Abstract
RNA polymerase (RNAP) may become arrested during transcript elongation when ternary complexes remain intact but further RNA synthesis is blocked. Using a combination of DNA and RNA footprinting techniques, we demonstrate that the loss of catalytic activity upon arrest of Escherichia coli RNAP is accompanied by an isomerization of the ternary complex in which the enzyme disengages from the 3′ end of the transcript and moves backward along the DNA with concomitant reverse threading of the intact RNA through the enzyme. The reversal of RNAP brings the active center to the internal RNA position and thereby it represents a step in factor-facilitated transcript cleavage. Secondary structure elements or the 5′ end of the transcript can prevent the isomerization by blocking the RNA threading. The described novel property of RNAP has far-reaching implications for the understanding of the elongation mechanism and gene regulation.
Keywords: DNA and RNA footprinting, elongation complex
Arrested ternary complexes of prokaryotic and eukaryotic RNA polymerases (RNAPs) retain the RNA but lose catalytic activity (1–4). DNA sites that provoke arrest do not show strong sequence similarity except that they are enriched in homopolymeric oligo(T) tracts that are 7–10 nucleotides long (4–6). However, transcription through certain regions in DNA is particularly predisposed to arrest. At such sites, the front end of the RNAP footprint appears to remain fixed on the template while RNA chain growth continues (5, 7–10). These unusual regions were called sites of discontinuous elongation, because normally RNAP footprints translocate monotonously along the template as each nucleotide is added to the RNA.
Being a potential block for RNAP passage through a transcription unit, arrest may affect gene expression by decreasing formation of full-sized transcripts, as documented in vitro for Escherichia coli RNAP transcribing the rrnB gene (3) and for eukaryotic RNA polymerase II (Pol II) transcribing histone H3.3, and several other genes (3, 4, 11). In addition, arrested complex formation represents a side pathway in normal termination, since a fraction of the ternary complexes at prokaryotic and intrinsic Pol II termination sites fall into an arrested state instead of dissociating from the template (4, 5).
Elongation factors TFIIS in eukaryotes and GreB in prokaryotes relieve arrest by inducing internal cleavage of the transcript (3, 6, 12–15). The 3′-proximal product of the cleavage may reach 10–20 nucleotides in length, whereas in active complexes the analogous products are only 2–3 nucleotides long (3, 6, 16, 17). The cleaved RNA fragments dissociate from the complex, which allows RNAP to resume elongation in the correct DNA–RNA register from the newly formed 3′ end. No movement of RNAP was detected upon transcript cleavage in arrested complexes (18), whereas in the majority of active complexes transcript cleavage caused upstream shifting of the enzyme (9, 19).
Many studies of arrest were performed with artificially halted individual elongation complexes (ECs) obtained by “walking” the RNAP along the DNA. Krummel and Chamberlin (7) showed that substitution of IMP for GMP in the 5′-proximal regions of short transcripts prevented arrest. A comparison of the deoxyribonuclease I footprints of the arrested and active inosine-containing complexes halted at the same template position showed that the front end of RNAP on the DNA did not move upon arrest (7). Applying the exonuclease III (ExoIII) footprinting technique, Nudler et al. (5, 9) reached the same conclusion. The data of the RNA/protein crosslinking and footprinting of the RNA revealed substantial changes of the RNA/protein interactions upon transcriptional arrest (20, 21).
Taken together, these data suggest that elongation arrest involves disengaging the active center from the 3′ end of the RNA (3, 8, 9, 22) and repositioning it over an internal phosphodiester bond of the transcript (14, 16). It is now widely accepted that this repositioning occurs through slippage of the active site along the transcript, either alone or together with the rear part of RNAP, while the front-end DNA-binding domain of the enzyme remains fixed on the template (8–10). This model is based on the idea of internal flexibility in RNAP, which is currently implicated in the explanation of the discontinuous phase of elongation as well (8–10). However, the alternative possibility, that the active-site relocation away from the 3′ end of the RNA could be caused by slippage of the transcript through the active site, has also been proposed (19). The experimental data on transcriptional arrest, although quite abundant, seem to be fragmentary, since they were obtained with different complexes and with RNAPs from different sources (from E. coli, yeast, or humans), which makes establishing a comprehensive picture of arrest difficult. Direct comparison of all the characteristics of the arrested complex with those of the same complex in the active state would provide a more extensive view of the arrest process.
In this paper, we used a combination of approaches, including solid-phase immobilized transcription, DNA and RNA footprinting, and GreB-induced RNA cleavage, to characterize the processes associated with the arrest of two ECs of E. coli RNAP. The major and unexpected finding is that arrest results from a previously undescribed capability of RNAP to move backward along the DNA and the RNA without breaking down the transcript, which has far-reaching implications for understanding the mechanisms and regulation of transcription.
MATERIALS AND METHODS
Transcription Template and Transcription Reactions.
The standard template for transcription was the PAGE-purified 386-bp DNA fragment carrying the T7A1 promoter, which was obtained by polymerase chain reaction (PCR) with the use of nonphosphorylated primers. The transcribed sequence of this fragment is … ATCGAGAGGG ACACGGCGAA TAGCCATCCC AATCGACACC… . RNAP, bearing a hexahistidine tag genetically fused to the carboxyl terminus of the β′ subunit, was purified and immobilized on Ni–nitrilotriacetate (NTA)-agarose (Qiagen) as described (23). All of the reactions were performed in transcription buffer (TB) containing 20 mM Tris·HCl at pH 7.9, 40 mM KCl, 5 mM MgCl2, and 1 mM 2-mercaptoethanol. The stable ternary complex, stalled at position +11, was obtained as described (5, 9) except the concentration of CpApUpC was 5 μM. The complex was then “walked” to the desired position as described (9). The transcripts were labeled by incorporation of [α-32P]NTP [New England Nuclear; 40 μCi of the NTP (3000 Ci/mmol); 1 Ci = 37 GBq] for 5 min. The homogeneous arrested complexes were purified by adding all four NTPs (500 μM each for a 5-min chase) followed by washing three times with TB containing 1 M KCl. All these and further procedures were performed at 25°C, unless otherwise indicated.
Potassium Permanganate and ExoIII Footprinting.
EC11 (the numerical index denotes the length of the transcript) was treated with 10 units of T4 polynucleotide kinase (New England Biolabs) and 50 μCi of [α-32P]ATP (4500 Ci/mmol; ICN Biomedicals, Costa Mesa, CA) for 10 min to label the DNA, washed with TB, and walked to the desired position. KMnO4 (1 mM final concentration) was added to 10 μl of the EC. The reaction was stopped after 5 min with 1 μl of 2-mercaptoethanol. After the complex was eluted from Ni-NTA-agarose with 50 mM EDTA, the DNA in the supernatant was precipitated in the presence of 10 μg of calf thymus DNA as a carrier, incubated in 100 μl of 10% piperidine for 15 min at 90°C, reprecipitated, and lyophilized. In this experiment and further experiments, the products were dissolved in gel loading buffer (50 mM EDTA/10 M urea) and separated on denaturing PAGE. A 10-μl aliquot of the EC was treated with ExoIII (1 unit; Boehringer Mannheim) for 5 min.
Cleavage of the RNA by RNases A and T1.
A 20-μl aliquot of the immobilized EC was incubated for 10 min with 20 ng of RNase A (Sigma), then 10 μl of the suspension and 5 μl of the supernatant were removed and combined with 3 μl of phenol. The pellet was washed five times with TB, and 3 μl of phenol was added. All samples were analyzed on denaturing PAGE. To obtain 5′-terminal truncation of the transcript, EC26 was treated with 10 units of RNase T1 (Sigma) in 10 μl of TB for 10 min and washed 10 times with TB. Then desired complexes were obtained as described above.
GreB-Induced RNA Cleavage Reaction.
GreB protein was purified as described (3). A 14-μl aliquot of the EC was treated with 40 ng of GreB for 10 min and the supernatant fraction was loaded on denaturing PAGE.
Enzymatic Phosphorylation of the 5′ End of the RNA in EC.
The RNA was labeled by incorporating [α-32P]CTP at position +12 of the transcript. The EC was then incubated with 10 units of T4 polynucleotide kinase (New England Biolabs) and 10 μM nonlabeled ATP in 10 μl of TB for 20 min.
Suppression of Arrest by Complementary Oligonucleotides.
Sequences of the oligonucleotides (Oligos Etc., Guileford, CT) used in this experiment were as follows (5′ → 3′): 1, CCGTGT; 2, GTGTCC; 3, GTCCCT; 4, CCCTCT; 5, CTCTCG; and 6, CTCGAT. The immobilized ECs that were at position +26 containing full-sized or truncated transcript were incubated in 10 μl of TB containing 300 μM KCl with different oligonucleotides (200 μM each). After 5 min, ATP and UTP were added to 5 μM final concentration each. After 10 min, the samples were chased to position +34 with 5 μM CTP for 2 min.
RESULTS
Arrest of Halted Ternary Complexes.
For the transcriptional assays, we employed histidine-tagged RNAP immobilized on Ni-NTA-agarose beads (23). The enzyme can be “walked” along the template by alternating NTP addition with pelleting and washing of the beads (5, 9, 10). The data of Fig. 1 indicate that about 70% of the ECs stalled at position +27 (EC27) of the template containing the T7A1 promoter (5, 9), and ≈30% of EC34 was unable to continue transcription after 20 min of halting at room temperature (lanes 4 and 6), whereas the control complex EC12 remained fully active (lane 2). The loss of activity was not caused by dissociation of the complexes, since they retained the RNA after the beads had been washed. The transcriptionally incompetent fractions of EC27 and EC34 were defined as arrested.
Figure 1.
Arrest of ECs. After a 20-min incubation RNA-labeled EC12, EC27, and EC34 (lanes 1, 3, and 5) were allowed to resume elongation with the addition of the indicated NTPs (5 μM each) for 5 min and then were washed with TB (lanes 2, 4, and 6).
Retreat of Arrested Complexes Along DNA.
To determine whether arrest was accompanied by structural transitions in EC27, we used ExoIII footprinting of RNAP in EC27. We monitored the state of the complex by testing the ability of the RNA to be extended. As can be seen from the RNA gel (Fig. 2A, lanes 1–4), most EC27 is active after 1 min at 25°C, about 70% of the complex becomes inactive after 20 min at 25°C, and most is inactivated after 20 min at 37°C. The ExoIII footprints of the complex prepared after different incubation times show that both the front and rear boundaries of RNAP retreat on DNA as EC27 undergoes the productive-to-arrested conversion (lanes 5–14). Fig. 2B illustrates that the same retreat accompanies the arrest of EC34 and brings both complexes to the location of productive EC12 (compare lanes 2, 4, and 6 and 7, 10, and 12). In this experiment, we obtained the arrested EC27 and EC34 in a homogeneous form suitable for the footprinting by incubating the complexes with the four NTPs and subsequently washing the beads to remove the active fractions that reached the end of the template and dissociated. The remaining complexes failed to resume elongation during a second incubation with 500 μM NTPs for 30 min, indicating the irreversibility of the arrest (data not shown). We used this approach in further experiments to purify the arrested fraction.
Figure 2.
Effect of arrest on RNAP position on the DNA. (A) (Left) Dependence of EC27 inactivation on time and temperature. EC27 was allowed to resume elongation with the addition of 5 μM ATP, CTP, and UTP for 5 min after stalling under the indicated conditions. (Center and Right) ExoIII digestion of the nontemplate and template DNA strands (showing the front-end and rear-end footprints, respectively) in EC27 prepared after the indicated time of stalling. (B) (Left) ExoIII digestion of the nontemplate and template DNA strands in the active EC12, EC20, EC27, and EC34 and in the arrested EC27 and EC34. The arrested complexes were isolated in homogeneous form by adding the four NTPs prior to the digestion. (Right) Scheme summarizing the data of ExoIII footprinting. Vertical black lines symbolize the DNA strands, asterisks indicate the 5′ labeling of the strands, and ovals indicate RNAP. (C) (Left) Potassium permanganate (KMnO4) footprinting of the nontemplate DNA strand in the same complexes. Arrows indicate signals that originated from modified single-strand thymidines involved into the transcriptional bubble. (Right) Scheme summarizing the data of KMnO4 footprinting. The symbols are the same as above. Hexagons symbolize the transcriptional bubble. The positions of reactive and nonreactive T residues in the nontemplate strand are shown by the solid and open circles, respectively.
The potassium permanganate footprinting of the nontemplate strand of DNA shows that the retreat of RNAP was accompanied by translocation of the transcriptional “bubble.” The forward movement of the bubble from EC12 to EC34 (Fig. 2C, lanes 1, 2, 3, and 5) was visualized by the appearance of permanganate-reactive unpaired thymidines at the leading edge of the bubble and their concomitant disappearance at the rear edge. However, in the arrested EC27 and EC34, the pattern of thymidine reactivity is indistinguishable from that in the productive EC12 (Fig. 2C, lanes 1, 4, and 6).
Reverse Threading of the RNA Through RNAP in the Arrested Complex.
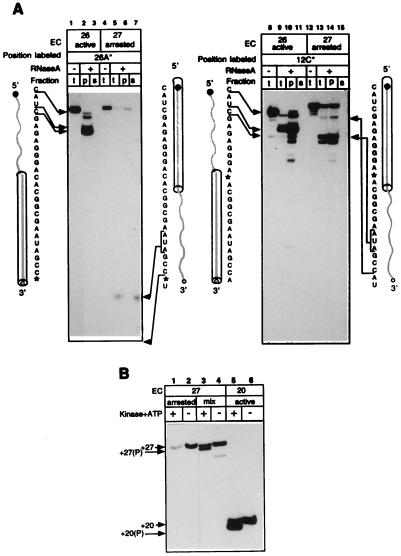
To test the state of the transcript in the retreated complexes, we digested EC27 with RNase A, which preferentially cleaves RNA after pyrimidine residues. Because stalled EC27 rapidly arrests, as a control we used EC26, which remained fully productive during the experiment. Fig. 3A Left documents the patterns of protection of the 3′-end-labeled transcripts. In the active EC26, the 5′ part of the RNA was extensively cleaved by the ribonuclease, whereas the 3′ part was completely protected (lanes 1 and 2). The long 3′-terminal products of the cleavage remained in the pelleted complex (lanes 2 and 3). Footprinting with RNase A did not allow determination of the exact size of the 3′-protected region, since there were no pyrimidines in the RNA between positions +3 and +12. Cleavage with RNase T1, which cleaves after G residues, demonstrated that the protection in EC26 ended at position +10G (data not shown). In the arrested EC27, the RNA footprint was remarkably different: the 5′ part of the RNA was fully protected, whereas the 3′ part was exposed to RNase A (lanes 4–6) and the short 3′ fragments were released into the supernatant (lane 7). The total label in lanes 6 and 7 decreased due to extensive cleavage of the RNA at position +25, which generated the dinucleotide products migrating at the front of the gel.
Figure 3.
Effect of arrest on the transcript arrangement in RNAP. (A) RNase A footprinting of the transcript in active EC26 and in arrested EC27. The RNA in the complexes was internally labeled at positions +26A or +12C. Each sample was treated with RNase A and fractionated by centrifugation into soluble (s) and matrix-associated (p) fractions before gel analysis. Sequences of the transcripts are shown alongside the autoradiograms, asterisks mark positions of labeling, and arrows show major cuts introduced into the RNA (bold shaded line) by RNase A. The cylinders represent the transcript segments protected by RNAP in the active and arrested complexes. Nonfractionated samples (t) are included as controls. (B) 5′-Terminal phosphorylation of the transcripts. RNA-labeled EC20 and EC27 (lanes 6 and 4) and arrested fraction of EC27 purified by chase (lane 2) were treated with T4 polynucleotide kinase in the presence of ATP (lanes 5, 3, and 1). The symbol (P) indicates the phosphorylated transcripts. Arrows indicate the mobility of phosphorylated and nonphosphorylated transcripts.
To delineate the extent of 5′-proximal protection in the arrested complex, we repeated the same experiment with the transcripts labeled at position +12C (Fig. 3A Right). The pattern of internally labeled transcript cleavage in the active EC26 was essentially the same as in the case of the 3′-terminal label (compare lanes 1–3 and 8–11). In the arrested EC27, RNase A generated 5′-terminal products 18–23 nucleotides long, revealing that at least 9 nucleotides of the 3′ end were exposed to the cleavage (lanes 12–14). The rearrangement of the RNA footprints after the arrest most likely indicates the reverse threading of the transcript through RNAP (as depicted by the schemes alongside the autoradiograms of Fig. 3A).
This conclusion was confirmed by enzymatic phosphorylation of the 5′-terminal hydroxyl group of RNA in RNAP using bacteriophage T4 polynucleotide kinase, which requires a transcript of at least 14–16 nucleotides for its action. The phosphorylation of shorter transcripts was blocked, presumably due to the protection of their 5′ ends by RNAP (data not shown). The addition of the terminal phosphate can be detected on the gel by an increased electrophoretic mobility of the RNA. Fig. 3B (lanes 5 and 3) shows that in productive EC20 and EC27, the RNA 5′ end was available for phosphorylation. However, in the arrested EC27 (from which the active fraction was removed by the chase), the transcript cannot be phosphorylated (lane 1). Thus, arrest of EC27 brought the 5′-proximal segment of the transcript into the enzyme, where it became unavailable for the kinase.
Rearrangement in the Pattern of GreB-Induced RNA Cleavage Associated with Transcriptional Arrest.
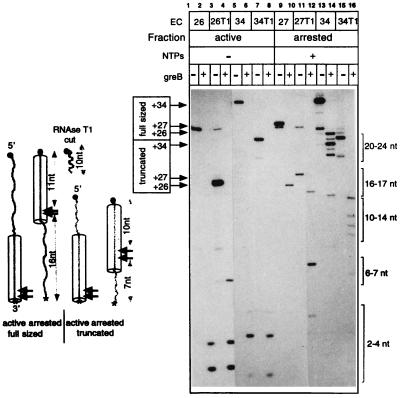
Much evidence argues that the internal transcript cleavage in ternary complexes is carried out by the catalytic center of RNAP and can be stimulated by factors GreA and GreB in prokaryotes and SII in eukaryotes (14, 15). Fig. 4 shows the result of GreB-induced cleavage of the 3′-labeled transcripts in active EC26 and EC34 or in the purified arrested EC27 and EC34. In both complexes in the active state, GreB primarily removed di- and tetranucleotides from the 3′ end of the transcript (lanes 2 and 6). The arrest led to the dramatic enlargement of the 3′-proximal products, consistent with the data obtained previously with eukaryotic RNAP II (16). The products became 16 and 21–24 nucleotides long in the arrested EC27 and EC34, respectively (lanes 10 and 14). To generate fragments of this size, the catalytic center had to cleave the RNA near position +11 (see the scheme in Fig. 4) and, therefore, it retreated in concert with the RNAP structural elements that determined the DNA and RNA footprints. Thus, the size of 3′-terminal products of the cleavage can indicate the position of retreated RNAP on the DNA.
Figure 4.
GreB-induced cleavage of the RNA in active and arrested complexes. (Right) Active EC26 and EC34 and homogeneous arrested EC27 and EC34 containing full-sized (lanes 1, 5, 9, and 13) or 5′-truncated transcripts (lanes 3, 7, 11, and 15) were labeled in the position +26A of the RNA (EC26, EC27, EC26T1, and EC27T1) or in position +34C (EC34 and 34T1; EC34T1 was obtained by walking with unlabeled EC26T1). The products of GreB cleavage were fractionated by centrifugation and the supernatants displayed the 3′-terminal fragments dissociated from the complexes. The arrows on the left side of the autoradiogram indicate the nontreated transcripts, and the brackets on the right side indicate the cleavage products. (Left) Scheme summarizing the results of the experiment. The large arrowheads show positions where GreB cleaves the RNA in the active and arrested complexes. The numbers indicate the size of corresponding segments of the transcripts derived from GreB-induced cleavage or from treatment with RNase T1. For other symbols, see Fig. 3.
The 5′ RNA End Stops Backward Movement of Arrested RNAP.
What determines the location of the retreated RNAP on the DNA? To test the role of the transcript in this process, we took advantage of the ability of RNase T1 to cut near the 5′ end of the RNA in EC26. Lanes 1 and 3 of Fig. 4 show that the RNase cleaves the transcript at position +10, creating the fully active truncated version EC26T1, which contains the 16-nucleotide RNA. After the RNase was removed by repeated washing, EC26T1 was walked to positions +27 and +34 (lanes 7, 11, and 15), where it formed arrested complexes as well as regular EC27 and EC34. The productive fractions of EC27T1 and EC34T1 were removed by chasing, and the arrested complexes were treated with GreB. Lanes 10 and 14 and 12 and 16 of Fig. 4 show that the 5′ truncation of the RNA generated a new pattern of the cleavage in both complexes. The cleavage produced a 16-nucleotide 3′ increment in arrested EC27, in contrast to a 7-nucleotide 3′-increment in arrested EC27T1. In arrested EC34 and EC34T1, the length of the 3′ increments is 21–24 nucleotides and 10–14 nucleotides, respectively. Estimating the position of the arrested polymerase by the size of the 3′-proximal products of the cleavage suggests that in all these cases the arrested enzyme stopped 10–12 nucleotides away from the 5′ end of the transcript. Thus, the 5′ end appears to be a factor limiting the distance at which the arrested complexes retreat.
Inhibition of Arrest by Oligonucleotides Complementary to the Transcript.
The role of the transcript in determining the position of the arrested complex on the DNA suggests that any obstacle in the RNA behind the RNAP molecule could stabilize the active state of the complex. That could explain the observation that oligonucleotides complementary to the RNA that were added to the transcription reaction inhibited arrest at some template positions (E. Nudler, personal communication).
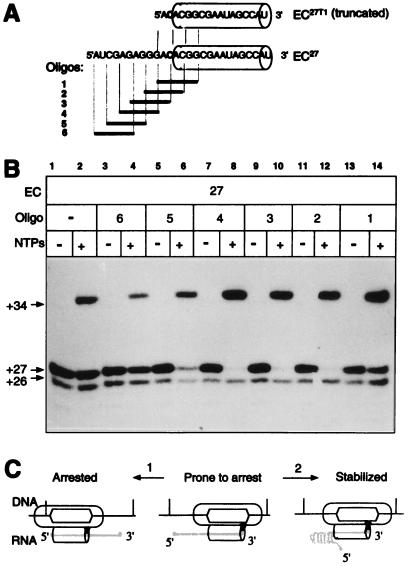
We used a set of hexameric oligonucleotides complementary to overlapping segments of the 5′ part of the RNA in EC27 to introduce such an obstacle (Fig. 5A). EC27 was synthesized and then chased to position +34 in the presence of each oligonucleotide. In the majority of ECs analyzed with different ribonucleases, the cut closest to the 3′ end could be introduced 14–16 nucleotides away from the tip of the transcript (unpublished observation). Such a shortening of the RNA affected neither the stability nor other properties of the EC, indicating that tight contacts between the enzyme and the transcript were limited to these 3′-proximal 14 nucleotides. Lanes 7–12 of Fig. 5B demonstrate that the oligonucleotides that were complementary to the short segment of the transcript immediately behind the enzyme efficiently suppressed the arrest. The truncation of the transcript with RNase T1, which took the target for the oligonucleotides away from the RNA sequence, abolished this effect (data not shown). In a separate experiment, the analogous oligonucleotides protected the corresponding site of the RNA in another EC from degradation with single-strand-specific ribonucleases (unpublished results). Therefore, the observed anti-arresting activity was most likely mediated by hybridization of the oligonucleotides to the RNA, which produced a duplex that interfered with the reverse threading of the transcript and the retreat of the RNAP molecule.
Figure 5.
Effect of oligonucleotides complementary to the 5′ part of the transcript on the efficiency of EC27 arrest and a model of transcriptional arrest. (A) Sequences of full-sized or truncated transcripts in EC27 and the set of overlapping complementary hexanucleotides (bold lines). The cylinders represent the zone of tight contacts between the enzyme and the product (see the text for detail). (B) EC27 was obtained from EC26 in the presence of the oligonucleotides and then chased to position +34. (C) A model of EC rearrangements during transcriptional arrest and the anti-arresting role of RNA secondary structure. The shaded triangle symbolizes the position of the active center (see the text for details).
DISCUSSION
Long-Distance Backward Translocation of EC.
The main observation of this work is the long-distance reverse translocation of E. coli RNAP along the DNA and the RNA without breakdown of the transcript. This retreat was accompanied by concomitant backward movement of the transcriptional bubble and resulted in formation of an arrested complex trapped in an upstream site with the 3′ end of the transcript intact and extruded from the enzyme (Fig. 5C, arrow 1). We demonstrated that, at least in the arrested complexes, the internal transcript cleavage was preceded by the reversal of the enzyme, which brought the active center to the internal RNA position, where it executed the cleavage.
The data of the present paper contrast our previous observations (see Introduction and refs. 5 and 10) that, at two other arresting sites, the front end of RNAP did not move upon the inactivation. However, in those experiments, the fraction of RNAP was defined as arrested according to its failure to resume transcription after 2–5 min of incubation with the substrates, whereas most recent studies have revealed that RNAP is only temporarily arrested at these sites and is able to resume full transcriptional competence upon prolonged (10- to 30-min) incubation with NTPs (unpublished results). Thus, in those cases inactivation of RNAP represents a phenomenon somewhat different from irreversible arrest of EC27 and EC34.
Our results are consistent with the observation of Krummel and Chamberlin (7) that DNase I footprints of two complexes that arrested at positions +14 and +20 in another transcription unit, despite their carrying transcripts of different length, were essentially identical to each other and to the footprint of the active complex halted on the same template at position +11. However, the comparison of footprints of the active forms of these complexes with their arrested counterparts led to the conclusion that catalytic inactivation of RNAP involved slipping backward of the rear part of RNAP only, whereas the front-end domain was supposed to remain fixed on the template, which apparently disagrees with the view of arrest presented here. We attribute this discrepancy to the fact that arrest of RNAP in these positions was prevented by substituting IMP for GMP in the upstream part of the transcripts. Substituting IMP for GMP may change the properties of ECs and especially their lateral stability (see below and ref. 19). Therefore, a direct comparison of active and arrested states of RNAP might not be possible because the active state of the GMP-containing complex (which was not analyzed in that work) may have a structure completely different from that of the IMP-containing complex.
Long-distance retreat of RNAP has been observed previously during processive pyrophosphorolysis and step-by-step cleavage of the RNA induced by GreA, GreB, and SII factors (6, 9, 19, 22, 24). However, in both cases, the retreat was accompanied by degradation of the transcript, so that its 3′ end was always located within the enzyme and engaged with the catalytic center; thus, the RNAP remained active. The principal novelty of the present result is that the complex is shown to move back and maintain the correct alignment of RNA·DNA base pairing preserving the intact transcript.
The observed sliding reaction opens the way to new ideas about mechanisms of gene regulation through RNA chain elongation. First, the principal role of the transcript both in maintaining the enzyme in the elongation-prone conformation and in determining the site where the backward sliding stops adds an extra regulatory potential to the phenomenon of transcriptional arrest. Like the truncation of the transcript (Fig. 4) and oligonucleotides that form the duplex with the transcript at its exit from RNAP (Fig. 5), RNA structure that can form when the transcript becomes long enough may stabilize RNAP in the active state or stop the retreat before the enzyme reaches the 5′ end (arrow 2 of Fig. 5C). Presumably, proteins bound to the nascent transcript may also suppress the arrest, either directly by reinforcing the interaction between the enzyme and the RNA, thus preventing RNAP from slipping backward, or indirectly by blocking the reverse threading of the RNA. Second, the one-dimensional mobility of stalled RNAP may affect our understanding of the basic elongation mechanism. Temporary arrest of the enzyme may involve back-and-forth oscillations of RNAP (unpublished results). They would explain the “compressed” DNA footprints of RNAP at the discontinuous phase of transcription, interpreted as an indication of the internal flexibility of the enzyme in the “inchworming” model of elongation (3, 8, 9, 10).
A Putative Mechanism Driving Backward Translocation of RNAP.
ECs were known to be predisposed to arrest at sites of discontinuous elongation (see Introduction and refs. 3, 5, and 7–10). We proposed earlier that at these sites, the transcript extension within the fixed enzyme leads to an accumulation of internal strain in the complex (5, 9). We further suggested that the strain could be the cause of disengagement of the catalytic center from the 3′ end of the transcript. However, here we demonstrate that the complexes representing monotonous (EC34) as well as discontinuous (EC27) phases of elongation are equally capable of falling into an arrested state, which argues against this idea.
Although the role of DNA in RNAP lateral stability was not analyzed in this work, we support the idea that nucleic acid rather than protein–nucleic acid interactions primarily determine the vector of lateral motion. Clearly, the shift of the bubble and the reverse threading of the transcript are associated with dramatic rearrangements in the geometry of DNA and RNA base pairing, so the retreated complex appears like an opened zipper—i.e., the chains of DNA and RNA are spread apart both behind and ahead of the lock that holds them together near the catalytic center. At each elementary step of the retreat, one DNA·DNA base pair must be separated at the rear edge of the transcriptional bubble and one RNA·DNA pair must be disrupted at the leading edge. Formation of one DNA·DNA base pair at the leading edge of the bubble and of one RNA·DNA base pair at the end of the DNA·RNA hybrid should compensate for this energy consumption. Since the calculated stability of all possible DNA·DNA and RNA·DNA base pairs varies significantly (25), there must be template positions where “zipping” of the DNA and concomitant extrusion of the RNA from the front end release energy. If in the same complex the sequence context at the rear edge of the bubble and at the end of the DNA·RNA hybrid also favors DNA·RNA instead of DNA·DNA pairing, RNAP retreat is propitious until the coupled acts of energy consumption and release balance each other. In this regard, relative weakness of dA·rU base paring versus dA·dT base paring at the leading edge of the bubble during transcription of homopolymeric oligo(T) tracts may explain why prokaryotic and eukaryotic RNAPs become efficiently arrested at these sequences in the nontemplate DNA strand (4–6). This notion can explain how the secondary structure of the transcript blocks the RNAP reversal by interfering with rehybridizing of the RNA and DNA strands at the upstream edge of the bubble. The inosines incorporated into RNA at the distance of 9–15 nucleotides from its 3′ end were shown to inhibit E. coli RNAP arrest (2, 7). This result may account for the fact that the substitution weakens DNA·RNA rehybridization, which should interfere with the intrusion of the RNA chain into the double-stranded DNA. Substituting IMP for GMP in the nascent transcript also facilitated long-distance retreat of RNAP caused by multiple-round cleavage of the RNA with GreA factor and by pyrophosphorolysis (19), which suggests that these types of movement are affected by similar variations in the strength of complementary interactions.
The above consideration implies that the 5′ end of the transcript blocks the reverse translocation of RNAP during arrest because the opportunity for rehybridization at the upstream RNA–DNA junction no longer persists. Thus, the distance of about 11 nucleotides from the 3′ end of the transcript, at which the retreating RNAP stops, seems to demarcate the branching point between DNA and RNA. Cleavage-induced backward translocation of E. coli RNAP and Pol II also stops about 10 nucleotides away from the 3′ end of the transcripts (6, 19). This result supports the idea of an extended DNA·RNA hybrid in ECs, which has been proposed on the basis of thermodynamic analysis of elongation (26). All of the above conclusions may be also drawn to explain why RNAP never slips forward, leaving the 3′ end of the RNA behind the catalytic center.
Acknowledgments
N.K. is in the Graduate Program of the Institute of Molecular Genetics, Russian Academy of Sciences, Moscow. We thank Alex Goldfarb and Vadim Nikiforov for supporting our research and helpful discussion. We are also grateful to Peter von Hippel and Elizabeth Kutter for critical reading of the manuscript. This work was supported by National Institutes of Health Grant GM49242 (to A. Goldfarb) and Russian Foundation for Fundamental Research Grant 96-04-49019 (to V. Nikiforov) and by the National Cancer Institute, under contract with ABL (M.K.).
ABBREVIATIONS
- RNAP
RNA polymerase
- EC
elongation complex
- ExoIII
exonuclease III
- NTA
nitrilotriacetate
References
- 1.Arndt K M, Chamberlin M J. J Mol Biol. 1990;213:79–108. doi: 10.1016/S0022-2836(05)80123-8. [DOI] [PubMed] [Google Scholar]
- 2.Krummel B, Chamberlin M J. J Mol Biol. 1992;225:221–234. doi: 10.1016/0022-2836(92)90917-9. [DOI] [PubMed] [Google Scholar]
- 3.Borukhov S, Sagitov V, Goldfarb A. Cell. 1993;72:459–466. doi: 10.1016/0092-8674(93)90121-6. [DOI] [PubMed] [Google Scholar]
- 4.Reines D, Chamberlin M J, Kane C M. J Biol Chem. 1989;264:10799–10809. [PubMed] [Google Scholar]
- 5.Nudler E, Kashlev M, Nikiforov V, Goldfarb A. Cell. 1995;81:351–357. doi: 10.1016/0092-8674(95)90388-7. [DOI] [PubMed] [Google Scholar]
- 6.Izban M G, Luse D S. Genes Dev. 1992;6:1342–1356. doi: 10.1101/gad.6.7.1342. [DOI] [PubMed] [Google Scholar]
- 7.Krummel B, Chamberlin M J. J Mol Biol. 1992;225:239–250. doi: 10.1016/0022-2836(92)90918-a. [DOI] [PubMed] [Google Scholar]
- 8.Chamberlin M J. Harvey Lect. 1995;88:1–21. [PubMed] [Google Scholar]
- 9.Nudler E, Goldfarb A, Kashlev M. Science. 1994;265:793–796. doi: 10.1126/science.8047884. [DOI] [PubMed] [Google Scholar]
- 10.Wang D, Meier T, Chan C L, Feng G, Lee D N, Landick R. Cell. 1995;81:341–350. doi: 10.1016/0092-8674(95)90387-9. [DOI] [PubMed] [Google Scholar]
- 11.Spencer C A, Groudine M. Oncogene. 1990;5:777–785. [PubMed] [Google Scholar]
- 12.Surratt C K, Milan S C, Chamberlin M J. Proc Natl Acad Sci USA. 1991;88:7983–7987. doi: 10.1073/pnas.88.18.7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reines D. In: Transcription. Conaway R, Conaway J W, editors. New York: Raven; 1993. pp. 263–278. [Google Scholar]
- 14.Rudd M D, Izban M G, Luse D S. Proc Natl Acad Sci USA. 1994;91:8057–8061. doi: 10.1073/pnas.91.17.8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Orlova M, Newlands J, Das A, Goldfarb A, Borukhov S. Proc Natl Acad Sci USA. 1995;92:4596–4600. doi: 10.1073/pnas.92.10.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Izban M G, Luse D S. J Biol Chem. 1993;268:12874–12855. [PubMed] [Google Scholar]
- 17.Izban M G, Luse D S. J Biol Chem. 1993;268:12864–12873. [PubMed] [Google Scholar]
- 18.Gu W, Powell W, Mote J, Jr, Reines D. J Biol Chem. 1993;268:25604–25616. [PMC free article] [PubMed] [Google Scholar]
- 19.Feng G, Lee D N, Wang D, Chen C, Landick R. J Biol Chem. 1994;269:22282–22294. [PubMed] [Google Scholar]
- 20.Markovtsov V, Mustaev A, Goldfarb A. Proc Natl Acad Sci USA. 1996;93:3221–3226. doi: 10.1073/pnas.93.8.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gu W, Wind M, Reines D. Proc Natl Acad Sci USA. 1996;93:6935–6940. doi: 10.1073/pnas.93.14.6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee D, Feng G, Landick R. J Biol Chem. 1994;269:22295–22303. [PubMed] [Google Scholar]
- 23.Kashlev M, Martin E, Polyakov A, Severinov K, Nikiforov V, Goldfarb A. Gene. 1993;130:9–14. doi: 10.1016/0378-1119(93)90340-9. [DOI] [PubMed] [Google Scholar]
- 24.Kassavetis G A, Geiduschek E P. Science. 1993;259:944–945. doi: 10.1126/science.7679800. [DOI] [PubMed] [Google Scholar]
- 25.Yager T D, von Hippel P. Biochemistry. 1991;30:1097–1118. doi: 10.1021/bi00218a032. [DOI] [PubMed] [Google Scholar]
- 26.Yager T D, von Hippel P. In: Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Washington, DC: Am. Soc. Microbiol.; 1987. pp. 1241–1275. [Google Scholar]