Abstract
RNase inhibitor (RI) binds with extraordinary affinity (Ki ≈ 10−13–10−16 M) to diverse proteins in the pancreatic RNase superfamily. In the present study, the structural basis for the recognition of two RI ligands, human angiogenin (Ang) and bovine RNase A, has been investigated by site-specific mutagenesis of human RI and Ang. The RI residues examined were those that appear to form strong contacts with RNase A in the crystal structure of the porcine RI·RNase A complex [Kobe, B. & Deisenhofer, J. (1995) Nature (London) 374, 183–186] that would not be replicated in the Ang complex. Ala substitutions of five of these residues (Glu-287, Lys-320, Glu-401, Cys-408, and Arg-457) were found to have little or no effect on binding of RNase A. In contrast, replacements of Tyr-434, Asp-435, and Tyr-437 and deletion of the C-terminal residue Ser-460 substantially weakened affinity for RNase A: the losses of binding energy associated with the mutations were 5.9, 3.6, 2.6, and 3.5 kcal/mol, respectively. Thus these four residues, which are neighbors in the tertiary structure, appear to constitute a “hot spot” for the RNase A interaction. However, only one of them, Asp-435, was equally important for binding of Ang; the Ki increases produced by mutations of the others were 20- to 93-fold smaller for Ang than for RNase A. Consequently, Tyr-434 plays a significant but lesser role in the Ang complex, whereas Tyr-437 and Ser-460 make only minor contributions. Ala mutations of four Ang residues (His-8, Gln-12, Asn-68, and Glu-108) that correspond to RI contacts on RNase A produced no major changes in affinity for RI. These findings indicate that RI uses largely different interactions to achieve its extremely tight binding of RNase A and Ang.
The interactions of RNase inhibitor (RI), a 50-kDa protein, with the various ≈14-kDa members of the pancreatic RNase superfamily provide a particularly rich and intriguing system for investigating the important problem of protein–protein recognition. RI complexes are among the tightest on record: dissociation constants for the seven ligands whose binding has been kinetically characterized extend from 10−13 to less than 10−15 M (1–5). The high affinities of RI for all of its ligands is remarkable in view of the modest degree of sequence identity that these proteins share. Human angiogenin (Ang) and bovine pancreatic RNase A have Ki values of 0.7 fM and 44 fM, respectively (1), and are 33% identical in sequence (6). Ang and human placental RNase have virtually indistinguishable Ki values (3) and yet are only 26% identical (7). Elucidation of the means by which RI achieves this versatility should shed considerable light on the nature of protein–protein interactions in general. It may also have implications for treatment of human diseases because some RI ligands are biological effectors that are thought to participate in various types of pathology. One of these, Ang, is a potent inducer of new blood vessel formation (8) that is critically involved in the establishment and growth of human tumors in athymic mice (9). RI inhibits the angiogenic activity of Ang (10), but is unlikely to have therapeutic utility because of its broad specificity, large size, and lability. Once the structural basis for RI action is known, it may be possible to design RI derivatives or mimics that do not suffer from these limitations.
The available functional and physical information on RI interactions suggests that RI recognizes its diverse targets in at least partially different ways (see ref. 11). Replacements of two poorly conserved residues, Arg-5 of Ang and Lys-7 of RNase A, greatly weaken RI binding (12, 13). Moreover, deletion of human RI (hRI) residues 315–371 attenuates binding to Ang 100 times more than it does to RNase A (14). Most importantly, the binding interface observed in the recently determined crystal structure of the complex of porcine RI (pRI) with RNase A (15) includes regions of RNase A that are highly variable among the different ligands. RI adopts a fold in which the 15 tandem homologous leucine-rich repeat units that comprise almost the entire molecule are arranged symmetrically in the shape of a horseshoe (ref. 16; see Fig. 1). When RNase A binds, one of its lobes fits into the central cavity of the horseshoe and another contacts the upper face. The resultant binding surface is large: it includes 28 RI residues contributed by 12 different repeat units and 24 RNase residues, also drawn from multiple segments of primary structure. Only eight of the RNase side-chains or main-chain elements that interact with RI are conserved in the other RI ligands whose three-dimensional structures are known [Ang (17); eosinophil-derived neurotoxin (18)].
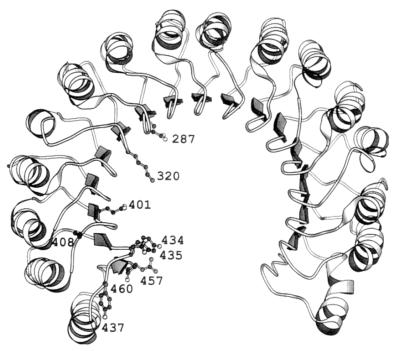
Figure 1.
Diagram of the structure of pRI in its complex with RNase A (15), drawn with molscript (30). The side chains are shown for contact residues whose hRI counterparts were mutated. hRI numbering is used.
In the present study, the structural basis for the tight binding of hRI to two of its ligands, Ang and RNase A, has been explored by site-specific mutagenesis. The primary guide for selection of mutation sites was the structure of the pRI·RNase A complex. hRI (19) and pRI (20) are 77% identical in sequence and all but one of the pRI residues involved in RNase A binding are conserved or conservatively replaced in hRI; Ki values for the two RI·RNase complexes are nearly identical (1, 2). We find that four residues in the C-terminal region of RI constitute a “hot spot” (21) for the RNase A interaction that may account for much of the binding energy of the complex. Three of these RI residues make lesser contributions to the Ang complex. Moreover, several Ang residues were examined that correspond to RI contacts on RNase A and they do not appear to play important roles. These findings support the view that RI utilizes a largely different set of interactions to achieve its extraordinarily tight binding of multiple RNase family proteins.
MATERIALS AND METHODS
Materials.
[14C(U)]-d-Glucose was purchased from DuPont/NEN. Recombinant human Ang, hRI, and the Ang mutant K40Q were produced as in earlier studies (22–24). Angiogenins were quantitated by amino acid analysis; RI concentrations were determined by inhibition of RNase A activity (25). The sources of other materials have been described (23, 25).
Mutagenesis and Production of Mutant Proteins.
Fragments of hRI or Ang genes containing the desired mutations were generated by PCR and cloned into the expression plasmids [pTRP-PRI (24) for RI and pAng3 (12) for Ang] by standard methods. DNA sequencing confirmed the presence of the mutations and the absence of any spurious changes. RI and Ang mutants were expressed in Escherichia coli strain W3110 cells transformed with the mutant plasmids and were purified as described (12, 14). SDS/PAGE analysis revealed that all mutants were >95% homogeneous and had the same size as the native proteins. The amino acid composition of each protein was consistent with its proposed structure.
[14C]Ang.
A synthetic gene for Ang (23) was cloned into the plasmid pET11 and expressed in E. coli strain BL21(DE3) cells grown in M9 medium containing [14C]glucose as the sole carbon source. [14C]Ang (2700 cpm/μg) was purified to homogeneity as described (12, 23).
Kinetics.
The enzymatic activity of RNase A was measured spectrophotometrically with CpA, CpG, or C>p as substrate in 0.1 M Mes (pH 6.0) containing 0.1 M NaCl and 10 μg/ml BSA at 25°C (14, 25). Dissociation rate constants for inhibitor–ligand complexes were determined by incubating the ligand with 1–1.5 molar equivalents of inhibitor for 10 min, then adding a scavenger for free inhibitor, and measuring the amount of free ligand present at various times (1). Incubations were performed at 25°C in 0.1 M Mes (pH 6.0) containing 0.1 M NaCl, 1 mM EDTA, dithiothreitol (5 mM with RNase A, 120 μM with Ang), and 100 μg/ml BSA. Dissociation of complexes was biphasic, with a relatively rapid release of up to 25% of the ligand followed by a slower dissociation of the remainder. Similar biphasic dissociation has been noted previously for RI complexes (1, 3, 12, 22, 26). Rate constants were calculated by treating the slower phase as a first-order process. The kinetic parameters for association of RI mutants with Ang were determined by stopped-flow spectrophotometry; association of RI mutants with RNase A and wild-type RI (wtRI) with Ang mutants was measured by examining the competition between RNase A and Ang or Ang mutant for RI or RI mutant (25). Most Ki values were calculated from the dissociation and association rate constants (1). The Ki value for the complex of the RI mutant Y434A with RNase A was obtained from the −[I] intercept of a plot of 1/v0 vs. [I] performed at a substrate (CpA) concentration well below Km; v0 is the initial rate of cleavage and [I] is the concentration of free inhibitor. The Ki for the D435A–RI·RNase A complex was calculated from the Ki for the D435A·Ang complex and the distribution of inhibitor between RNase A and Ang measured at equilibrium (14).
RESULTS AND DISCUSSION
Selection of Potential RI Contact Residues in Ang for Mutation.
The initial approach used for investigating the structural basis for the tight binding of RI to both Ang and RNase A was to examine the roles of Ang residues that are structurally analogous to those in RNase that serve as contacts in the pRI·RNase A complex (15). Presumably these amino acids would provide a substantial portion of the binding energy if RI recognizes the two ligands in similar ways. However, they are relatively few in number (see ref. 11). Of the 24 contact residues on RNase A, 4 are deleted in Ang and have no structural equivalents, another 8 have nonconservative replacements, and 2 are main-chain elements that are fairly distant from their Ang counterparts.
Ten contact residues on RNase A are retained or conservatively replaced in Ang. The importance of three of them—Arg-31, Lys-40, and His-114 of Ang—has already been assessed by mutagenesis. Lys-40 is clearly a key contact: its replacement by Gln decreases the binding energy by 4.2 kcal/mol, suggesting that it forms a hydrogen bond with an acidic residue on RI (27). Chemical modification data indicate that the corresponding lysine in RNase A is also involved in RI binding (28). In contrast, His-114 of Ang has a minor role (26) and Arg-31 makes no contribution (12). Another of the Ang residues under consideration, Ala-106, is also probably unimportant because its RNase A counterpart is involved only in two van der Waals contacts with RI. Leu-35 and Ile-42 of Ang correspond to Leu-35 and Val-43 of RNase A, which form nonoptimal hydrogen bonds with RI through their main-chain atoms. The contributions of such interactions to the Ang·RI complex cannot be assessed readily by Ang mutation.
The four remaining Ang residues—His-8, Gln-12, Asn-68, and Glu-108—were considered to be suitable targets for examination. His-8 occupies a position similar to that of Lys-7 in RNase A, which forms a hydrogen bond with RI. Replacement of this lysine by methylcysteine decreases the pRI binding energy by 2.4 kcal/mol (13). Gln-12, Asn-68, and Glu-108 are equivalent to Gln-11, Asn-71, and Glu-111 in RNase A, each of which also hydrogen bonds to the inhibitor. The importance of these residues for the RI·RNase A interaction has not yet been determined.
Effects of Ang Mutations on RI Binding.
We have examined the inhibitor binding roles of His-8, Gln-12, Asn-68, and Glu-108 in Ang by characterizing the interactions of the corresponding Ala mutants (H8A, Q12A, N68A, and E108A) with hRI. As in previous studies with Ang derivatives (1, 12, 26, 27), Ki values could not be determined directly due to the extreme tightness of binding and were instead calculated from the dissociation and association rate constants (Table 1). Mutation of His-8 produced a 4.6-fold increase in Ki that reflects primarily a change in the dissociation rate constant. Substitutions of Gln-12 and Asn-68 increased the Ki value by only 60% and 20%, respectively, and replacement of Glu-108 improved it slightly. Association of the last derivative was twice as rapid as for unmodified Ang. The minor effects of these four mutations, together with the earlier results detailed above, suggest that interactions of Ang residues that are structurally equivalent to RI contact residues in RNase A make only a small contribution to the overall binding energy and, therefore, that Ang and RNase A bind to RI in largely different ways.
Table 1.
Kinetic constants for binding of Ang mutants to RI
| Ligand | kd × 107, s−1 | ka × 10−8, M−1·s−1 | Ki × 1015, M | Ki,mut* Ki,A | ΔΔG,†kcal/mol |
|---|---|---|---|---|---|
| Ang‡ | 1.4 | 2.8 | 0.5 | — | — |
| H8A | 4.5 | 2.0 | 2.3 | 4.6 | 0.9 |
| Q12A | 2.0 | 2.4 | 0.83 | 1.6 | 0.3 |
| N68A | 2.3 | 3.8 | 0.61 | 1.2 | 0.2 |
| E108A | 1.6 | 5.5 | 0.29 | 0.6 | −0.3 |
Natural RI was used. kd and ka are the dissociation and association rate constants, respectively (1, 27). Ki vales represent kd/ka.
*Ki for mutant protein divided by Ki for Ang.
Difference in free energies of binding Ang and mutant proteins, calculated fro the equation ΔΔG = −RTln (Ki,A/Ki,mut). ΔG for the native complex is −20.8 kcal/mol.
Selection of RI Residues for Mutation.
The specific nature of the differences in the modes of interaction of Ang and RNase A with RI was investigated by mutating hRI, focusing particularly on residues that appear to form strong contacts with RNase A that would not be duplicated in the Ang complex. Twenty-eight RI residues are seen to interact with RNase A in the crystal structure of the pRI–ligand complex. Four lie within the segment 1–93 that has been shown to provide only 3% of the binding energy (29), and it is therefore unlikely that any of these is important. No biochemical evidence pertinent to the roles of other individual residues has been reported, although the results of a deletion mutagenesis study on hRI (14) indicate that interactions with the C-terminal segment 372–460 may be crucial for both RNase A and Ang binding. Seven residues within this region and two from other parts of hRI were selected for mutation (Table 2; Fig. 1). The pRI equivalents of four of these (Glu-287, Lys-320, Glu-401, and Arg-457) bind oppositely charged amino acids in RNase A that are not conserved in Ang. The porcine counterparts of Tyr-434 and Tyr-437 interact extensively with regions of RNase A that are only partially maintained in Ang; moreover, the Ang residues that correspond to the RNase contacts with Tyr-437 are known (Asn-68, Glu-108, His-114) or suspected (Ala-106) to have little functional importance. Cys-408 is analogous to Cys-404 in pRI, which makes a 2.8-Å hydrogen bond with a loop of RNase A that is not present in Ang. The C-terminal serine (460 in hRI) forms hydrogen bonds with Lys-7 and Gln-11 of RNase A. As shown above, the Ang counterpart of Lys-7, His-8, contributes much less to binding strength than does the lysine, and Gln-12 of Ang does not appear to form any significant interactions (Table 1).
Table 2.
RI residues selected for mutation
| RI residue | Contacts in RNase A | Corresponding Ang residues |
|---|---|---|
| E287 | K91 | P88 |
| K320 | E86 | L83 |
| E401 | R39 | P38 |
| C408 | K66 | — |
| Y434 | L35, R39, K41, P42 | L35, P38, K40, D41 |
| D435 | K41, V43 | K40, I42 |
| Y437 | N67, Q69, N71, A109, E111, H119 | —, —, N68, A106, E108, H114 |
| S460 | K7, Q11 | H8, Q12 |
In contrast to the other RI residues mutated, Asp-435 was considered to be a likely major contact for Ang. This expectation was not based directly on the structure of the RI·RNase A complex, but rather on a model (not shown) of the Ang complex obtained by superposition of the atomic coordinates of Ang (1ANG-PDB) onto those of RNase A in its RI complex (Quanta; Molecular Simulations, Waltham, MA). In this model, the carboxylate of Asp-435 is close to the ɛ-amino group of the Ang residue Lys-40, which, as already noted, is thought to form a strong hydrogen bond with RI. Moreover, no other potential hydrogen-bonding partner on RI lies in the vicinity of Lys-40. Although this Asp–Lys hydrogen bond is not observed in the RI·RNase complex (where the corresponding lysine makes only van der Waals contacts with RI), the orientation of these residues may well have been altered by the presence of a sulfate ion that contacts both of them.§
Expression and Purification of RI Mutants.
All RI residues selected were mutated to Ala, with the exception of the C-terminal residue Ser-460, which was deleted to eliminate any potential contacts by its α-carboxylate group. Mutant proteins were expressed in E. coli and purified by RNase A-Sepharose chromatography. The yields were similar to that obtained for wtRI (1–1.8 mg per 8 liters of bacterial culture) in all cases except for Y434A. Only 80 μg of this mutant were recovered, suggesting that replacement of Tyr-434 greatly weakens binding to the RNase affinity ligand. Since wtRI binds much more tightly to Ang than to RNase A, Ang-Sepharose was used for isolating a second preparation of Y434A, with a resultant 12-fold increase in yield.
Kinetics of Dissociation of RNase A Complexes with RI Mutants.
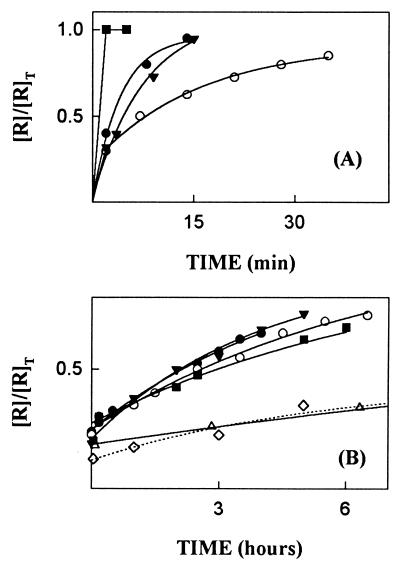
Earlier studies showed that when single-site mutations produced large decreases in protein–protein binding affinity they were mainly due to effects on the dissociation rate constant (kd) (12, 27, 31, 32). Therefore, RI mutants were screened initially by measuring this parameter. The ligand used was RNase A because the residues mutated were for the most part selected on the basis of their predicted importance for binding this protein. Dissociation was followed by assaying the enzymatic activity of RNase A released from the complex at various times after the addition of excess Ang as a scavenger for free RI mutant. The kd value for the complex of RNase A with wtRI obtained by this method was 1.2 × 10−5·s−1, corresponding to a t½ of 16 h. Eight of the nine mutants dissociated more quickly (Fig. 2). The most dramatic change was observed with Y434A, which was fully released during the mixing time of the assay (<30 s). Dissociation of the D435A complex was also too rapid to allow determination of a value for kd by this method. Y437A and the Ser-460 deletion mutant [des(460)] dissociated less quickly, but still much faster than wtRI (t½ = 15 min and 5 min, respectively). E287A, K320A, E401A, and R457A were released 3- to 5-fold more rapidly than RI (t½ = 4.6, 4.9, 3.6, and 3.3 h, respectively), and C408A dissociated at about the same rate as RI (t½ = 22 h). Additional kinetic studies were performed with the four mutants—Y434A, D435A, Y437A, and des(460)—that had substantially increased dissociation rates.
Figure 2.
Dissociation of the complexes of RNase A with RI mutants. [R]T and []R] are the total and free concentrations of RNase, respectively, at the times indicated. (A) Y434A (▪), D435A (•), Y437A (○), and des(460) (▾). (B) E287A (•), K320A (▾), E401A (○); C408A (▵); R457A (▪); wtRI (⋄). Additional data points were obtained for C408A and wtRI at times beyond those shown.
Kinetics of Dissociation of Ang Complexes with RI Mutants.
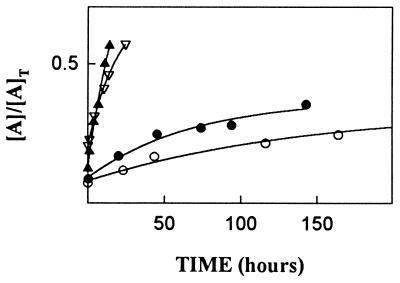
The standard method for examining dissociation of complexes of native RI with Ang and Ang mutants has been to use HPLC to follow the release of free Ang after addition of excess RNase A as a scavenger for free RI (1). However, pilot experiments revealed that this approach would not be suitable for analysis of most of the present RI mutants because the replacements affect binding of RNase A more drastically than that of Ang, and thus preclude the use of RNase A as an efficient scavenger. As an alternative method, Ang was biosynthetically labeled with 14C in E. coli and the radiolabeled protein was used to form the initial complex with RI mutants. Unlabeled Ang (20 eq) was then added as a scavenger for free inhibitor, and at various times free and bound [14C]Ang were separated by Mono S chromatography (1) and quantitated by scintillation counting. The kd value for the complex of [14C]Ang with wtRI was 1.1 × 10−7·s−1 (t½ = 73 days). All four mutant RI complexes dissociated more rapidly (Fig. 3). The largest changes were found for the Y434A and D435A complexes, which had half-lives of 26 h and 14 h, respectively—i.e., 66-fold and 127-fold shorter than for the wtRI complex. The half-lives of the Y437A and des(460) complexes were 26 days and 13 days, respectively. The magnitudes of the kd increases for all mutants except D435A were much smaller than those observed for the corresponding RNase A complexes.
Figure 3.
Dissociation of the complexes of Ang with the RI mutants Y434A (▿), D435A (▴), Y437A (○), and des(460) (•). [A]T and [A] are the total and free concentrations of Ang at the times indicated, respectively.
Kinetics of Association of Ang with RI Mutants.
Association of Ang with RI mutants was monitored by performing stopped-flow measurements of changes in tryptophan fluorescence that accompany complex formation (25). All mutant RI complexes exhibited fluorescence enhancements similar to that for the native complex (≈50%). The dependence of the pseudo-first-order rate constant (kobs) for this fluorescence increase on Ang concentration in all cases was consistent with the two-step mechanism described previously for association of Ang and native RI (25). In this mechanism, rapid formation of a loose complex is followed by a slower isomerization to the tight complex. Values for the dissociation constant of the initial complex (K1), the rate constant for conversion of this complex to the tight complex (k2), and the apparent second-order rate constant for association [ka (≈k2/K1)] are obtained from plots of 1/kobs vs. 1/[Ang], which yield straight lines with intercepts at 1/k2 and −1/K1 and a slope equal to 1/ka. These parameters were altered significantly for Y434A, D435A, and des(460) (Table 3). The ka values for the first two complexes were decreased by 3.5-fold and 2.8-fold, respectively. K1 values for these derivatives appeared to be changed by even greater factors that were then partially offset by increases in k2. However, it should be noted that these individual parameters could not be quantitated as accurately as the k2/K1 ratio owing to limitations of the stopped-flow instrument, which cannot measure kobs values more than ≈100 s−1. K1 and k2 values for des(460) were both decreased by 2- to 3-fold, yielding a ka value only 30% lower than for wtRI.
Table 3.
Kinetic parameters for the association of Ang with RI mutants
| RI | K1, μM | k2, s−1 | k2/K1, M−1·s−1 |
|---|---|---|---|
| wt | 0.29 | 80 | 2.8 × 108 |
| Y434A | 2.80 | 213 | 7.6 × 107 |
| D435A | 1.50 | 154 | 1.0 × 108 |
| Y437A | 0.34 | 69 | 2.0 × 108 |
| Des460 | 0.14 | 26 | 1.9 × 108 |
Kinetics of Association of RNase A with RI Mutants.
No significant change in fluorescence occurs upon formation of the RI·RNase A complex. The rate constants for association of the complexes of the RI mutants Y437A and des(460) with RNase A were therefore determined by examining the competition between RNase and Ang for inhibitor (1, 14). Both mutants were found to associate somewhat less rapidly than wtRI: values for ka were 2.8 × 108 (Y437A) and 2.5 × 108 M−1·s−1 [des(460)], compared with 3.8 × 108 M−1·s−1 for RI. Rapid complex dissociation precluded the use of this approach for Y434A and D435A.
Dissociation Constants.
Ki values for the complexes of Ang with the four RI mutants and the complexes of RNase A with Y437A and des(460) were calculated from the rate constants for dissociation and association. The dissociation constants for the complexes of RNase with Y434A and D435A were determined by other methods because the individual rate constants could not be quantitated. Assays measuring the effects of Y434A on RNase A-catalyzed CpA cleavage showed that concentrations of inhibitor well above that of enzyme (0.1 nM) produced only partial inhibition. Thus it was possible to obtain the Ki from a standard kinetic plot of 1/v0 vs. [I]. Binding of D435A to RNase A was considerably tighter than for Y434A, and the Ki for this complex was determined by measuring the distribution of RI mutant between Ang and RNase at equilibrium. Mixtures of Ang, 1.6 molar equivalents of D435A, and 100, 250, or 375 eq of RNase were incubated for 4 days (>7 half-lives of dissociation of either complex). The Ki was then calculated from the concentrations of free and bound ligands. The values obtained from the three experiments were in excellent agreement.
The Ki values for the complexes of RNase A with the four mutants examined (Table 4) are all substantially higher than for the wtRI complex. The increases range from 84-fold for Y437A to 23,000-fold for Y434A, corresponding to losses in binding free energy of 2.6 to 5.9 kcal/mol. Thus each of the sites mutated appears to be important for complex stability. However, only one of these residues, Asp-435, makes a similar contribution to binding of Ang (Table 4). Mutations of Tyr-437 and Ser-460 produce relatively small increases in the Ki for Ang (4.1-fold and 8.5-fold, respectively). The effect of Tyr-434 replacement on affinity for Ang is much larger, but still nearly two orders of magnitude less than with RNase A. Consequently, the Y434A, Y437A, and des(460) mutants have greatly enhanced specificity for Ang vs. RNase A: their Ki values for Ang are lower than those for RNase A by factors of 7500, 1630, and 3640, respectively, compared with a factor of 80 for wtRI.
Table 4.
Kinetic constants for complexes of RI mutants
| RI | kd s−1 | ka, M−1·s−1 | Ki, M | Ki,mut*Ki,wt | ΔΔG† kcal/cal |
|---|---|---|---|---|---|
| RNase A complexes | |||||
| wt | 1.2 × 10−5 | 3.8 × 108 | 3.1 × 10−14 | ||
| Y434A | 7.2 × 10−10 | 23,000 | 5.9 | ||
| D435A | 1.5 × 10−11 | 470 | 3.6 | ||
| Y437A | 7.4 × 10−4 | 2.8 × 108 | 2.6 × 10−12 | 84 | 2.6 |
| Des460 | 2.9 × 10−3 | 2.5 × 108 | 1.2 × 10−11 | 387 | 3.5 |
| Ang complexes | |||||
| wt | 1.1 × 10−7 | 2.8 × 108 | 3.9 × 10−16 | ||
| Y434A | 7.3 × 10−6 | 7.6 × 107 | 9.6 × 10−14 | 246 | 3.3 |
| D435A | 1.4 × 10−5 | 1.0 × 108 | 1.4 × 10−13 | 358 | 3.5 |
| Y437A | 3.1 × 10−7 | 2.0 × 108 | 1.6 × 10−15 | 4.1 | 0.8 |
| Des460 | 6.3 × 10−7 | 1.9 × 108 | 3.3 × 10−15 | 8.5 | 1.3 |
Ki for mutant RI divided by Ki for wtRI.
Difference in free energies of binding wt and mutant RIs, calculated from the equation ΔΔG = −RTln(Ki,wt/Ki,mut). ΔG values for the native RNase A and Ang complexes are −18.4 kcal/mol and −21.0 kcal/mol, respectively.
Dissociation of Complexes of Ang Lys-40 Mutants with D435A–RI.
We investigated the existence of a proposed hydrogen bond between Asp-435 of RI and Lys-40 of Ang, the key residues for the RI–Ang interaction identified by mutagenesis, by determining the rate of dissociation of the Ang mutant K40Q from its complex with D435A–RI. The half-life was found to be 2.8 h, nearly identical to that measured for the complex of K40Q with wtRI (3.4 h; ref. 27). This provides strong evidence for the hydrogen bond and suggests, moreover, that Asp-435 does not interact with any Ang residues other than Lys-40. However, it is unclear whether the converse is also true because D435A dissociates from its complex with K40Q several-fold more rapidly than from its complex with native Ang (t½ = 14 h). This difference implies either that the Lys-40 amino group contacts some additional RI residue or that the presence of the Gln-40 amide group in K40Q is sterically disruptive.
These alternatives were distinguished by examining the rate of dissociation of a K40G Ang mutant from RI. The half-life (15.5 h) was nearly identical to that for the complex of native Ang with D435A–RI. This supports the second explanation and suggests that Lys-40 does not form important interactions with RI residues other than Asp-435. Thus, it would appear that the extensive contacts between the alkyl portion of the RNase A residue Lys-41 and the phenyl ring of Tyr-430 seen in the pRI complex (15) are not replicated in the Ang complex or that they contribute little to the strength of that complex. Consistent with this view, measurements of the dissociation rate of the K40G–Ang·Y434A–RI complex demonstrate that the effects of the K40G and Y434A mutations are independent. The half-life of this mutant–mutant complex is ≈10 min—i.e., 100-fold and 200-fold faster than for the complexes of K40G–Ang and Y434–RI with their unmutated partners, respectively. These increases in dissociation rate are comparable to those produced by each individual mutation.
Conclusions.
The present mutagenesis study reveals major differences in the molecular interactions of RI with two of its ligands, RNase A and Ang. Comparison of the structures of Ang and RNase A suggested that only few Ang residues are potentially capable of forming important contacts similar to those of their counterparts in RNase A. Individual mutations of four of these (His-8, Gln-12, Asn-68, and Glu-108) were shown here to produce relatively minor changes in binding affinity, and replacements of two others (Arg-31 and His-114) were found previously to have little or no effect (12, 26). Indeed, only a single Ang residue (Lys-40) that is analogous to an RI contact on RNase A has been demonstrated to have a major functional role (27). These findings indicate, albeit indirectly, that RI recognizes Ang and RNase in largely different ways.
More direct evidence in support of this conclusion is provided by the differential effects of some RI mutations on RNase A and Ang binding. Substitutions of three of the four RI residues examined here in detail—Tyr-434, Tyr-437, and Ser-460—produced 20- to 93-fold larger increases in the Ki values for RNase A than for Ang (Table 4). Consequently, interactions with Tyr-437 and Ser-460 appear to play a minor role in the Ang complex but contribute substantially to binding of RNase A. Tyr-434 is particularly critical for RNase recognition: its replacement by Ala decreases the binding energy of the complex by 5.9 kcal/mol—i.e., 32% of the total.¶ Although this residue is also important for Ang binding, its contribution is much smaller. Only Asp-435 plays similarly large roles in the two complexes.¶
The pRI equivalents of Tyr-434, Asp-435, and Tyr-437 lie on a single loop, which is close to the C-terminal serine (Fig. 1). All of these residues contact the active site region of RNase A, and the importance of their human analogues can for the most part be rationalized in terms of interactions observed in the crystal structure (Table 2). The side-chain OH of Tyr-430 (434 in hRI) hydrogen bonds to Leu-35 of RNase and the phenyl ring forms many van der Waals contacts with Pro-42, Arg-39, and the catalytic residue Lys-41. As a consequence, this amino acid is almost completely buried in the complex, whereas in the free inhibitor one face is exposed to solvent (15, 16). Additional binding energy may derive from the major reorientation of the RNase residue Arg-39 that is necessary for the ligand to accommodate the Tyr side chain. The pRI counterpart of Tyr-437 interacts extensively with the purine binding site of RNase A: its side chain hydrogen bonds to both Asn-71 and Glu-111 and makes van der Waals contacts with these and other residues. Ser-456 (460 in hRI) hydrogen bonds through its γ-OH and α-carboxylate to Lys-7 and Gln-11, components of two of the phosphate-binding subsites of RNase A. As noted above, the interaction with Lys-7 contributes 2.4 kcal/mol to pRI binding (13). The somewhat larger energy loss upon deletion of Ser-460 may indicate that Lys-7 is even more important for binding hRI or that the contact with Gln-11 in RNase A provides additional energy. Alternatively, removal of Ser-460 may weaken the interactions of neighbors such as Tyr-434.
The residue equivalent to Asp-435 in the pRI·RNase A crystal structure forms van der Waals contacts with Lys-41 and His-119, and a hydrogen bond with the carbonyl oxygen of Val-43 that can occur only if the Asp carboxylate is protonated. Such interactions seem unlikely to account for the key contribution of this residue at the pH of 6 used for kinetic determinations. We propose instead that in solution Asp-435 hydrogen bonds to the ɛ-amino group of Lys-41 and that this interaction is prevented in the crystal by the bound sulfate. This hypothesis is based on the demonstrated existence of the corresponding Asp–Lys hydrogen bond in the RI·Ang complex (see above) and the nearly identical effects of mutating this Asp on Ang and RNase binding.
The small changes in affinity for Ang associated with the replacement of Tyr-437 and deletion of Ser-460 in RI are consistent with the RI·RNase A crystal structure and effects of the Ang mutations reported here (Table 1) and in previous studies. Thus the Ang residues (Asn-68 and Glu-108) that correspond to the hydrogen-bonding partners for Tyr-437 in RNase A do not contribute to binding strength, perhaps indicating that they are not positioned similarly to Asn-71 and Glu-111 in the RNase complex. Minor interactions with His-114 (26) may then account for the 4-fold decreased affinity of Y437A. The loss of binding energy due to deletion of Ser-460 approximates the sum of the losses from mutating its putative hydrogen-bonding partners, His-8 and Gln-12. The effect of the Tyr-434 mutation on Ang binding appears to be more complex. Two of the RNase A residues that interact with this tyrosine (Arg-39 and Pro-42) are not conserved in Ang, and the counterpart of another (Lys-41) does not form any significant contacts with it. It is therefore reasonable that Tyr-434 is less important for binding of RNase A than that of Ang. At the same time, it is not clear how this residue makes its 3.3 kcal/mol contribution to the stability of the Ang complex.
The RI residues Tyr-434, Asp-435, Tyr-437, and Ser-460 appear to constitute a hot spot within the binding interface of the RNase A complex. Five additional RI residues examined by mutagenesis—Glu-287, Lys-320, Glu-401, Cys-408, and Arg-457—make only small contributions, estimated to be <1 kcal/mol on the basis of changes in kd, although all of them were expected to form hydrogen bonds or charge–charge interactions with RNase (15). These results therefore suggest that a major portion of the binding energy resides in a small region of the molecule. It should be noted, however, that the apparent energetic contributions of individual residues are frequently not fully additive, and that their sum can in fact greatly exceed the total energy (21, 31). It is therefore possible that other regions of RI yet to be examined may be of considerable importance.
Clearly, a large fraction of the binding energy for the Ang complex remains to be accounted for, since the contributions of the four RI residues examined, even if completely additive, would provide only 43% of the total. Part of this additional energy presumably derives from contacts for the Ang residues Arg-5 and Arg-32, whose replacements decrease the free energy of binding by 2.4 kcal/mol and 0.8 kcal/mol, respectively (12). Arg-5 would be expected to bind to the C-terminal region of RI, not far from the four residues examined here, whereas Arg-32 would interact near the N terminus. Crystallographic studies on the Ang·RI complex, together with mutational studies based on this structure, should ultimately identify the interactions that contribute the remainder of the binding energy and provide further insights into the different strategies used by RI to bind so extraordinarily tightly to its various ligands.
Acknowledgments
We thank J. Tversky, C. Roh, and J. Brito for excellent technical assistance; Dr. B. Kobe for the atomic coordinates of the pRI·RNase A complex; Dr. W. E. Brown for providing the pET11-Ang expression plasmid; Dr. D. J. Strydom for amino acid analyses; Dr. K. R. Acharya for Fig. 1; and Drs. D. S. Auld, J. F. Riordan, and B. L. Vallee for helpful discussions. This work was supported by the National Institutes of Health (Grant HL-52096).
ABBREVIATIONS
- RI
RNase inhibitor
- Ang
angiogenin
- hRI
human RI
- pRI
porcine RI
- wtRI
wild-type recombinant hRI
- des(460)
recombinant hRI in which Ser-460 is deleted
Footnotes
The complex was crystallized from 0.6 M ammonium sulfate. Kinetic parameters for RI interactions are determined in the absence of sulfate.
The thermal denaturation profiles for the RI mutants Y434A and D435A are nearly identical to that of wtRI (data not shown). Thus neither mutation appears to have a significant effect on the overall integrity of RI.
References
- 1.Lee F S, Shapiro R, Vallee B L. Biochemistry. 1989;28:225–230. doi: 10.1021/bi00427a031. [DOI] [PubMed] [Google Scholar]
- 2.Vicentini A M, Kieffer B, Matthies R, Meyhack B, Hemmings B A, Stone S R, Hofsteenge J. Biochemistry. 1990;29:8827–8834. doi: 10.1021/bi00489a046. [DOI] [PubMed] [Google Scholar]
- 3.Shapiro R, Vallee B L. Biochemistry. 1991;30:2246–2255. doi: 10.1021/bi00222a030. [DOI] [PubMed] [Google Scholar]
- 4.Bond M D, Strydom D J, Vallee B L. Biochim Biophys Acta. 1993;1162:177–186. doi: 10.1016/0167-4838(93)90145-h. [DOI] [PubMed] [Google Scholar]
- 5.Boix E, Wu Y, Vasandani V M, Saxena S K, Ardelt W, Ladner J, Youle R J. J Mol Biol. 1996;257:992–1007. doi: 10.1006/jmbi.1996.0218. [DOI] [PubMed] [Google Scholar]
- 6.Strydom D J, Fett J W, Lobb R R, Alderman E M, Bethune J L, Riordan J F, Vallee B L. Biochemistry. 1985;24:5486–5494. doi: 10.1021/bi00341a031. [DOI] [PubMed] [Google Scholar]
- 7.Beintema J J, Hofsteenge J, Iwama M, Morita T, Ohgi K, Irie M, Sugiyama R H, Schieven G L, Dekker C A, Glitz D G. Biochemistry. 1988;27:4530–4538. doi: 10.1021/bi00412a046. [DOI] [PubMed] [Google Scholar]
- 8.Fett J W, Strydom D J, Lobb R R, Alderman E M, Bethune J L, Riordan J F, Vallee B L. Biochemistry. 1985;24:5480–5486. doi: 10.1021/bi00341a030. [DOI] [PubMed] [Google Scholar]
- 9.Olson K A, Fett J W, French T C, Key M E, Vallee B L. Proc Natl Acad Sci USA. 1995;92:442–446. doi: 10.1073/pnas.92.2.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shapiro R, Vallee B L. Proc Natl Acad Sci USA. 1987;84:2238–2241. doi: 10.1073/pnas.84.8.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shapiro R, Riordan J F, Vallee B L. Nat Struct Biol. 1995;2:350–354. doi: 10.1038/nsb0595-350. [DOI] [PubMed] [Google Scholar]
- 12.Shapiro R, Vallee B L. Biochemistry. 1992;31:12477–12485. doi: 10.1021/bi00164a026. [DOI] [PubMed] [Google Scholar]
- 13.Neumann U, Hofsteenge Protein Sci. 1994;3:248–256. doi: 10.1002/pro.5560030209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee F S, Vallee B L. Biochemistry. 1990;29:6633–6638. doi: 10.1021/bi00480a012. [DOI] [PubMed] [Google Scholar]
- 15.Kobe B, Deisenhofer J. Nature (London) 1995;374:183–186. doi: 10.1038/374183a0. [DOI] [PubMed] [Google Scholar]
- 16.Kobe B, Deisenhofer J. Nature (London) 1993;366:751–756. doi: 10.1038/366751a0. [DOI] [PubMed] [Google Scholar]
- 17.Acharya K R, Shapiro R, Allen S C, Riordan J F, Vallee B L. Proc Natl Acad Sci USA. 1994;91:2915–2919. doi: 10.1073/pnas.91.8.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mosimann S C, Newton D L, Youle R J, James M N G. J Mol Biol. 1996;260:540–552. doi: 10.1006/jmbi.1996.0420. [DOI] [PubMed] [Google Scholar]
- 19.Lee F S, Fox E A, Zhou H-M, Strydom D J, Vallee B L. Biochemistry. 1988;27:8545–8553. doi: 10.1021/bi00423a007. [DOI] [PubMed] [Google Scholar]
- 20.Hofsteenge J, Kieffer B, Matthies R, Hemmings B A, Stone S R. Biochemistry. 1988;27:8537–8544. doi: 10.1021/bi00423a006. [DOI] [PubMed] [Google Scholar]
- 21.Clackson T, Wells J A. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 22.Shapiro R, Fox E A, Riordan J F. Biochemistry. 1989;28:1726–1732. doi: 10.1021/bi00430a045. [DOI] [PubMed] [Google Scholar]
- 23.Shapiro R, Harper J W, Fox E A, Jansen H-W, Hein F, Uhlmann E. Anal Biochem. 1988;175:450–461. doi: 10.1016/0003-2697(88)90569-6. [DOI] [PubMed] [Google Scholar]
- 24.Lee F S, Vallee B L. Biochem Biophys Res Commun. 1989;160:115–120. doi: 10.1016/0006-291x(89)91628-8. [DOI] [PubMed] [Google Scholar]
- 25.Lee F S, Auld D S, Vallee B L. Biochemistry. 1989;28:219–224. doi: 10.1021/bi00427a030. [DOI] [PubMed] [Google Scholar]
- 26.Shapiro R, Vallee B L. Biochemistry. 1989;28:7401–7408. doi: 10.1021/bi00444a038. [DOI] [PubMed] [Google Scholar]
- 27.Lee F S, Vallee B L. Biochemistry. 1989;28:3556–3561. doi: 10.1021/bi00434a061. [DOI] [PubMed] [Google Scholar]
- 28.Blackburn P, Gavilanes J G. J Biol Chem. 1980;255:10959–10965. [PubMed] [Google Scholar]
- 29.Hofsteenge J, Vicentini A M, Stone S R. Biochem J. 1991;275:541–543. doi: 10.1042/bj2750541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 31.Schreiber G, Fersht A R. Biochemistry. 1993;32:5145–5150. doi: 10.1021/bi00070a025. [DOI] [PubMed] [Google Scholar]
- 32.Cunningham B C, Wells J A. J Mol Biol. 1993;234:554–563. doi: 10.1006/jmbi.1993.1611. [DOI] [PubMed] [Google Scholar]