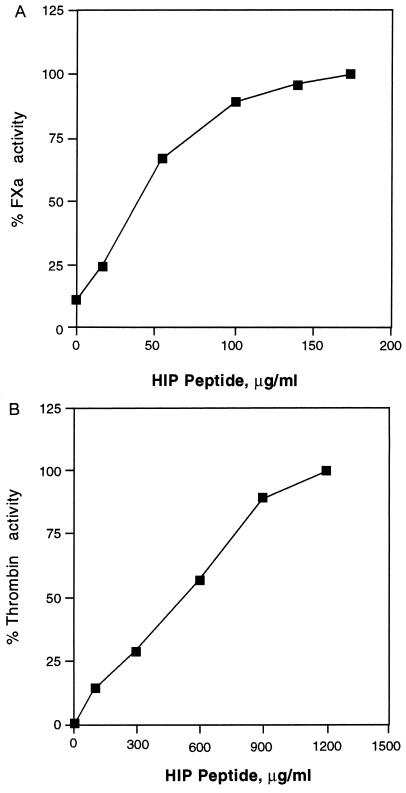
Based on the above observations, it was hypothesized that HIP peptide competes with AT-III for Hep binding and might modulate the anticoagulant activity of the polysaccharide. To test this, both FXa- and thrombin-dependent coagulation assays were used. As shown in Fig. 4A, the addition of HIP peptide in the presence of the AT-III–Hep complex restored FXa-dependent activity in a concentration-dependent and saturable manner. Full restoration of FXa activity was achieved at a HIP peptide concentration of 150 μg/ml. A scrambled sequence of this peptide (CKDKTKPRAKQAAAKK) had no significant effect on activity. Furthermore, HIP peptide alone at the same or higher concentrations in the absence of FXa did not result in any color reaction (data not shown). Therefore, HIP peptide appears to functionally compete with AT-III for Hep binding and inhibit the formation of Hep–AT-III–FXa complexes. Similarly, thrombin activity was restored by HIP peptide. Again, this effect was HIP peptide concentration-dependent up to 1 mg/ml (Fig. 4B). Furthermore, purified HIP (S.L. and D.D.C., unpublished work) also was able to restore, in a dose-dependent fashion, FXa activity in the presence of AT-III and maximally inhibitory doses of Hep (0.07 unit/ml). Collectively, these results demonstrate that HIP and HIP peptide effectively antagonize AT-III–Hep complex actions in blood coagulation events.
Figure 4.
HIP peptide antagonizes Hep action in FXa- and thrombin-dependent coagulation assays. The FXa- and thrombin-dependent chromogenic reaction was performed as described. Data are expressed as the percentage of the AT-III–Hep complex inhibitable enzymatic activity in each assay. All reactions were performed in the presence of 0.1 unit/ml of AT-III and 0.07 unit/ml Hep and the indicated HIP peptide concentrations. Results are means of duplicate determinations with SDs <15%. (A) FXa activity. (B) Thrombin activity.
DISCUSSION
Our results demonstrate that HIP peptide specifically recognizes AT-III-binding Hep species and modulates blood coagulation activities. Previous studies by Bae et al. (16) have shown specific recognition of Hep motifs by a synthetic peptide corresponding to amino acids 123–139 of AT-III. This is the first demonstration of a non-AT-III-derived synthetic peptide recognizing a specific structural motif in Hep. The apparent high affinity between HIP peptide and HA-Hep is particularly remarkable considering the simple structure of the 16-aa peptide. Protein structure-predicting algorithms predict that HIP peptide would occur in an α-helical domain of the intact protein. Interestingly, biophysical studies indicate that helix-forming peptides optimally promote interactions with Hep, although interactions with particular Hep motifs were not identified in these studies (17). Therefore, studies of the interaction of native and modified HIP peptide with Hep may provide important information on the structure–function relationships of interacting proteins and Hep motifs.
The interaction between AT-III and Hep has been studied extensively (18, 19). A unique pentasaccharide sequence in Hep is specifically recognized by AT-III (1, 3, 15). Since the procedure used to generate Hep oligosaccharides destroys glucosaminyl residues at the reducing terminus, the present studies indicate that HIP peptide also recognizes a motif of five to six saccharide units. Furthermore, Hep oligosaccharides that bind to HIP peptide with high affinity also bind to AT-III with high affinity. If the motif in Hep required for high-affinity binding to HIP peptide is indeed larger than a pentasaccharide, it appears that the additional monosaccharide unit also facilitates AT-III binding, since HA-Hep represents a subset of HA-Hep–AT-III with particularly high affinity for the protease inhibitor. Furthermore, the anticoagulant activity of HA-Hep is at least 10- to 40-fold higher than that of unfractionated Hep. Uncertainty exists in the exact degree in increase in specific activity of HA-Hep, since this fraction is larger than unfractionated Hep, tending to further increase this value. On the other hand, larger Hep chains may contain more than one high-affinity binding site, which might lower the specific activity when considered on a per-site basis. In any case, HA-Hep is substantially enriched for anticoagulant activity and with regard to affinity for AT-III–Sepharose.
Cell surface-expressed HIP has been suggested to be involved in Hep/HS-dependent cell–cell or cell–extracellular matrix interactions (7, 8). Results of the current study suggest another possible function of HIP, namely, modulation of blood coagulation activities. Since both purified HIP and HIP peptide compete with AT-III for Hep binding and modulate coagulation events, it is likely HIP also competes with AT-III for Hep binding in vivo. HIP is expressed by most human epithelial cells and cell lines as well as normal human umbilical vein vascular endothelial cells (7) and is not secreted to an appreciable extent (8). Therefore, under normal situations, cell surface HIP can participate in cell–cell or cell–matrix interactions of the expressing cell. When tissue injury occurs, HIP may be released locally into the injured area, where it competes with AT-III for HS/Hep binding and neutralizes the anticoagulant AT-III–HS/Hep complex. Consequently, a higher activity of coagulant proteases would be available at sites of injury. Moreover, HIP also may cooperate with other adhesion proteins to facilitate cell binding to the injured endothelium or epithelium through its HS/Hep-binding function. Collectively, these actions might accelerate the recovery of the injured area. Blood coagulation has long been a major area of research, and new factors continue to be identified (2, 4, 5, 20, 21). The evidence that HIP and HIP peptide antagonize Hep action and modulate coagulation events suggests that HIP might be a novel modulator of the coagulation cascade.
Acknowledgments
We thank Dr. Mimi DeSouza, Margaret French, David Hoke, Dr. Andrew Jacobs, JoAnne Julian, Ruth Pimental, Drs. Gloria Regisford and Scott Smith, Gulnar Surveyor, and Xinhui Zhou for their helpful comments and critical reading of the manuscript. We thank Ms. Sharron Kingston for her excellent secretarial assistance in preparation of this manuscript and Mrs. Karen Hensley for her excellent graphics work. The University of Texas M.D. Anderson Cancer Center Core Facility for Peptide Synthesis is supported by National Institutes of Health/National Cancer Institute Grant CA-16672. This work was supported by National Institutes of Health Grant HD25235 (D.D.C.).
ABBREVIATIONS
- Hep
heparin
- HS
heparan sulfate
- AT-III
antithrombin-III
- HIP
Hep/HS-interacting protein
- HA- and LA-Hep
subsets of Hep that bind HIP with high and low affinity, respectively
- FXa
factor Xa
- FPLC
fast protein liquid chromatography
- RT-Hep
run-through Hep
- bFGF
basic fibroblast growth factor
References
- 1.Lindahl U, Backstrom G, Thunberg L, Ledeer I G. Proc Natl Acad Sci USA. 1980;77:6551–6555. doi: 10.1073/pnas.77.11.6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Esmon C T, Owen W G. Proc Natl Acad Sci USA. 1981;78:2249–2252. doi: 10.1073/pnas.78.4.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casu B, Oreste P, Torri G, Zoppetti G, Choay J, Lormeau J-C, Petitou M, Sinay P. Biochem J. 1981;197:599–609. doi: 10.1042/bj1970599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Esmon C T, Esmon N L, Harris K W. J Biol Chem. 1982;257:7944–7947. [PubMed] [Google Scholar]
- 5.Tollefsen D M. In: Heparin: Chemical and Biological Properties, Clinical Applications. Lane D A, Lindahl U, editors. London: Arnold; 1989. pp. 256–273. [Google Scholar]
- 6.Kjellen L, Lindahl U. Annu Rev Biochem. 1991;60:443–475. doi: 10.1146/annurev.bi.60.070191.002303. [DOI] [PubMed] [Google Scholar]
- 7.Liu S, Smith S E, Julian J, Rohde L H, Karin N J, Carson D D. J Biol Chem. 1996;271:11817–11823. doi: 10.1074/jbc.271.20.11817. [DOI] [PubMed] [Google Scholar]
- 8.Rohde L H, Julian J, Babaknia A, Carson D D. J Biol Chem. 1996;271:11824–11830. doi: 10.1074/jbc.271.20.11824. [DOI] [PubMed] [Google Scholar]
- 9.Chang C D, Meienhofer J. Int J Pept Protein Res. 1978;11:246–249. doi: 10.1111/j.1399-3011.1978.tb02845.x. [DOI] [PubMed] [Google Scholar]
- 10.Höök M, Bjork I, Hopwood J, Lindahl U. FEBS Lett. 1976;66:90–93. doi: 10.1016/0014-5793(76)80592-3. [DOI] [PubMed] [Google Scholar]
- 11.Shively J E, Conrad H E. Biochemistry. 1976;15:3932–3942. doi: 10.1021/bi00663a005. [DOI] [PubMed] [Google Scholar]
- 12.Bitter T, Muir H. Anal Biochem. 1962;4:330–334. doi: 10.1016/0003-2697(62)90095-7. [DOI] [PubMed] [Google Scholar]
- 13.Maccarana M, Casu B, Lindahl U. J Biol Chem. 1993;268:23898–23905. [PubMed] [Google Scholar]
- 14.Faham S, Hileman R E, Fromm J R, Linhardt R J, Rees D C. Science. 1996;271:1116–1120. doi: 10.1126/science.271.5252.1116. [DOI] [PubMed] [Google Scholar]
- 15.Atha D H, Stephens A W, Rosenberg R D. Proc Natl Acad Sci USA. 1984;81:1030–1034. doi: 10.1073/pnas.81.4.1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bae J, Desai U R, Pervin A, Aldwell E E O, Weiler J M, Linhardt R J. Biochem J. 1994;301:121–129. doi: 10.1042/bj3010121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferran D S, Sobel M, Harris R B. Biochemistry. 1992;31:5010–5016. doi: 10.1021/bi00136a014. [DOI] [PubMed] [Google Scholar]
- 18.Davie E W, Fujikawa K, Kisiel W. Biochemistry. 1991;30:10363–10370. doi: 10.1021/bi00107a001. [DOI] [PubMed] [Google Scholar]
- 19.Bourin M-C, Lindahl U. Biochem J. 1993;289:313–330. doi: 10.1042/bj2890313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suzuki K, Nishioka J, Hashimoto S. J Biol Chem. 1983;258:163–168. [PubMed] [Google Scholar]
- 21.Hirsch J. N Engl J Med. 1991;324:1565–1574. doi: 10.1056/NEJM199105303242206. [DOI] [PubMed] [Google Scholar]