Abstract
Cytohesin-1, a protein abundant in cells of the immune system, has been proposed to be a human homolog of the Saccharomyces cerevisiae Sec7 gene product, which is crucial in protein transport. More recently, the same protein has been reported to be a regulatory factor for the αLβ2 integrin in lymphocytes. Overexpression of human or yeast ADP-ribosylation factor (ARF) genes rescues yeast with Sec7 defects, restoring secretory pathway function. ARFs, 20-kDa guanine nucleotide-binding proteins initially identified by their ability to stimulate cholera toxin ADP-ribosyltransferase activity and now recognized as critical components in intracellular vesicular transport, exist in an inactive cytosolic form with GDP bound (ARF-GDP). Interaction with a guanine nucleotide-exchange protein (GEP) accelerates exchange of GDP for GTP, producing the active ARF-GTP. Both soluble and particulate GEPs have been described. To define better the interaction between ARF and Sec7-related proteins, effects of cytohesin-1, synthesized in Escherichia coli, on ARF activity were evaluated. Cytohesin-1 enhanced binding of 35S-labeled guanosine 5′-[γ-thio]triphosphate [35S]GTP[γS] or [3H]GDP to ARF purified from bovine brain (i.e., it appeared to function as an ARF-GEP). Addition of cytohesin-1 to ARF3 with [35S]GTP[γS] bound, accelerated [35S]GTP[γS] release to a similar degree in the presence of unlabeled GDP or GTP[γS] and to a lesser degree with GDP[βS]; release was negligible without added nucleotide. Cytohesin-1 also increased ARF1 binding to a Golgi fraction, but its effect was not inhibited by brefeldin A (BFA), a drug that reversibly inhibits Golgi function. In this regard, it differs from a recently reported BFA-sensitive ARF-GEP that contains a Sec7 domain.
A human cDNA clone (B2–1) was isolated by Liu and Pohajdak (1), using subtractive hybridization (natural killer cells − T helper Jurkat cells). The protein product of the B2–1 gene, expressed at high levels in natural killer cells and peripheral T lymphocytes and at very low levels in purified monocytes and several cultured cell lines, was initially believed to be a human homolog of the Saccharomyces cerevisiae Sec7 protein (Sec7p), a crucial component in protein transport between and from compartments of the yeast Golgi (2). The two proteins differ considerably, however, in size and cellular distribution. More recently a Jurkat cell cDNA that encodes the same protein as B2–1 was isolated using a yeast two-hybrid system with the intracellular domain of integrin β2 as bait (3). It was named cytohesin-1, and seems to play an important role in the regulation of the adhesive function of αLβ2 integrin (3). Cytohesin-1 contains two structural motifs already known in other proteins, a large central Sec7 domain of ≈200 amino acids, which has been recognized to date in four other sequences, some of unknown function (4), and, in the carboxyl-terminal region, a ≈100-amino acid (3) pleckstrin homology (PH) domain; PH domains in several proteins are involved in signal transduction processes (5). Overexpression of a cytohesin-1 Sec7 domain fusion protein in Jurkat cells markedly increased cell binding to the extracellular domain of ICAM-1, and direct interaction of the Sec7 domain with the cytoplasmic domain of β1 integrin was demonstrated in vitro (3).
ADP-ribosylation factor (ARF) proteins have been implicated in vesicular membrane trafficking in several intracellular compartments, including endoplasmic reticulum, Golgi, endosomes, and nuclear envelope. They are believed to initiate vesicle formation in mammalian cells by recruiting clathrin or COP I coat complexes to the donor membrane (6). Evidence for interactions of ARF and the Sec7 gene product comes from analysis of double-mutant combinations in yeast. Cells harboring both arf1-null and sec7–1 mutations exhibited synthetic defects and arf1− together with sec21, ypt1, or bet2 mutations were synthetically lethal (7). More recently it was reported that overexpression of human or yeast ARF gene products rescued Sec7-defective cells, restoring secretory pathway function in an allele-specific manner (8).
ARFs are 20-kDa GTP-binding proteins discovered as activators of cholera toxin-catalyzed ADP-ribosylation of the α-subunit of the adenylyl cyclase-stimulatory G protein (Gsα), other proteins, and guanidino compounds such as agmatine (6). ARFs are active when GTP or the nonhydrolyzable analogue guanosine 5′-[γ-thio]triphosphate (GTP[γS]), but not GDP or ATP, is bound. Hydrolysis of bound GTP to GDP, with assistance of a GTPase-activating protein, results in inactive ARF-GDP. Conversion of ARF-GDP to ARF-GTP is promoted by a guanine nucleotide-exchange protein (GEP). Cytosolic (9) as well as particulate (10) GEPs that accelerate GTP binding and, thereby, ARF activation have been described. Inhibition of GEP activity by brefeldin A (BFA), a fungal metabolite that reversibly causes apparent disintegration of Golgi in cells, has been reported by several groups. A BFA-sensitive ARF-GEP that contains a Sec7 domain was recently purified (11). Two soluble GEPs isolated earlier were not inhibited by BFA (9, 12).
We report here that cytohesin-1, the protein product of the B2–1 gene, acts as a GEP that accelerates [35S]GTP[γS] and [3H]GDP binding to purified native ARF3 and induces ARF binding to Golgi membranes via a mechanism insensitive to BFA.
EXPERIMENTAL PROCEDURES
Materials
A line of transformed human lymphocytes from peripheral blood (CRL 1773) was purchased from American Type Culture Collection. Tri Reagent from Sigma was used for isolation of total RNA. All reagents for reverse transcription–PCR (RT-PCR) and cloning were from Boehringer Mannheim. For high level expression and protein purification, QIAexpressionist from Qiagen (Chatsworth, CA) was used. [35S]GTP[γS] (1000–1500 Ci/mmol; 1 Ci = 37 GBq) and [3H]GDP (32.7 Ci/mmol) were from New England Nuclear. Sources of other materials are noted in earlier publications (10).
Methods
RT-PCR.
The RT reaction was performed using 2 μg of poly(A)+RNA with specific primers in the coding region (AGC TTC GGC CGG CTA CCC GGA GAG, 520–542) and in the 3′-untranslated region of B2–1 cDNA (GGG AGA AGG GTG GGC AGT TAT CAC TGG GG, 1420–1450). PCR primers of sequences ATG GAG GAG GAC GAC AGC TAC and TCA GTG TCG CTT CGT GGA GGA GAC in the 5′ and 3′ regions, respectively, were used to amplify a full-length cDNA using Pwo DNA polymerase with 35 cycles of 94°C, 1 min/45°C, 1 min/72°C, 2 min. A second PCR was performed to introduce KpnI and HindIII restriction sites at 5′ and 3′ ends, respectively.
Expression of Protein Product of B2–1 Gene.
The B2–1 cDNA was subcloned in plasmid pQE30 using KpnI and HindIII restriction sites in the same reading frame as the 6×His tag. The B2–1/pQE30 expression construct was transfected into the M15 host strain carrying the pREP4 repressor plasmid, and transformants were selected on plates containing both ampicillin and kanamycin. A single colony was transferred to 100 ml of Luria–Bertani medium containing 150 μg/ml ampicillin and 30 μg/ml kanamycin. When cultures reached a density (OD600) of 0.5, expression of the protein was induced by the addition of 2 mM isopropyl-β-d-thiogalactopyranoside followed by incubation for 4 hr at 37°C before cells were harvested by centrifugation (4000 × g, 5 min). The cells were incubated with lysozyme (1 μg/mg of cell protein) for 20 min on ice in buffer A (50 mM sodium phosphate, pH 7.8/200 mM NaCl/10 μg/ml each aprotinin, leupeptin, and pepstatin/0.5 mM phenylmethanesulfonyl fluoride/0.5 mM amino ethyl+benzene sulfonyl fluoride) and lysed by three cycles of freezing and thawing. The lysate was centrifuged at 14,000 × g for 20 min at 4°C. Ni2+-nitrilo-triacetic acid resin, previously equilibrated in buffer A, was added to the supernatant according to instructions of the manufacturer. After incubation at 4°C with gentle agitation for 2 hr, the resin was transferred to a column and washed with 20 volumes of buffer containing 50 mM sodium phosphate, 200 mM NaCl, and 10% glycerol (pH 6.0). Protein was eluted with buffer A containing 0.1 M imidazole and stored in small portions at −20°C. All experiments were replicated at least twice with two different preparations of His-tagged cytohesin-1.
Preparation of ARF and Golgi Fraction.
Preparations of native ARF1 and ARF3 from bovine brain cytosol (13), recombinant myristoylated ARF5 (14), and Golgi fraction from rat brain (15) have been reported.
Assay for GEP Activity.
The GEP assay was based on stimulation of [35S]GTP[γS] binding to ARF by a published procedure (9). Data presented are means of values from duplicate assays that differed <10%.
RESULTS AND DISCUSSION
As shown in Fig. 1, a portion of the protein product of the B2–1 gene has extensive similarity to the sequence described by Shevell et al. as a Sec7 domain (4). Sec7 domains have been identified thus far in only four other proteins: yeast Sec7; Arabidopsis EMB30 (4); an anonymous C. elegans ORF, K06H7 (16); and a BFA-sensitive ARF-GEP (11). The amino acid sequences of Sec7 domains in B2–1, Sec7, and EMB30 are more similar to each other (42–47%) than any of them is to K06H7 (Table 1). At the nucleotide level, B2–1 cDNA is ≈72% identical (270 nt) to a partial cDNA clone (EST01394) isolated from a brain library (17). Thus, the existence of a family of cytohesin-1-like proteins seems probable. Both EMB30 and Sec7 cDNAs encode large proteins of 1451 and 2009 amino acids, respectively. Deletion of the yeast Sec7 gene is lethal. Its inactivation causes defects in protein glycosylation and secretion, reflecting a defect in transport between Golgi compartments (8). In contrast to Sec7, EMB30 is not required for cell viability (4), and the functional significance of the similarity between the two protein products remains unclear. Other proteins with Sec7 domains are much smaller than Sec7 itself and do not necessarily function within the Golgi (e.g., cytohesin-1), which serves to modulate the adhesive behavior of integrin αLβ2 in lymphocytes (3).
Figure 1.
Alignment of sequences of Sec7 domains in related proteins. Sec7 domains of cytohesin-1 (B2–1; amino acids 62–249), Sec7 (amino acids 827-1017), EMB30 (amino acids 557–752), and Caenorhabditis elegans cosmid K06H7 (K06H7) sequences were aligned using clustal (pc gene). The Sec7 domains of B2–1 and KO6H7 contain 188 amino acids; Sec7, 189; and EMB30, 196.
Table 1.
Percentage identity of amino acid sequences of Sec7 domains of four proteins
| B2-1 | Sec7 | EMB30 | |
|---|---|---|---|
| Sec7 | 47.0 | ||
| EMB30 | 45.7 | 42.3 | |
| K06H7 | 39.3 | 34.4 | 34.1 |
Sec7 domains of B2-1 and K06H7 each contain 188 amino acids; Sec7, 189; and EMB30, 196.
To define better the role of Sec7-related proteins in vesicular transport, the protein product of the B2–1 gene was expressed in Escherichia coli and its ability to regulate ARF activity was investigated. His-tagged cytohesin-1 in the lysed bacteria was completely soluble and, after affinity purification, appeared on SDS/PAGE as a single band of ≈46 kDa (data not shown). Protein yields were 15–50 mg/l of culture, and the purity was ≈90%.
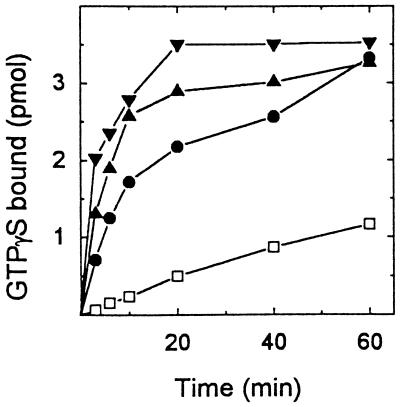
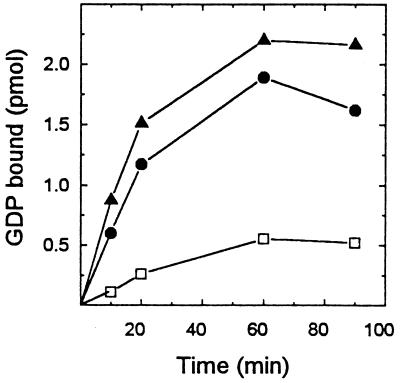
[35S]GTP[γS] binding to native ARF3 (ARF3) was increased in a concentration-dependent manner by His-tagged cytohesin-1 (Fig. 2). The affinity-purified protein increased [35S]GTP[γS] binding almost 6-fold to 0.28 mol/mol ARF3. Maximal binding was achieved in 20 min with 0.24 μg of the recombinant protein. The supernatant fraction from noninduced cells had little effect on [35S]GTP[γS] binding. Binding of [3H]GDP, which reached a maximum of 0.17 mol [3H]GDP/mol ARF3 after 60 min with 0.2 μg of cytohesin-1 (Fig. 3), also was accelerated by cytohesin-1, but apparently less so than was [35S]GTP[γS] binding.
Figure 2.
Effect of cytohesin-1 on [35S]GTP[γS] binding to ARF3. Native ARF3 (0.25 μg, ≈12.5 pmol) was incubated with zero (□), 0.06 (•), 0.12 (▴), or 0.24 μg (▾) of cytohesin-1 and 4 μM [35S]GTP[γS] (2 × 106 cpm) in 100 μl containing 20 mM Tris (pH 8.0), 10 mM imidazole, 100 mM NaCl, 10 μg of phosphatidylserine, 40 μg of BSA, 10 mM DTT, and 3.1 mM MgCl2 at 36.5°C for the time indicated, then at 0°C for a total of 60 min. Proteins were collected on nitrocellulose filters for radioassay of bound GTP[γS]. GTP[γS] bound to samples without ARF3 or cytohesin-1 after 60 min at 0°C was 0.1 pmol, to ARF3 was 0.14 pmol, and to cytohesin-1 alone was 0.11 pmol. After 60 min at 0°C, 12.5 pmol of ARF3 plus 0.06 μg, 0.12 μg, or 0.24 μg of cytohesin-1 bound 0.99, 1.2, and 1.53 pmol, respectively. Control values have been subtracted from the data presented.
Figure 3.
Effect of cytohesin-1 on [3H]GDP binding to ARF3. Samples of ARF3 (15 pmol) were incubated with zero (□), 0.06 (•), or 0.2 μg (▴) of cytohesin-1; 2 μM [3H]GDP (7.5 × 105 cpm); and 4 mM MgCl2 in 100 μl of the medium described in Fig. 2, at 36°C for the indicated time before proteins with bound [3H]GDP were collected on nitrocellulose for radioassay. Samples without ARF3, with or without cytohesin-1, bound 0.06–0.08 pmol; these control values have been subtracted from the data presented.
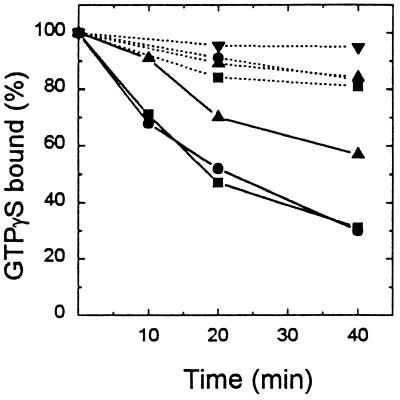
After binding of [35S]GTP[γS] to ARF3 in the presence of a low concentration of MgCl2, with EDTA and phosphatidylserine, there was no significant release during subsequent incubation in the absence of cytohesin-1 (Fig. 4). When cytohesin-1 was present, rates of release of [35S]GTP[γS] were similar with 50 μM unlabeled GDP or GTP[γS] reaching 50% in 20 min. Release was considerably slower with 50 μM GDP[βS], as had been observed in studies of a ≈55-kDa GEP purified from spleen cytosol (12), and was negligible without added nucleotide.
Figure 4.
Effect of cytohesin-1 on release of bound GTP[γS] from ARF3. Samples of native ARF3 (1.5 μg, 75 pmol) were incubated with 4 μM [35S]GTP[γS] (2 × 106 cpm), as described (12), in 100 μl containing 20 mM Tris (pH 8.0), 50 mM NaCl, 20 μg of phosphatidylserine, 50 μg of BSA, 12 mM DTT, 0.65 mM MgCl2, and 1 mM EDTA for 40 min at 36.5°C. Tubes were placed in ice and 20-μl samples (15 pmol of ARF3) were transferred to tubes containing 50 μM unlabeled GTP[γS] (▪), GDP (•), GDP[βS] (▴), or no nucleotide (▾), with (solid line) or without (dotted line) cytohesin-1 (0.5 μg) and other additions, as indicated in Fig. 2, in a final volume of 100 μl. After incubation for the indicated time, samples were filtered for radioassay of protein-bound [35S]GTP[γS]. One hundred percent binding (3.1–3.2 pmol) was the mean of values from samples of ARF3 ± cytohesin-1 and unlabeled nucleotide kept at 0°C for 60 min. Without ARF3, 0.03–0.05 pmol of [35S]GTP[γS] was bound; these control values have been subtracted from the data presented.
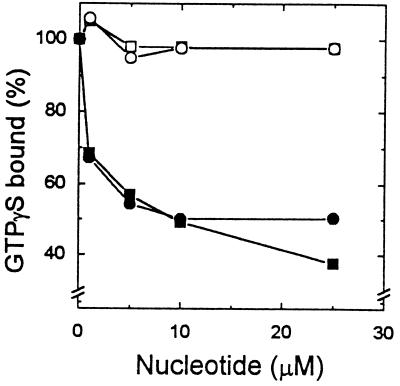
The effect of concentration of GTP[γS] and GDP on release of bound [35S]GTP[γS] from native ARF3 is shown in Fig. 5. During incubation of ARF3-[35S]GTP[γS] and cytohesin-1 with 1 μM GTP[γS] or GDP for 40 min at 36.5°C, 33% of bound [35S]GTP[γS] was released. These dissociation experiments revealed a difference in the behavior of cytohesin-1 and the ≈55-kDa GEP from spleen cytosol (12). In the presence of the latter, [35S]GTP[γS] release was apparently faster with GTP[γS] than it was with GDP (12), which is similar to earlier reports of the exchange of guanine nucleotide bound to S. cerevisiae Ras2p catalyzed by Cdc25p (18).
Figure 5.
Effect of nucleotide concentration on release of bound GTP[γS] from ARF3 with or without cytohesin-1. ARF3 (1.5 μg, 75 pmol) was incubated with 4 μM [35S]GTP[γS], as described in Fig. 4, and cooled. Samples (20 μl) were then incubated with (▪, •) or without (□, ○) cytohesin-1 (0.5 μg), and the indicated concentration of GTP[γS] (○, •) or GDP (□, ▪) for 40 min at 36.5°C before assay of protein-bound [35S]GTP[γS]. One hundred percent binding (3.1–3.2 pmol) was the mean of values for ARF3 ± cytohesin-1 and unlabeled nucleotide incubated at 0°C for 60 min. Control values (0.06–0.08 pmol bound in the absence of ARF3) have been subtracted.
Cytohesin-1, like previously characterized ARF-GEPs (9–12), failed to activate recombinant myristoylated ARF5 (data not shown). It is unclear why no GEP for a class II (ARFs 4 and 5) or class III (ARF6) ARF has been found. Perhaps those reactions have characteristics or requirements sufficiently different that they have not been detected in the assays used thus far. It is intriguing that two different Sec7 mutant yeast alleles were rescued by expression of human ARF4 (8). The explanation for the failure of human ARF5, which is more similar to ARF4 than either is to other known mammalian or yeast ARFs, to replace ARF4 is unclear, as ARF5 protein could not be measured in those studies (8).
Inhibition of GEP activity by BFA causes apparent disintegration of Golgi structure in cells (19, 20). Because activation of class I ARFs increases their binding to Golgi membranes, the effects of cytohesin-1 on ARF association with Golgi and of BFA on this process were investigated. In the same experiments, a GEP partially purified from brain cytosol by chromatography on Sepharose CL-6B was used as a positive control for BFA inhibition. As shown in Table 2, [35S]GTP[γS] binding to Golgi in the presence of ARF1 was increased by His-tagged cytohesin-1, but that increase was unaffected by 2 μg of BFA; whereas the increment in [35S]GTP[γS] binding caused by the Sepharose CL-6B-purified GEP was inhibited ≈75% by the same amount of BFA, as previously reported (11).
Table 2.
Effect of BFA on GTP[γS] binding to ARF1 and Golgi in the presence of cytohesin-1 or ARF-GEP
| Additions | [35S]GTP[γS] bound, pmol
|
|
|---|---|---|
| No BFA | 2 μg BFA | |
| None | 1.1 | 1.3 |
| Cytohesin-1 | 4.3 | 4.2 |
| ARF-GEP | 2.1 | 1.6 |
ARF1 (1 μg, 50 pmol) was incubated with Golgi membrane (12.5 μg of protein) in Tris buffer (pH 8.0) containing 1 mM ATP, 5 mM creatine phosphate, 1 unit of creatine phosphokinase, 3.3 mM MgCl2, and 64 mM NaCl without or with cytohesin-1 (1 μg) or partially purified BFA-sensitve ARF-GEP (20 μg) from Sepharose CL-6B chromatography (11), and 2 μg of BFA in 0.2% EtOH or vehicle alone in a volume of 90 μl at 21°C for 10 min. After addition of 4 μM [35S]GTP[γS], samples were incubated for 40 min at 36.5°C before assay of bound [35S]GTP[γS]. Control values (1.0–1.1 pmol bound in the absence of ARF with or without cytohesin-1, ARF-GEP, and/or BFA) have been subtracted.
Several recent observations suggest that Sec7-related proteins and ARFs play a key role in vesicular membrane trafficking in several intracellular compartments. The observation of allele-specific suppression of temperature-sensitive Sec7 mutants by yeast ARFs provides evidence that Sec7p and yeast ARFs 1 and 2 interact functionally in vesicular transport (8). It may be that at least two different families of Sec7-related proteins are involved in the regulation of ARF activity: larger proteins such as the products of Sec7-like genes (11), and the smaller proteins (e.g., cytohesin-1) that participate in different processes. The demonstration that cytohesin-1 is a GEP for ARF3 and ARF1 (class I ARFs), along with evidence for the existence of other members of a cytohesin family, permits speculation that the latter could be GEPs specific for different ARF proteins. The Sec7 domain could be essential for the ARF-GEP activity, whereas the PH domain could be characteristic for a specific ARF. It has been similarly suggested that all proteins containing Dbl homology and PH domains in tandem are guanyl nucleotide-exchange factors for members of the Rho family of proteins (21).
On the other hand, it has not been shown that the Sec7 domain itself possesses GEP activity, whereas its interaction with and effect on function of β2 integrin has been directly demonstrated (3). Although it seems quite unlikely that the same structural unit serves as an ARF-GEP and a β2 integrin regulator, the designated Sec7 domain is certainly large enough to accommodate more than one functional sequence. Elucidation of the physical and functional consequences of interactions of ARF with specific cytohesin domains and their relationships to cytohesin–integrin interaction are clearly of major interest and importance.
Acknowledgments
We thank Ms. Carol Kosh for expert secretarial assistance.
ABBREVIATIONS
- ARF
ADP-ribosylation factor
- GEP
guanine nucleotide-exchange protein
- [35S]GTP[γS]
guanosine 5′-[γ-thio]triphosphate
- BFA
brefeldin A
Note Added in Proof.
ARF-GEP activity of the Sec7 domain of an ≈47-kDa BFA-insensitive human protein termed ARNO (ARF nucleotide-binding site-opener) has now been demonstrated (22). This protein also contains a PH domain, which was shown to be responsible for the stimulation of GEP activity by phosphatidylinositol bisphosphate (22). Larger yeast proteins [Gea1 and Gea2 (guanine nucleotide exchange on ARF)] analogous to ARNO, which are 50% identical and contain Sec7 domains, were also described (23). Gea1 displayed BFA-sensitive ARF-GEP activity (23).
References
- 1.Liu L, Pohajdak B. Biochim Biophys Acta. 1992;1132:75–78. doi: 10.1016/0167-4781(92)90055-5. [DOI] [PubMed] [Google Scholar]
- 2.Franzusoff A, Schekman R. EMBO J. 1989;8:2695–2702. doi: 10.1002/j.1460-2075.1989.tb08410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kolanus W, Nagel W, Schiller B, Zeitlmann L, Godar S, Stockinger H, Seed B. Cell. 1996;86:233–242. doi: 10.1016/s0092-8674(00)80095-1. [DOI] [PubMed] [Google Scholar]
- 4.Shevell D E, Leu W M, Gillmor C S, Xia G, Feldmann K A, Chua N H. Cell. 1994;77:1051–1062. doi: 10.1016/0092-8674(94)90444-8. [DOI] [PubMed] [Google Scholar]
- 5.Musacchio A, Gibson T, Rice P, Thompson J, Saraste M. Trends Biochem Sci. 1993;18:343–348. doi: 10.1016/0968-0004(93)90071-t. [DOI] [PubMed] [Google Scholar]
- 6.Moss J, Vaughan M. J Biol Chem. 1995;270:12327–12330. doi: 10.1074/jbc.270.21.12327. [DOI] [PubMed] [Google Scholar]
- 7.Stearns T, Willingham M C, Botstein D, Kahn R A. Proc Natl Acad Sci USA. 1990;87:1238–1242. doi: 10.1073/pnas.87.3.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deitz S B, Wu C, Silve S, Howell K E, Melançon P, Kahn R A, Franzusoff A. Mol Cell Biol. 1996;16:3275–3284. doi: 10.1128/mcb.16.7.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsai S-C, Adamik R, Moss J, Vaughan M. Proc Natl Acad Sci USA. 1994;91:3063–3066. doi: 10.1073/pnas.91.8.3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Randazzo P, Yang Y C, Rulka C, Kahn R A. J Biol Chem. 1993;268:9555–9563. [PubMed] [Google Scholar]
- 11.Morinaga N, Tsai S-C, Moss J, Vaughan M. Proc Natl Acad Sci USA. 1996;93:12856–12860. doi: 10.1073/pnas.93.23.12856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsai S-C, Adamik R, Moss J, Vaughan M. Proc Natl Acad Sci USA. 1996;93:305–309. doi: 10.1073/pnas.93.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsai S-C, Noda M, Adamik R, Chang P P, Chen H-C, Moss J, Vaughan M. J Biol Chem. 1988;263:1768–1772. [PubMed] [Google Scholar]
- 14.Haun R S, Tsai S-C, Adamik R, Moss J, Vaughan M. J Biol Chem. 1992;267:7064–7068. [PubMed] [Google Scholar]
- 15.Tsai S-C, Adamik R, Haun R S, Moss J, Vaughan M. J Biol Chem. 1993;268:10820–10825. [PubMed] [Google Scholar]
- 16.Wilson R, Alnscough R, Anderson K, Baynes C, Berks M, et al. Nature (London) 1994;368:32–38. doi: 10.1038/368032a0. [DOI] [PubMed] [Google Scholar]
- 17.Adams M D, Dubnick M, Kerlavage A R, Moreno R, Kelley J M, Utterback T R, Nagle J W, Fields C, Venter J C. Nature (London) 1992;355:632–634. doi: 10.1038/355632a0. [DOI] [PubMed] [Google Scholar]
- 18.Hanley S A, Broach J R. J Biol Chem. 1994;269:16541–16548. [PubMed] [Google Scholar]
- 19.Donaldson J G, Finazzi D, Klausner R D. Nature (London) 1992;360:350–352. doi: 10.1038/360350a0. [DOI] [PubMed] [Google Scholar]
- 20.Helms J B, Rothman J E. Nature (London) 1992;360:352–354. doi: 10.1038/360352a0. [DOI] [PubMed] [Google Scholar]
- 21.Glaven J A, Whitehead I P, Nomanbhoy T, Kay R, Cerione R A. J Biol Chem. 1996;271:27374–27381. doi: 10.1074/jbc.271.44.27374. [DOI] [PubMed] [Google Scholar]
- 22.Chardin P, Paris S, Antonny B, Robineau S, Béraud-Dufour S, Jackson C L, Chabre M. Nature (London) 1996;384:481–484. doi: 10.1038/384481a0. [DOI] [PubMed] [Google Scholar]
- 23.Peyroche A, Paris S, Jackson C L. Nature (London) 1996;384:479–481. doi: 10.1038/384479a0. [DOI] [PubMed] [Google Scholar]