Abstract
We previously have presented evidence for prominent structural changes in helices F and G of bacteriorhodopsin during the photocycle. These changes were determined by carrying out electron diffraction analysis of illuminated two-dimensional crystals of wild-type bacteriorhodopsin or the Asp-96 → Gly mutant that were trapped at a stage in the photocycle after light-driven proton release, but preceding proton uptake from the aqueous medium. Here, we report structural analysis of the long-lived O intermediate observed in the photocycle of the Leu-93 → Ala mutant, which accumulates after the release and uptake of protons, but before the reisomerization of retinal to its initial all-trans state. Projection Fourier difference maps show that upon illumination of the Leu-93 → Ala mutant, significant structural changes occur in the vicinity of helices C, B, and G, and to a lesser extent near helix F. Our results suggest that (i) all four helices that line the proton channel (B, C, F, and G) participate in structural changes during the late stages of the photocycle, and (ii) completion of the photocycle involves significant conformational changes in addition to those that are associated with steps in proton transport.
Bacteriorhodopsin is a light-driven proton pump (1). Each cycle of proton transport is initiated by the all-trans to 13-cis photoisomerization of retinal and completed with the thermal re-isomerization of retinal and return of the protein to its initial conformation. A number of distinct intermediates that occur in the course of the photocycle have been identified by spectroscopic methods (2–4). Site-specific mutagenesis studies (5) and selection of transport-negative mutants (6) have identified key residues that play important roles in the formation and decay of these spectroscopic intermediates. Electron cryomicroscopic studies of two-dimensional crystals have resulted in the determination of a model for the structure of bacteriorhodopsin at atomic resolution (7, 8), allowing a structural interpretation of the spectroscopic and biochemical experiments.
To understand chemical aspects of the molecular mechanism of proton transport, it is also necessary to determine structural changes in the protein at different stages of the photocycle. A combination of neutron (9), x-ray (10–13), and electron diffraction (14, 15) experiments have begun to provide such information. Two general strategies have been used in these experiments. In one, structural changes have been observed by collecting diffraction data from wild-type bacteriorhodopsin where the photocycle has been slowed down by lowering the temperature, changing the pH, adding chaotropic reagents, or combining two or more of these variables. In the other strategy, mutants that have pronounced kinetic defects in specific stages of the photocycle have been used to trap and structurally characterize the corresponding intermediates. To a first approximation, the magnitude and nature of the structural changes determined by the different methods and by the different approaches are in good agreement with each other.
In previous electron diffraction experiments (14), we determined structural changes in the photocycle of wild-type bacteriorhodopsin (at 5°C) and the D96G¶ mutant (at 25°C). The experiments were carried out with the aid of a plunge-freeze apparatus, which allowed two-dimensional crystals of bacteriorhodopsin to be frozen in liquid ethane at different times after illumination with a flash of light. Crystals of both wild-type and the D96G mutant were trapped at a time in the photocycle that was subsequent to the release of a proton from the Schiff base, but before the uptake of a proton from the external aqueous medium. Under the conditions of our experiments, time-resolved visible spectroscopic experiments have shown that the M intermediate predominantly was accumulated in both wild-type bacteriorhodopsin (16) and the D96G mutant (17). Diffraction patterns recorded from the frozen crystals were processed to construct Fourier projection maps of the differences between the structures of the trapped intermediates and that of unilluminated bacteriorhodopsin. The most prominent structural changes observed in the photocycle of the D96G mutant were in the vicinity of helices F and G and were interpreted as an ordering of the cytoplasmic end of helix G and an outward tilt of the cytoplasmic end of helix F. Similar changes also were observed in the photocycle of wild-type bacteriorhodopsin, although the features in the vicinity of helix F were less pronounced in comparison to those near helix G.
The above observations raise an interesting question with respect to the conformational changes in the late stages of the photocycle. Does completion of the photocycle simply involve reversal of the changes observed in crystals of illuminated wild-type bacteriorhodopsin and the D96G mutant? Alternatively, because the protein-catalyzed thermal reisomerization of retinal must occur at a later stage in the photocycle, are there other distinct structural changes in the photocycle? One approach to addressing this question is to investigate light-driven conformational changes under conditions where the photocycle is interrupted at a stage subsequent to proton release and uptake, but before retinal reisomerization. As discussed below, the L93A mutant is an excellent candidate for such studies.
Replacement of Leu-93 by Ala (but not by Val) is known to result in a 250-fold decrease in the rate of completion of the photocycle due to the accumulation of a long-lived O intermediate (18). The kinetic defect in the photocycle of the L93A mutant occurs at a stage after the completion of proton release and uptake, and retinal reisomerization is kinetically the rate-limiting step in the photocycle of this mutant (19). We therefore have used the L93A mutant to determine structural changes at the retinal reisomerization stage of the photocycle. We show here that difference Fourier maps of light-induced protein conformational changes in the L93A mutant are different from those previously observed for the D96G mutant. The most prominent new feature is in the vicinity of helix C and is consistent with an outward movement of this helix. Combined with the results from structural changes in the photocycle of wild-type bacteriorhodopsin and the D96G mutant, the new findings suggest that all four helices that line the proton channel participate in structural changes during the photocycle, and that completion of the photocycle involves significant conformational changes in addition to those that are associated with steps in proton transport.
MATERIALS AND METHODS
Preparation of Crystalline Patches of Bacteriorhodopsin.
Purple membranes containing either wild-type bacteriorhodopsin or the L93A mutant were isolated from Halobacterium salinarium as described (20). The isolated membranes, which were typically 0.5–1.0 μm in size, then were fused in the presence of octyl glucoside and dodecyltrimethylammonium chloride as described (21) to obtain larger (average size ≈5 μm) crystalline patches.
Preparation of Specimens for Microscopy.
Samples were processed for electron microscopy essentially as described by Subramaniam et al. (14). Briefly, fused membranes were deposited on carbon-coated electron microscopic grids and blotted with Whatman paper to remove excess liquid. The fully hydrated grids (typically at ≈6°C) then were plunged into liquid ethane either after a dark adaptation period of typically 3 min, or after a flash of light (λ > 570 nm) to initiate the photocycle. The duration of the light flash was ≈1 msec, preventing the formation of secondary photointermediates resulting from illumination of the O intermediate (significant accumulation of the O intermediate is only observed at times ≫≈1 msec after illumination). The intensity was sufficient to excite most (>80%) of the bacteriorhodopsin molecules on the grid. Because only a maximum of ≈50% conversion into the O intermediate is observed under saturating illumination (19), the flash illumination conditions used here result in a higher concentration of the O intermediate on the grid than expected under steady-state illumination. The delay time between the flash and the freezing step was 5 sec in all of the experiments with the L93A mutant. Under these conditions, decay of the M intermediate is complete, and the O intermediate is fully accumulated (19). The grids were transferred immediately from liquid ethane into liquid nitrogen, and all subsequent manipulations of the sample were carried out at liquid nitrogen temperatures.
Recording of Diffraction Patterns and Construction of Fourier Difference Maps.
The specimens were analyzed using a Philips CM-12 microscope operating at 120 kV. Diffraction patterns were collected either on Kodak SO-163 film or with the aid of a charge-coupled device detector (22). Fourier maps determined using data recorded using film or the charge-coupled device camera showed the same features (S.S., unpublished observations). The patterns then were processed using previously described methods (21). Difference Fourier maps were obtained using phases for bacteriorhodopsin reported in Henderson et al. (7).
RESULTS
Crystals of both wild-type bacteriorhodopsin and the L93A mutant diffracted to a resolution of better than 3.5 Å. Averaged intensities from reflections with z* values ranging from −0.007 to 0.007 Å−1 were used to construct difference Fourier maps in projection. The experiments presented here address two questions: (i) does the replacement of Leu-93 by Ala perturb the structure of the protein? and (ii) what is the nature of light-induced structural changes in the L93A mutant, and how do they differ from those observed previously with wild-type bacteriorhodopsin and the D96G mutant?
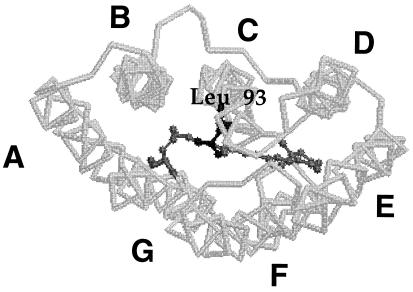
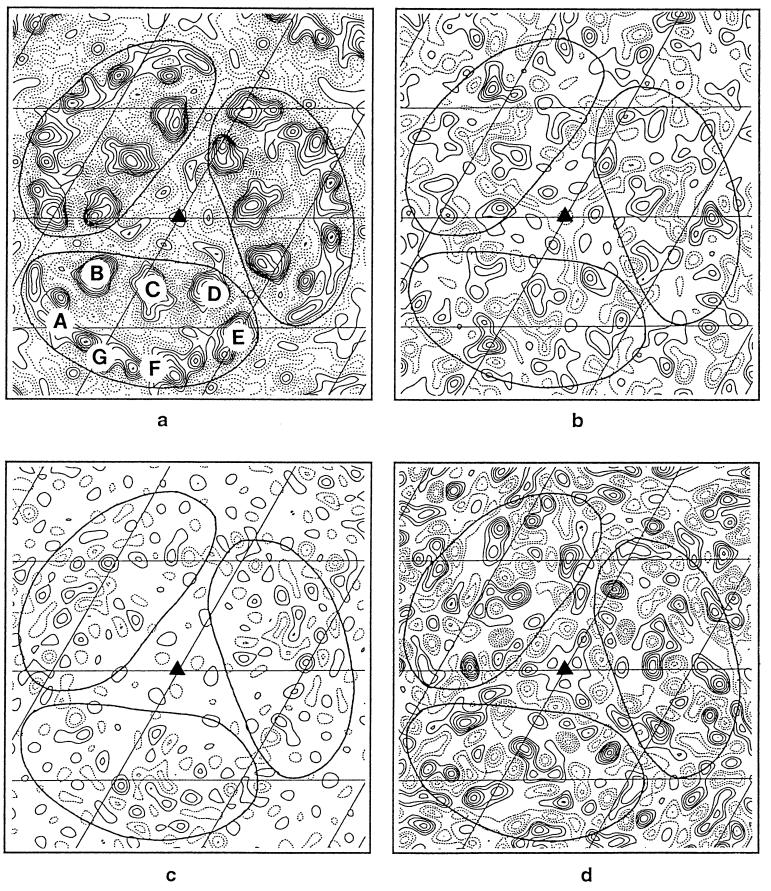
Fig. 1 shows a backbone drawing of the structure of bacteriorhodopsin as viewed from the cytoplasmic side, identifying the location of the seven transmembrane helices. A similar view of a projection electron density map of a trimer of wild-type bacteriorhodopsin is shown in Fig. 2a. To determine whether the replacement of Leu-93 by Ala alters the structure of bacteriorhodopsin, a difference map was calculated by comparing unilluminated crystals of the L93A mutant (at pH 5.2, 6°C in 50 mM potassium phosphate buffer) with unilluminated crystals of wild-type bacteriorhodopsin. As shown in Fig. 2b, significant density peaks are observed in the vicinity of helices C and G, and to a lesser extent near helix B. To establish whether these peaks were significantly above the noise level in the projection map, patterns obtained from unilluminated specimens of the L93A mutant were randomly divided into two sets, and a control difference map was constructed (Fig. 2c). Comparison of the maps in Fig. 2 b and c shows that the peaks in b are typically about two contour levels above the average noise level of the control difference map (c), indicating that the replacement of Leu-93 by Ala introduces a detectable structural change in the protein.
Figure 1.
Model for the atomic structure of bacteriorhodopsin showing the location of Leu-93. The view is from the cytoplasmic side of the membrane.
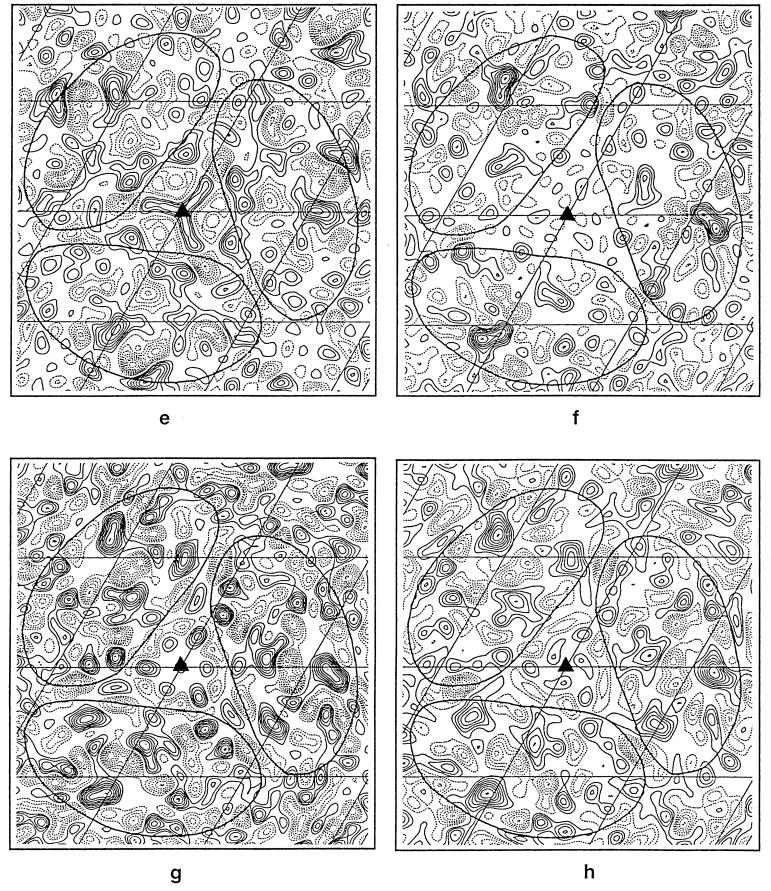
Figure 2.
Two-dimensional projection Fourier maps at 3.5-Å resolution. The solid triangles in each map indicate the locations of the threefold axes, and the grid bars are spaced at 20.81 Å, which is one-third of the unit cell. (a) Wild-type bacteriorhodopsin at 3.5-Å resolution. The seven helices are identified. (b) Difference map of structural changes in unilluminated samples of the L93A mutant (at 6°C, pH 5.2) and wild-type bacteriorhodopsin obtained using 29 crystals of the L93A mutant and 9 wild-type crystals. (c) Control difference map obtained using 15 unilluminated crystals of the L93A mutant used above into two random sets of 8 and 7 crystals. (d) Difference map of structural changes in the L93A mutant 5 sec after illumination at 6°C, pH 5.2, obtained using 28 illuminated and 29 unilluminated L93A mutant crystals. (e) Difference map of structural changes in the D96G mutant 20 msec after illumination at 25°C, pH 8.5, obtained using 8 illuminated and 12 unilluminated D96G crystals. (f) Difference map of structural changes in wild-type bacteriorhodopsin 20 msec after illumination at 5°C, pH 7.0, obtained using 7 illuminated and 9 unilluminated wild-type crystals. (g) Difference map of structural changes in the L93A mutant 5 sec after illumination at 6°C, pH 5.2, obtained using 28 illuminated crystals of the L93A mutant and 9 unilluminated wild-type crystals. (h) Difference map of structural changes in the L93A mutant 5 sec after illumination at 6°C, pH 7.0, obtained using 18 illuminated crystals of the L93A mutant and 9 unilluminated wild-type crystals. The solid lines seen in all the maps have been drawn to show the approximate boundaries of individual molecules in the trimer. All difference maps (b–h) have been contoured at the same interval, which, in turn, is 1/10 of the contour interval used for the projection map shown in Fig. 2a. The difference maps in e and f are the same as those previously published by Subramaniam et al. (14) except for the difference in scale factor.
Next, we determined light-induced structural changes in the L93A mutant. Fig. 2d shows a projection map comparing the difference between samples of the L93A mutant 5 sec after illumination (at pH 5.2, 6°C) with unilluminated samples of the L93A mutant. Under these conditions, the O intermediate decays with a t1/2 of ≈12 sec and therefore is accumulated in high yield (19). The largest changes are observed in the vicinity of helices C, G, and B, with slightly less pronounced changes near helix F. The peak near helix C is a pair of closely apposed negative- and positive-density peaks, and its location approximately coincides in projection with the location of the Leu-93 side chain. The direction of the largest-density gradient points outward from helix C, suggesting a net outward movement of this helix in the photocycle of the L93A mutant. The positive peaks on helices B and G are surrounded by regions of negative density, but are not associated with negative peaks as clearly as the peak near helix C. The map obtained with the L93A mutant is thus different from the map previously observed (14) with illuminated crystals of the D96G mutant (Fig. 2e), and in wild-type bacteriorhodopsin (Fig. 2f). Interestingly, in the L93A map (Fig. 2d), the peaks observed near helices F and G are at a location similar to the locations previously observed in the late stages of the photocycle in the D96G mutant (Fig. 2e).
These results demonstrate that the structure of the L93A mutant shows small, but significant, perturbations in the vicinity of helices B, C, and G even in the absence of illumination. Upon illumination, a conformational change occurs. It is worth emphasizing that the most prominent structural changes in unilluminated samples of the L93A mutant are at a location similar to those locations observed in illuminated samples. However, the extent of the changes observed upon illumination are at least 2 times as large as the structural difference observed with unilluminated crystals. The difference map shown in Fig. 2g, which compares illuminated L93A crystals to unilluminated wild-type bacteriorhodopsin crystals, provides an estimate of the net structural change in the O intermediate of the L93A mutant. As expected from the result shown in Fig. 2 b and d, the most pronounced features of this map are in the vicinity of helices C, G, and B, with a minor set of peaks near helix F.
The experiments reported above with the L93A mutant were carried out at pH 5.2, because lower pH values favor accumulation of the O intermediate state in wild-type bacteriorhodopsin (16), and also because our previous work (19, 23) had shown that the O intermediate is accumulated in high yield at lower pH values. To directly compare structural changes in the L93A mutant to those previously observed with wild-type bacteriorhodopsin and the D96G mutant at physiological pH, we also obtained a set of difference Fourier maps for the L93A mutant upon illumination at pH 7. As shown in Fig. 2h, the difference map comparing illuminated samples of the L93A mutant with unilluminated samples of wild-type bacteriorhodopsin shows features that are essentially similar to the corresponding map at pH 5.2 (Fig. 2g).
DISCUSSION
The results presented here provide a glimpse of conformational changes in the photocycle of the L93A mutant and show that significant protein conformational changes must occur either during or after the reisomerization of retinal. The most significant new feature in the L93A difference maps is the identification of structural changes in the vicinity of helix C. The pattern of positive and negative peaks is consistent with a net outward movement of this helix in the final stages of the photocycle.
An important question to be addressed is whether the structural changes observed in the L93A mutant are relevant to those that occur in wild-type bacteriorhodopsin. In general, one can envision two classes of explanations to account for a difference between structural changes observed in wild-type bacteriorhodopsin and a site-specific mutant: (i) the structural changes in the mutant are unique and different from those that occur in the wild-type protein, or that (ii) the sequence of intermediates and structural changes in the mutant are similar to those in wild-type bacteriorhodopsin, but different intermediates are kinetically accumulated in the mutant. It is not yet possible to discriminate unequivocally between these two scenarios, but an analysis of the spectroscopic and biochemical properties of the L93A mutant allows a more critical examination of these possibilities.
The visible absorption maximum of the L93A mutant in its light-adapted state is ≈542 nm (19), which is ≈30 nm lower than that of wild-type bacteriorhodopsin. The difference in λmax suggests that the retinal-binding pocket is altered in the L93A mutant, and the structural difference observed by electron diffraction analysis (Fig. 2b) supports this hypothesis. While this difference in the initial conformation of the L93A mutant suggests caution in extending the structural changes observed in the mutant to those that occur in wild-type bacteriorhodopsin, proton pumping experiments show conclusively that the L93A mutant functions as a vectorial proton pump like wild-type bacteriorhodopsin (18). Further, spectroscopic experiments show that the transient absorbance traces observed at different wavelengths can be explained using the same kinetic model and sequence of intermediates used for the wild-type protein (19, 23). Thus, even though the initial conformation of the mutant is different, it is reasonable to expect that both the mutant and wild-type bacteriorhodopsin would undergo a similar series of changes to vectorially transport protons across the membrane. On the basis of the above considerations, we favor the possibility that the structural changes observed in the mutant are relevant to the changes in wild type. However, unlike wild-type bacteriorhodopsin, the L93A mutation kinetically decouples proton uptake from retinal reisomerization, thereby allowing accumulation of an intermediate that normally would not accumulate in the photocycle of wild-type bacteriorhodopsin.
Another important question related to the above discussion is the nature of the connection between the structure of the O intermediate in the wild-type photocycle and the O intermediate in the mutant. In the wild-type photocycle, retinal is reisomerized to a twisted all-trans configuration (24) in the O intermediate. In contrast, retinal is in the 13-cis configuration in the long-lived O intermediate in the L93A mutant, and the reisomerization to the all-trans state occurs with decay of the O intermediate (19). Second, the amplitude of the O intermediate in the L93A mutant is approximately temperature-independent, in marked contrast to the decrease in amplitude observed in the case of wild-type bacteriorhodopsin. Furthermore, the activation energy for the transition from the O intermediate to the initial bacteriorhodopsin state in the mutant is ≈77 kJ/mol, which is ≈18 kJ/mol higher than that observed for wild-type bacteriorhodopsin. Together, these considerations argue that the long-lived O intermediate observed in the photocycle of the L93A mutant is different from the twisted, all-trans O intermediate that is populated in the late stages of the photocycle of wild-type bacteriorhodopsin. Thus while the structural changes observed here for the L93A mutant may well occur transiently during the course of the wild-type photocycle, they are not likely to reflect the conformation of the O intermediate in the wild-type photocycle.
It is interesting to speculate on the possible interpretation of the structural changes observed with the L93A mutant, especially in context of the structural work on other mutants. Electron, x-ray, and neutron diffraction experiments with the D96G (or D96N) mutants all are consistent with a conformational change occurring at helix F and G during the photocycle under conditions where the M intermediate is populated. Infrared spectroscopic studies (25) have suggested that although the optical spectroscopic features of this intermediate most resemble those of the M intermediate, the conformation of the peptide backbone bears a greater similarity to that of the N intermediate of the wild-type photocycle. Studies of light-driven structural changes in the F171C mutant by x-ray diffraction (13) and in the F219L mutant by electron diffraction (15) mutants show essentially identical features near helices F and G under conditions where an N-like intermediate is populated as determined by both optical and vibrational spectroscopic methods. A role for the movement of helix F at the N intermediate stage also has been proposed on the basis of the slower rate of decay of the N intermediate (and therefore of proton uptake) when the mobility of the cytoplasmic end of helix F was restricted by photo cross-linking with benzophenone (26). Irrespective of the nomenclature of the intermediates trapped in the different mutants, one can conclude from the above experiments that the structural changes in the vicinity of helices F and G are associated with the stage of the photocycle preceding proton uptake from the medium.
Under the conditions used to trap the O intermediate of the L93A mutant, the proton uptake step is completed, but retinal is not yet reisomerized to the all-trans state (19). The location of the features observed near helix C in Fig. 2 coincide with the location of Leu-93 in projection, consistent with the suggestion that the contact of Leu-93 with retinal is a key element in mediating conformational changes in the final stages of the photocycle (18, 19). In addition to the helix C feature, the maps show evidence for structural changes near helices B, G, and F, which are the three other helices lining the pathway of proton transport (7). The structural changes near helix B are in approximately the same location as the peaks seen in previous experiments where a significant amount of the M intermediate was accumulated (11, 14). Our observations support the conclusion from spin-labeling experiments (27) that there are likely to be conformational changes at the cytoplasmic end of helix C well after the formation of the M intermediate. However, because changes are observed in helices B, C, F, and G, it is clear that the structural changes on helices B and G are not likely to be limited to only the first phase of the conformational switch (13).
The data presented here were obtained using diffraction patterns obtained from untilted specimens. The next phase of this work will require collection of diffraction patterns from tilted specimens to obtain a three-dimensional map of structural changes in the photocycle of the L93A mutant. Structural studies of untilted and tilted specimens of the L93A, D96N double mutant are also likely to provide further insights into conformational changes in the late stages of the photocycle. Spectroscopic studies with this double mutant have shown that the decay of both M and O intermediates is slowed down, and conditions to selectively accumulate one or the other intermediate have been identified (23). Finally, electron diffraction analysis of the long-lived O intermediate in the recently described E204Q mutant (28) also will provide further valuable insights into structural changes that occur in the retinal reisomerization stage of the photocycle.
Acknowledgments
This work was supported by grants to S.S. from the National Eye Institute and the Searle Scholars Program/Chicago Community Trust.
Footnotes
Bacteriorhodopsin mutants are designated using the one-letter abbreviations for the amino acids. The amino acid to the left of the residue number is the original amino acid, and the amino acid on the right points to the substituting amino acid.
References
- 1.Oesterhelt D, Stoeckenius W. Proc Natl Acad Sci USA. 1973;70:2853–2857. doi: 10.1073/pnas.70.10.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ebrey T G. In: Thermodynamics of Membrane Receptors and Channels. Jackson M, editor. Boca Raton, FL: CRC; 1993. pp. 353–387. [Google Scholar]
- 3.Lanyi J K. Biochim Biophys Acta. 1993;1183:241–261. doi: 10.1016/0005-2728(93)90226-6. [DOI] [PubMed] [Google Scholar]
- 4.Lozier R H, Bogomolni R A, Stoeckenius W. Biophys J. 1975;15:955–963. doi: 10.1016/S0006-3495(75)85875-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khorana H G. J Biol Chem. 1988;263:7439–7442. [PubMed] [Google Scholar]
- 6.Soppa J, Otomo J, Straub J, Tittor J, Meessen S, Oesterhelt D. J Biol Chem. 1989;264:13049–13056. [PubMed] [Google Scholar]
- 7.Henderson R, Baldwin J M, Ceska T A, Zemlin F, Beckmann E, Downing K H. J Mol Biol. 1990;213:899–929. doi: 10.1016/S0022-2836(05)80271-2. [DOI] [PubMed] [Google Scholar]
- 8.Grigorieff N, Ceska T, Downing K H, Baldwin J M, Henderson R. J Mol Biol. 1996;259:393–421. doi: 10.1006/jmbi.1996.0328. [DOI] [PubMed] [Google Scholar]
- 9.Dencher N A, Dresselhaus D, Zaccai G, Büldt G. Proc Natl Acad Sci USA. 1989;86:7876–7879. doi: 10.1073/pnas.86.20.7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koch M H J, Dencher N, Oesterhelt D, Plöhn H-J, Rapp G, Büldt G. EMBO J. 1991;10:521–526. doi: 10.1002/j.1460-2075.1991.tb07978.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakasako M, Kataoka M, Amemiya Y, Tokunaga F. FEBS Lett. 1991;292:73–75. doi: 10.1016/0014-5793(91)80837-s. [DOI] [PubMed] [Google Scholar]
- 12.Kataoka M, Kamikubo H, Tokunaga F, Brown L S, Yamazaki Y, Maeda A, Sheves M, Needleman R, Lanyi J K. J Mol Biol. 1994;243:621–638. doi: 10.1016/0022-2836(94)90037-x. [DOI] [PubMed] [Google Scholar]
- 13.Kamikubo H, Kataoka M, Varo G, Oka T, Tokunaga F, Needleman R, Lanyi J K. Proc Natl Acad Sci USA. 1996;93:1386–1390. doi: 10.1073/pnas.93.4.1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Subramaniam S, Gerstein M, Oesterhelt D, Henderson R. EMBO J. 1993;12:1–8. doi: 10.1002/j.1460-2075.1993.tb05625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vonck J. Biochemistry. 1996;35:5870–5878. doi: 10.1021/bi952663c. [DOI] [PubMed] [Google Scholar]
- 16.Varo G, Lanyi J K. Biochemistry. 1991;30:5016–5022. doi: 10.1021/bi00234a025. [DOI] [PubMed] [Google Scholar]
- 17.Tittor J, Soell C, Oesterhelt D, Butt H, Bamberg E. EMBO J. 1989;8:3477–3482. doi: 10.1002/j.1460-2075.1989.tb08512.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Subramaniam S, Greenhalgh D A, Rath P, Rothschild K J, Khorana H G. Proc Natl Acad Sci USA. 1991;88:6873–6877. doi: 10.1073/pnas.88.15.6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delaney J K, Schweiger U, Subramaniam S. Proc Natl Acad Sci USA. 1995;92:11120–11124. doi: 10.1073/pnas.92.24.11120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oesterhelt D, Stoeckenius W. Methods Enzymol. 1974;31:667–678. doi: 10.1016/0076-6879(74)31072-5. [DOI] [PubMed] [Google Scholar]
- 21.Baldwin J, Henderson R. Ultramicroscopy. 1984;14:319–336. [Google Scholar]
- 22.Faruqi A R, Andrews H N, Henderson R. Nucl Instrum Methods Phys Res. 1995;367A:408–412. [Google Scholar]
- 23.Delaney J K, Subramaniam S. Biophys J. 1996;70:2366–72. doi: 10.1016/S0006-3495(96)79803-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith S O, Pardoen J A, Mulder P P J, Curry B, Lugtenburg J, Mathies R. Biochemistry. 1983;22:6141–6148. [Google Scholar]
- 25.Sasaki J, Schichida Y, Lanyi J K, Maeda A. J Biol Chem. 1992;267:20782–20786. [PubMed] [Google Scholar]
- 26.Brown L S, Varo G, Needleman R, Lanyi J K. Biophys J. 1995;69:2103–2111. doi: 10.1016/S0006-3495(95)80081-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steinhoff H-J, Mollaaghababa R, Altenbach C, Hideg K, Krebs M, Khorana H G, Hubbell W. Science. 1994;266:105–107. doi: 10.1126/science.7939627. [DOI] [PubMed] [Google Scholar]
- 28.Brown L S, Sasaki J, Kandori H, Maeda A, Needleman R, Lanyi J K. J Biol Chem. 1995;270:27122–27126. doi: 10.1074/jbc.270.45.27122. [DOI] [PubMed] [Google Scholar]