Abstract
We studied CD8 T cell responses against HIV-1, cytomegalovirus, Epstein–Barr virus, and influenza in 128 subjects and demonstrate that polyfunctional CD8 T cell responses, also including IL-2 production and Ag-specific proliferation, are predominantly driven by virus epitopes restricted by HLA-B alleles. Interestingly, these protective CD8 T cells are equipped with low-avidity T cell receptors (TCRs) for the cognate virus epitope. Conversely, HLA-A-restricted epitopes are mostly associated with “only effector” IFN-γ-secreting, with cytotoxicity, and with the lack of IL-2 production and Ag-specific proliferation. These CD8 T cells are equipped with high-avidity TCR and express higher levels of the T cell exhaustion marker PD-1. Thus, the functional profile of the CD8 T cell response is strongly influenced by the extent to which there is stimulation of polyfunctional (predominantly restricted by HLA-B) versus only effector (restricted by HLA-A) T cell responses. These results provide the rationale for the observed protective role of HLA-B in HIV-1-infection and new insights into the relationship between TCR avidity, PD-1 expression, and the functional profile of CD8 T cells.
Keywords: HLA genotype, PD-1 expression, functional profile, T cell avidity
Certain functions, such as the proliferation capacity and the secretion of IL-2, appear to be associated with effective T cell responses (1–6). On the basis of the analysis of IL-2 and IFN-γ, three functionally distinct populations of Ag-specific CD4 T cells (single IL-2, dual IL-2/IFN-γ, and single IFN-γ) and two functionally distinct populations of CD8 T cells (dual IL-2/IFN-γ and single IFN-γ) have been identified (1, 2, 5–9), and the presence of IL-2-secreting T cells was consistently associated with the Ag-specific proliferation capacity (1, 4–6). Recently, the term “polyfunctional” has been used to define T cell responses that, in addition to typical effector functions such as secretion of IFN-γ, TNF-α, and MIP-1β and cytotoxic activity, comprise distinct T cell populations also able to secrete IL-2 and that retain Ag-specific proliferation capacity (7). The term “only effector” defines T cell responses/populations able to secrete cytokines such as IFN-γ, TNF-α, and MIP-1β and endowed with cytotoxic activity but lacking IL-2 and proliferation capacity (7). Of interest, several studies have demonstrated that polyfunctional, and not only effector, T cell responses were associated with protective antiviral immunity (1–3, 5–9).
Several studies have clearly demonstrated the importance of HLA genotype in influencing the evolution of HIV and the progression of HIV-associated disease. In particular, certain HLA-B alleles are most closely associated with nonprogressive disease and low viral load or disease and have a dominant involvement on the clinical course of HIV-associated disease (10, 11).
Because polyfunctional CD8 T cell responses are associated with protective antiviral immunity and nonprogressive HIV disease and because HLA-B influences the outcome of HIV disease, it was of interest to investigate the relationship between HLA-B and polyfunctional responses to determine whether HLA-B influenced the generation of polyfunctional CD8 T cell responses. To test the hypothesis, we performed a comprehensive four-digit characterization of HLA genotype and of virus-specific CD8 T cell responses against HIV-1, CMV, EBV, and influenza (Flu) in 128 subjects comprising 69 HIV-negative, 50 HIV-1-infected subjects with chronic progressive infection, and 9 HIV-1-infected subjects with nonprogressive disease.
Results
CD8 T cell responses were studied in a cohort of 50 HIV-1-chronically infected subjects with progressive disease 1 year after initiation of antiviral therapy. The 50 subjects had gag-specific CD8 T cell responses. We performed four-digit HLA genotype, and it is important to mention that the proportion of HLA alleles associated with low relative hazards of progression (10) in the study population was not increased over what are the expected natural frequencies in a white Caucasian population (2 of 50 with HLA-B*5701 and 6 of 50 with HLA-B*2705). Furthermore, the long-term nonprogressors (LNTPs) were not a subset of the main population studied.
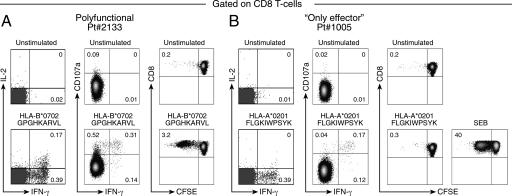
The initial epitope characterization was performed on the basis of potential epitopes inferred from the HLA genotype (12, 13). HLA restrictions and gag-derived peptides inducing specific responses are shown in supporting information (SI) Table 1. On the basis of this analysis, 20 subjects showed polyfunctional CD8 T cell responses, and 30 subjects showed only effector responses. Representative examples of typical polyfunctional CD8 T cell responses, as indicated by the presence of dual IL-2/IFN-γ, single IFN-γ, proliferating, and CD8 T cells with degranulation activity, and of only effector CD8 T cells, as indicated by the presence of single IFN-γ and CD8 T cells with degranulation activity but lacking proliferation capacity and IL-2 secretion, are shown in Fig. 1.
Fig. 1.
Functional profile of HIV-1-specific CD8 T cell populations. (A) Representative flow cytometry profiles of a polyfunctional response defined by IL-2 and IFN-γ secretion, degranulation activity (as measured by CD107a mobilization), and proliferation capacity (as measured by CFSE dilution). (B) Representative flow cytometry profiles of an only effector CD8 T cell response defined by IFN-γ secretion and degranulation activity and by the lack of IL-2 secretion and of proliferation capacity. Staphylococcal enterotoxin serotype B (SEB) was used as positive control.
CD8 T cell responses against 82 gag epitopes and 39 peptide-HLA associations were identified. The number of responses induced by individual gag epitopes and their frequency of response are shown in SI Table 2. The HLA restrictions and epitope mapping were experimentally assessed using HLA matched and mismatched BCL in 16 of the 39 gag-specific peptide–HLA associations identified (SI Fig. 7). The HLA class I genotype of the patients and of the HLA-matched and mismatched BCL used to confirm HLA restrictions are shown in SI Table 3. Epitopes were randomly selected. The HLA restrictions inferred from the literature were experimentally confirmed (12, 13).
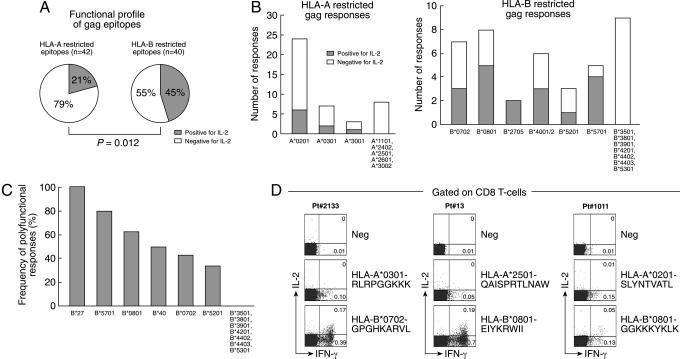
We then investigated the association between HLA-A/B alleles and both only effector and polyfunctional CD8 T cell responses. The large majority (33 of 42, 79%) of epitopes restricted by HLA-A were associated with only effector CD8 T cell responses compared with only 9 of 42 (21%) epitopes associated with polyfunctional responses (Fig. 2A). Of interest, ≈45% of epitopes restricted by HLA-B were associated with polyfunctional CD8 T cell responses, and 55% were associated with only effector CD8 T cell responses (Fig. 2A). The differences in the association between HLA-A and HLA-B and polyfunctional CD8 T cell responses were highly significant (P = 0.012).
Fig. 2.
Relationship between HLA-A and HLA-B and the functional profile of HIV-1-specific CD8 T cell populations. (A) Proportion of HIV-1 gag epitopes restricted by HLA-A and HLA-B inducing polyfunctional (positive for IL-2) and effector (negative for IL-2) CD8 T cell responses. (B) Association between HLA-A and HLA-B genotypes and polyfunctional CD8 T cell responses. (C) Hierarchy within HLA-B alleles in their association with polyfunctional responses. (D) Analysis of HLA-A restricted only effector and HLA-B restricted polyfunctional gag-specific CD8 T cell responses within the same blood samples. IL-2 and IFN-γ secretion was assessed in three patients selected on the basis of the simultaneous presence of polyfunctional and only effector responses restricted by HLA-B and HLA-A alleles, respectively.
We then determined whether there was a different distribution of HLA alleles within subjects with polyfunctional and only effector responses and a preferential association between certain HLA alleles and polyfunctional CD8 T cell responses. There were not significant differences in the distribution of HLA-A alleles. With regard to HLA-B alleles, HLA-B27 was significantly enriched in subjects with polyfunctional responses and HLA-B35 was significantly enriched in those with only effector responses (SI Fig. 8).
In-depth analysis of the total 82 gag-specific responses showed that amongst the 42 responses restricted by HLA-A, the polyfunctional gag-specific CD8 T cell responses were restricted by HLA-A*0201 (n = 6), HLA-A*0301 (n = 2), and HLA-A*3001 (n = 1) (Fig. 2B); among the 40 responses restricted by HLA-B, the polyfunctional gag-specific CD8 T cells were restricted by a panel of HLA-B genotypes, including HLA-B*0702 (n = 3), HLA-B*0801 (n = 5), HLA-B*2705 (n = 2), HLA-B*4001/2 (n = 3), HLA-B*5201 (n = 1), and HLA-B*5701 (n = 4) (Fig. 2B). Although HLA-B alleles, such as HLA-B57 and -B27, which have been shown to be associated with slow disease progression to AIDS (10), were associated with polyfunctional responses, it is important to underscore that they are not solely responsible for the preferential association between HLA-B and polyfunctional responses. In fact, if HLA-B57 and -B27 were not included in the analysis, then the difference between HLA-A and HLA-B regarding the association with polyfunctional responses was lost (P = 0.59). Similarly, the removal of other HLA-B alleles from the analysis led to a loss of association between HLA-B and polyfunctional responses as well (P = 0.13). Therefore, the data do not support the effects of specified single HLA-B alleles and rather indicate a broader, more generic effect of HLA-B group alleles and the presence of a hierarchy within HLA-B alleles in their association with polyfunctional responses (Fig. 2C).
In further support of this hypothesis, we analyzed HLA-A- and HLA-B-restricted CD8 T cell responses for polyfunctionality in the same blood sample in three patients. We consistently observed that HLA-B-restricted epitopes induced polyfunctional CD8 T cell responses, whereas HLA-A restricted epitopes only effector CD8 T cell responses (Fig. 2D).
A slight difference in viral load and not in CD4 T cell counts between the 20 subjects with polyfunctional and the 30 subjects with only effector CD8 T cell responses (4.54 ± 0.12 vs. 4.86 ± 0.11 Log RNA copies per milliliter, P = 0.04; 588 ± 60 vs. 515 ± 59 CD4 T cells per microliter, P = 0.2, respectively) was present before therapy. However, virus replication was effectively suppressed in both groups after antiviral therapy.
Taken together, these results indicate that HLA-B genotype influenced the generation of polyfunctional profile of CD8 T cells. They also confirmed that suppression of viral load was not sufficient for the generation of polyfunctional responses because only HLA-B responses, and not HLA-A-restricted responses, were associated with polyfunctional responses within the same patient.
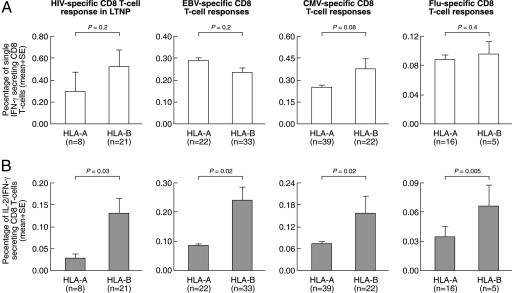
We then investigated the importance of HLA-B in (i) shaping the CD8 T cell response in HIV-1-infected individuals with nonprogressive disease, i.e., LTNPs, and (ii) in virus-specific CD8 T cell responses other than HIV. A large number of virus-specific (HIV-1-specific in LTNPs, CMV-, EBV- and Flu-specific) CD8 T cell responses restricted by HLA-A (n = 85) or HLA-B (n = 81) were then investigated (CMV-, EBV- and Flu-specific epitopes are listed in SI Table 4). After Ag-specific stimulation, the percentage of dual IL-2/IFN-γ-secreting cells, but not of single IFN-γ-secreting cells, was significantly higher in the epitopes restricted by HLA-B compared with HLA-A in all four models of virus-specific CD8 T cell response (Fig. 3). These results indicate that HLA-B has a general role in governing the generation and the magnitude of polyfunctional CD8 T cell responses.
Fig. 3.
Influence of HLA-B on CD8 T cell responses against CMV, EBV, Flu, and HIV-1 in LTNP. Cumulative data on the percentage of single IFN-γ and dual IL-2/IFN-γ CD8 T cells after stimulation with HIV-1-, CMV-, EBV-, and Flu-derived peptides restricted by HLA-A and HLA-B are shown. Cumulative percentage data are expressed as the mean ± SE.
We then characterized eventual differences in the responsiveness of CD8 T cell populations restricted by HLA-A and HLA-B. For these purposes, we investigated the T cell receptor (TCR) avidity for the cognate epitope of polyfunctional and only effector CD8 T cell populations. The TCR avidity was determined in CMV- and EBV-specific CD8 T cells obtained from HIV-negative healthy donors and in HIV-1-specific CD8 T cells obtained from subjects with progressive disease.
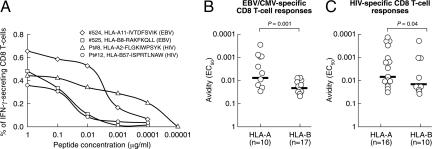
TCR avidity was initially analyzed in HLA-A vs. HLA-B restricted CD8 T cell responses. Subject nos. 524 and 525 had an EBV-specific CD8 T cell response against the IVTDFSVIK and RAKFKQLL peptides restricted by HLA-A11 and by HLA-B8, respectively. Patients no. 8 and no. 12 had an HIV-1-specific CD8 T cell response against the FLGKIWPSYK and ISPRTLNAW peptides restricted by HLA-A2 and by HLA-B57, respectively. Following Ag-specific stimulation with decreasing peptide con centration and assessment of total IFN-γ secreting CD8 T cells, there was a rapid loss in the percentage of RAKFKALL- and ISPRTLNAW-specific CD8 T cells restricted by HLA-B8 and HLA-B57 compared with IVTDFSVIK- and FLGKIWPSYK-specific CD8 T cells restricted by HLA-A11 and HLA-2 (Fig. 4A). The significant difference between CD8 T cell responses restricted by HLA-A and HLA-B in the avidity for the cognate epitopes was confirmed in a larger number of responses (Fig. 4 B and C).
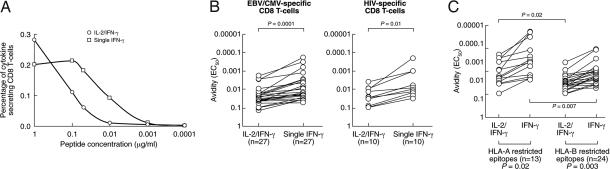
Fig. 4.
TCR avidity of HLA-A- and HLA-B-restricted responses. (A) Representative examples of the responsiveness of HLA-A and HLA-B restricted EBV- and HIV-1-specific CD8 T cell responses after stimulation with decreasing peptides concentrations. Two EBV-derived peptides restricted by HLA-A11 and -B8 and two HIV-1-derived peptides restricted by HLA-A2 and -B57 were used. (B and C) Cumulative data for EBV/CMV (B) and HIV-1 (C) are shown. The EC50 corresponds to the peptide concentration needed to achieve a half-maximal response.
We then analyzed TCR avidity on the two functionally distinct CD8 T cell populations, i.e., dual IL-2/IFN-γ and single IFN-γ (representative subject no. 525 is shown). As shown in Fig. 5, after Ag-specific stimulation with decreasing peptide concentration and assessment of IL-2 and IFN-γ, there was a rapid loss in the percentage of dual IL-2/IFN-γ compared with single IFN-γ-secreting CD8 T cell populations (Fig. 5A). The significant difference between IL-2/IFN-γ and single IFN-γ in the avidity for the cognate epitopes were confirmed for 27 CMV/EBV-specific responses in healthy subjects (Fig. 5B). A similar difference was observed for HIV-1-specific responses (Fig. 5B). We then compared the TCR avidity of IL-2/IFN-γ and single IFN-γ CD8 T cell populations on the basis of the HLA restriction. Of interest, the EC50 of both IL-2/IFN-γ and single IFN-γ-secreting virus-specific CD8 T cell populations recognizing virus epitopes restricted by HLA-B was significantly higher compared with that of epitopes restricted by HLA-A (Fig. 5C).
Fig. 5.
TCR avidity of single IFN-γ and dual IL-2/IFN-γ CD8 T cells. (A) Representative example (subject no. 525) of the responsiveness of the single IFN-γ and dual IL-2/IFN-γ CD8 T cells after stimulation with decreasing peptide concentrations of one HLA-B8 restricted EBV-derived peptide (RAKFQLL). (B) Cumulative data on the TCR avidity of EBV/CMV- and HIV-1-specific single IFN-γ and dual IL-2/IFN-γ CD8 T cells. (C) Cumulative data on the TCR avidity of virus-specific (HIV-1+CMV+EBV) single IFN-γ and dual IL-2/IFN-γ CD8 T cells recognizing HLA-A- and HLA-B-restricted epitopes. The EC50 corresponds to the peptide concentration needed to achieve a half-maximal response.
Taken together, these results indicate that overall virus-specific CD8 T cell populations recognizing virus epitopes restricted by HLA-B are equipped with lower avidity TCR for the cognate epitopes compared with those recognizing epitopes restricted by HLA-A.
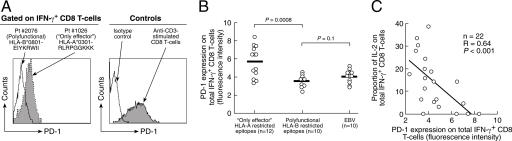
We have also analyzed the expression of programmed death 1 (PD-1) that has been recently shown to be selectively up-regulated by exhausted CD8 T cells during chronic virus infections (14–17). PD-1 expression was assessed in HLA-B restricted polyfunctional versus HLA-A-restricted only effector HIV-1-specific CD8 T cell populations. PD-1 expression was significantly up-regulated in HLA-A-restricted only effector compared with HLA-B-restricted polyfunctional CD8 T cell populations (Fig. 6 A and B). Representative examples and cumulative data are shown (Fig. 6 A and B). PD-1 expression on HLA-B-restricted polyfunctional HIV-1-specific CD8 T cell populations were similar to those observed in EBV-specific CD8 T cell populations from HIV-negative subjects (Fig. 6B). Of interest, there was a significant negative correlation between the proportion of HIV-1-specific IL-2-secreting CD8 T cell populations and PD-1 expression (Fig. 6C).
Fig. 6.
PD-1 expression. (A) Representative flow cytometry profiles of PD-1 expression on HLA-B restricted polyfunctional and HLA-A restricted only effector HIV-1-specific CD8 T cells. PD-1 expression was assessed on the total IFN-γ secreting CD8 T cells. PD-1 expression on CD8 T cells after anti-CD3 stimulation was used as control. (B) Cumulative data on the expression of PD-1 in the total IFN-γ secreting CD8 T cells in HIV-infected subjects with HIV-1-specific HLA-B and HLA-A restricted responses and in HIV-negative subjects with EBV-specific responses are also shown. (C) Correlation between the proportion of HIV-1-specific IL-2 secreting CD8 T cells and PD-1 expression.
Discussion
The results reported here provide insights into the delineation of the factors influencing the functional diversity of Ag-specific CD8 T cells and demonstrate that the HLA genotype is an important factor in shaping the functional profile of CD8 T cells. They also help to shed light on the protective role of HLA-B genotype in virus infections (10, 11) and on the relationship between TCR avidity for the cognate epitope and effective/protective response in chronic virus infection.
The influence of HLA-B in the selection of polyfunctional CD8 T cell responses is strongly supported by two findings: (i) the generality of the effect of HLA-B to four RNA and DNA viruses and (ii) the observation that HIV-1-specific HLA-A restricted CD8 T cell responses were only effector and HLA-B-restricted were polyfunctional CD8 T cell responses within the same patients. Furthermore, the recent observation of the increased magnitude of HLA-B-restricted HIV-1-specific CD8 T cell responses (18) can be extended to polyfunctional CMV-, EBV-, and Flu-specific CD8 T cell responses, which were significantly greater when virus-specific epitopes were restricted by HLA-B than HLA-A alleles.
The mechanisms by which HLA-B alleles also induce polyfunctional CD8 T cell responses rather than only effector responses, as it is mostly the case for HLA-A, and why they confer protection from HIV-associated disease are unknown. Our present findings demonstrating that these responses were predominantly induced by epitopes restricted by HLA-B may provide the rationale for explaining the protective role of HLA-B reported in virus infection such as HIV (10, 11) and more recently HCV (19). In this regard, it is currently debated whether the protective effect is limited to few alleles such as HLA-B57 and HLA-B27 or whether it is a broader effect of HLA-B alleles. Our observations regarding polyfunctional responses to HIV-1, EBV, CMV, and Flu are not explained simply by the effect of specified single HLA-B alleles but by a broader, more generic effect of HLA-B group alleles. Although certain HLA-B alleles, such as HLA-B27 and HLA-B57, associated with low relative hazard to AIDS, were enriched in subjects with polyfunctional gag-specific CD8 T cell responses, these responses were not sufficient to support the significant difference between HLA-A and HLA-B alleles in selecting for polyfunctional CD8 T cell responses. As previously shown for the association between HLA-B alleles and low relative hazard to AIDS (11), the present results indicate a hierarchy within HLA-B alleles in influencing the generation of polyfunctional CD8 T cell responses. HLA-B alleles such as HLA-B57 and -B27 were associated with polyfunctional responses in ≈100% of cases, HLA-B7, -B40, and -B8 were associated with polyfunctional responses in ≈50%, and HLA-B52 was associated with polyfunctional responses in ≈30% of cases.
Our TCR avidity results provide new insights into understanding the relationship between TCR avidity of CD8 T cells, their functional profile, and disease control in chronic virus infections. A recent study (18) failed to show a difference in TCR avidity between HLA-A- and HLA-B-restricted epitopes in HIV-1 infection. However, there was no functional characterization of only effector and polyfunctional responses. Our results indicate that polyfunctional CD8 T cell populations are equipped with low-avidity TCR compared with only effector CD8 T cell populations. However, it is worth mentioning that, to measure TCR avidity, we have used reference strain sequences of the epitopes for stimulation. Therefore, we cannot exclude that, in some instances, the peptide sequences used do not perfectly correspond to the sequences of the circulating virus in vivo. However, we have shown the same phenomenon also for responses to other virus infections in which sequence changes and partial escape from recognition are highly unlikely and in which it is also unlikely that changes in the epitope sequences of the circulating virus would occur more frequently for HLA-B- than for HLA-A-restricted epitopes.
Because polyfunctional CD8 T cell responses have been clearly shown to be associated with better disease control in chronic virus infections in humans and mice, these results may appear to be somewhat in conflict with the current paradigm of higher avidity corresponding to greater efficacy of the CD8 T cell response (20–22). This paradigm is predominantly based on the observation that higher avidity cytotoxic CD8 T cells have superior antitumor activity and mediate effective antiviral activity during the acute phase of viral infection (20–22).
It is not our intention to challenge the relationship between higher avidity CD8 T cells and superior antitumor and antiviral activity. However, we want to propose that the superiority of higher avidity CD8 T cells may be substantially undermined under conditions of Ag persistence and high levels of viral load. This hypothesis is supported by two observations: (i) higher functional avidity of CD8 T cells is most strongly associated with escape mutations (23–25) in both SIV and HIV infections, and (ii) higher avidity CD8 T cells are more susceptible to activation-induced cell death and exhaustion (26, 27). Our results indicate that only effector CD8 T cell populations were equipped with high-avidity TCR and expressed higher PD-1 levels, further supporting our hypothesis that these cells are at risk of exhaustion and thus that their antiviral activity may be substantially impaired.
Polyfunctional CD8 T cell populations were equipped with low-avidity TCR and expressed low PD-1 levels. Although their intrinsic antiviral activity is likely inferior to the higher avidity CD8 T cell populations, it is possible that, under conditions of Ag persistence and high Ag load, they may be more suitable for mediating at least partial viral control. Because of being equipped with low-avidity TCR, polyfunctional CD8 T cell populations are less susceptible to exhaustion and less efficient in driving escape mutants. In support of the hypothesis that low-avidity TCR polyfunctional CD8 T cells may be more efficient under conditions of Ag persistence, recent studies dealing with antitumor activity during tumor persistence showed that tumor control was associated with cytotoxic CD8 T cells equipped with low-avidity TCR (28) and the accumulation of low-avidity antimelanocortin receptor 1 CD8 T cells in benign skin lesions of melanoma-related depigmentation (29). Similarly, polyfunctional CD8 T cells are invariably found in chronic virus infections with effective virus control, such as HIV-1 infection in LTNP, CMV, and EBV infections. Furthermore, IL-2/IFN-γ-secreting CD8 T cells have been shown to mediate effective antiviral activity comparable with typical effector CD8 T cells in the LCMV model (30).
Therefore, notwithstanding the intrinsic superior antiviral activity of higher avidity TCR CD8 T cells, low-avidity TCR polyfunctional CD8 T cells may be more fit in providing disease control during chronic virus infection.
Finally, these results may provide insights into the development of strategies for the design of T cell vaccines that may select predominantly for the generation of polyfunctional CD8 T cell responses.
Materials and Methods
Study Groups.
Fifty subjects with progressive chronic HIV-1 infection were recruited in this study. All patients were enrolled in clinical trials, receiving antiviral therapy with nucleoside and protease inhibitors (31). At baseline (before the initiation of antiviral therapy), the median CD4 T cell count was 411 cells per microliter, and the median HIV-1 plasma viremia was 4.67 Log RNA copies per milliliter. One year after initiation of antiviral therapy, median CD4 T cell count were 621 cells per microliter, and median plasma viremia was 20 HIV-1 RNA copies per milliliter. A modified PCR assay with a limit of detection of 20 copies (32) was used in patients with levels of viremia <50 copies. Nine HIV-1-infected patients with nonprogressive disease, i.e., LTNP, as defined by documented HIV-1 infection since >14 years, stable CD4 T cell counts >500 cells per microliter (median: 1,302), and plasma viremia <1,000 HIV-1 RNA copies per milliliter (median: 17) were also included. In addition, blood from 69 HIV-negative subjects was obtained either from the local blood bank (Lausanne, Switzerland) or from laboratory coworkers. The studies were approved by the Institutional Review Board of the Centre Hospitalier Universitaire Vaudois and informed consent was obtained from each patient.
Synthetic Peptides.
All of the peptides used in this study were HPLC purified (>80% purity). Fine epitope mapping was performed on the predicted HLA genotype according to the Los Alamos database (12, 13) and on the HLA class I genotype of the patients. On the basis of these analyses, 32 HLA class I restricted gag epitopes were identified (SI Table 2). Furthermore, a set of peptides (n = 28) most frequently recognized in CMV, EBV, and Flu infection were used to assess the functional profile of virus-specific CD8 T cell responses (33, 34).
Detection of IFN-γ and IL-2 Secretion (ICS).
Cryo-preserved blood mononuclear cells (1–2 × 106) were stimulated overnight as described in ref. 6. At the end of the stimulation period, cells were washed, permeabilized, and then stained with CD8-PerCP-Cy5.5, CD4-FITC, IFN-γ-APC, and IL-2-PE (BD Pharmingen, San Diego, CA). For PD-1 expression, cells were stained with anti-PD-1-FITC, CD8-PerCP-Cy5.5, and IFN-γ-APC antibodies (BD Pharmingen). Data were acquired on a FACScalibur or on an LSRII and were analyzed using CellQuest and DiVa softwares (BD Pharmingen), respectively. The number of lymphocyte-gated events ranged between 105 and 106 in the flow cytometry experiments shown. With regard to the criteria of positivity of ICS, the background in the unstimulated controls never exceeded 0.01% to 0.02%. To be considered positive, an ICS had to have a background <20% of the total percentage of cytokine-positive cells in the stimulated samples.
Functional Avidity.
The stimulation was performed as described in ref. 6. The functional avidity of responses was assessed by performing limiting peptide dilutions and determining the peptide concentration required to induce half-maximal responses in in vitro assays (18). Peptides were added in serial dilutions ranging from 1 μg/ml to 1 pg/ml to ICS assays as described above. Both IFN-γ- and IL-2-secreting CD8 T cells were measured. EC50 was determined as the peptide concentration needed to achieve a half-maximal response.
Degranulation Activity (CD107a Mobilization).
Cryo-preserved blood mononuclear cells (1–2 × 106) were stimulated for 6 h in 1 ml of complete media (RPMI + 10% FBS) in the presence of Golgistop (1 μl/ml; BD Pharmingen), Golgiplug (1 μl/ml; BD Pharmingen), αCD28 Ab (0.5 μg/ml; BD Pharmingen), αCD107a-PE (pretittered volume; BD Pharmingen) and 1 μg/ml peptide as described in ref. 7. SEB stimulation (200 ng/ml) served as positive control. At the end of the stimulation period, cells were washed, permeabilized, and stained as described in ref. 7. The following Abs were used in combination: CD14-FITC, CD16-FITC, CD19-FITC, CD69-PerCP, IFN-γ-APC, CD3-ECD, and CD8-Pacific blue.
Ex Vivo Proliferation Assay.
Ag-specific proliferation was determined using 5,6-carboxyfluorescein succinimidyl ester (CFSE) (Molecular Probes, Eugene, OR) as described in ref. 6. At day 5, cells were harvested and stained with CD4-PerCP-Cy5.5 (BD Pharmingen) and CD8-APC (BD Pharmingen). Cells were fixed with CellFix (BD Pharmingen) and acquired on an LSRII (BD Pharmingen).
HLA Class I Typing.
Four-digit HLA class I genotyping was performed by direct sequencing methods as described in ref. 35. The data were analyzed and alleles were assigned using Assign-SBT version 3.5 (Conexio Genomics, Applecross, Australia).
Statistical Analysis.
Statistical analysis was done using GraphPad (San Diego, CA) Prism version 3.0 and Excel (Microsoft, Redmond, WA). Statistical significance (P values) of the results was calculated by two-tailed t test. A two-tailed P value of <0.05 was considered significant. The correlations among variables were tested by least-squares regression analysis. The association between HLA-A/B alleles and both only effector and polyfunctional CD8 T cell responses on the 82 gag epitopes was determined by χ2 test (α = 0.05).
Supplementary Material
Acknowledgments
This work was supported by Swiss National Science Foundation Research Grant FN 3100-66788.
Abbreviations
- Flu
influenza virus
- LTNP
long-term nonprogressors
- TCR
T cell receptor.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0707570104/DC1.
References
- 1.Harari A, Petitpierre S, Vallelian F, Pantaleo G. Blood. 2004;103:966–972. doi: 10.1182/blood-2003-04-1203. [DOI] [PubMed] [Google Scholar]
- 2.Harari A, Vallelian F, Meylan PR, Pantaleo G. J Immunol. 2005;174:1037–1045. doi: 10.4049/jimmunol.174.2.1037. [DOI] [PubMed] [Google Scholar]
- 3.Pantaleo G, Koup RA. Nat Med. 2004;10:806–810. doi: 10.1038/nm0804-806. [DOI] [PubMed] [Google Scholar]
- 4.Iyasere C, Tilton JC, Johnson AJ, Younes S, Yassine-Diab B, Sekaly RP, Kwok WW, Migueles SA, Laborico AC, Shupert WL, et al. J Virol. 2003;77:10900–10909. doi: 10.1128/JVI.77.20.10900-10909.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Younes SA, Yassine-Diab B, Dumont AR, Boulassel MR, Grossman Z, Routy JP, Sekaly RP. J Exp Med. 2003;198:1909–1922. doi: 10.1084/jem.20031598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zimmerli SC, Harari A, Cellerai C, Vallelian F, Bart PA, Pantaleo G. Proc Natl Acad Sci USA. 2005;102:7239–7244. doi: 10.1073/pnas.0502393102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Betts MR, Nason MC, West SM, De Rosa SC, Migueles SA, Abraham J, Lederman MM, Benito JM, Goepfert PA, Connors M, et al. Blood. 2006;107:4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harari A, Dutoit V, Cellerai C, Bart PA, Du Pasquier RA, Pantaleo G. Immunol Rev. 2006;211:236–254. doi: 10.1111/j.0105-2896.2006.00395.x. [DOI] [PubMed] [Google Scholar]
- 9.Pantaleo G, Harari A. Nat Rev Immunol. 2006;6:417–423. doi: 10.1038/nri1840. [DOI] [PubMed] [Google Scholar]
- 10.Carrington M, O'Brien SJ. Annu Rev Med. 2003;54:535–551. doi: 10.1146/annurev.med.54.101601.152346. [DOI] [PubMed] [Google Scholar]
- 11.Kiepiela P, Leslie AJ, Honeyborne I, Ramduth D, Thobakgale C, Chetty S, Rathnavalu P, Moore C, Pfafferott KJ, Hilton L, et al. Nature. 2004;432:769–775. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- 12.Frahm N, Brander C. [Accessed December 20, 2005];HIV Molecular Immunology Database. 2005 Available at www.hiv.lanl.gov/content/immunology/index.html.
- 13.Korber BTE, Brander C, Haynes BF, Koup R, Moore JP, Walker BD, Watkins DI. [Accessed December 20, 2005];Theoretical Biology and Biophysics, T-10. 2005 Available at www.t10.lanl.gov/index.shtml.
- 14.Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, Freeman GJ, Ahmed R. Nature. 2006;439:682–687. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- 15.Day CL, Kaufmann DE, Kiepiela P, Brown JA, Moodley ES, Reddy S, Mackey EW, Miller JD, Leslie AJ, DePierres C, et al. Nature. 2006;443:350–354. doi: 10.1038/nature05115. [DOI] [PubMed] [Google Scholar]
- 16.Petrovas C, Casazza JP, Brenchley JM, Price DA, Gostick E, Adams WC, Precopio ML, Schacker T, Roederer M, Douek DC, Koup RA. J Exp Med. 2006;203:2281–2292. doi: 10.1084/jem.20061496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trautmann L, Janbazian L, Chomont N, Said EA, Gimmig S, Bessette B, Boulassel MR, Delwart E, Sepulveda H, Balderas RS, et al. Nat Med. 2006;12:1198–11202. doi: 10.1038/nm1482. [DOI] [PubMed] [Google Scholar]
- 18.Bihl F, Frahm N, Di Giammarino L, Sidney J, John M, Yusim K, Woodberry T, Sango K, Hewitt HS, Henry L, et al. J Immunol. 2006;176:4094–4101. doi: 10.4049/jimmunol.176.7.4094. [DOI] [PubMed] [Google Scholar]
- 19.Neumann-Haefelin C, McKiernan S, Ward S, Viazov S, Spangenberg HC, Killinger T, Baumert TF, Nazarova N, Sheridan I, Pybus O, et al. Hepatology. 2006;43:563–572. doi: 10.1002/hep.21049. [DOI] [PubMed] [Google Scholar]
- 20.Dutoit V, Rubio-Godoy V, Dietrich PY, Quiqueres AL, Schnuriger V, Rimoldi D, Lienard D, Speiser D, Guillaume P, Batard P, et al. Cancer Res. 2001;61:5850–5856. [PubMed] [Google Scholar]
- 21.Alexander-Miller MA. Immunol Res. 2005;31:13–24. doi: 10.1385/IR:31:1:13. [DOI] [PubMed] [Google Scholar]
- 22.Snyder JT, Alexander-Miller MA, Berzofskyl JA, Belyakov IM. Curr HIV Res. 2003;1:287–294. doi: 10.2174/1570162033485230. [DOI] [PubMed] [Google Scholar]
- 23.Leslie A, Price DA, Mkhize P, Bishop K, Rathod A, Day C, Crawford H, Honeyborne I, Asher TE, Luzzi G, et al. J Immunol. 2006;177:4699–4708. doi: 10.4049/jimmunol.177.7.4699. [DOI] [PubMed] [Google Scholar]
- 24.Vogel TU, Friedrich TC, O'Connor DH, Rehrauer W, Dodds EJ, Hickman H, Hildebrand W, Sidney J, Sette A, Hughes A, et al. J Virol. 2002;76:11623–11636. doi: 10.1128/JVI.76.22.11623-11636.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Connor DH, Allen TM, Vogel TU, Jing P, DeSouza IP, Dodds E, Dunphy EJ, Melsaether C, Mothe B, Yamamoto H, et al. Nat Med. 2002;8:493–499. doi: 10.1038/nm0502-493. [DOI] [PubMed] [Google Scholar]
- 26.Van Parijs L, Abbas AK. Science. 1998;280:243–248. doi: 10.1126/science.280.5361.243. [DOI] [PubMed] [Google Scholar]
- 27.Molldrem JJ, Lee PP, Kant S, Wieder E, Jiang W, Lu S, Wang C, Davis MM. J Clin Invest. 2003;111:639–647. doi: 10.1172/JCI16398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morgan DJ, Kreuwel HT, Fleck S, Levitsky HI, Pardoll DM, Sherman LA. J Immunol. 1998;160:643–651. [PubMed] [Google Scholar]
- 29.Wankowicz-Kalinska A, Mailliard RB, Olson K, Graham F, Edington H, Kirkwood JM, Martinek S, Das PK, Storkus WJ. Melanoma Res. 2006;16:165–174. doi: 10.1097/01.cmr.0000198452.03957.73. [DOI] [PubMed] [Google Scholar]
- 30.Wherry EJ, Teichgraber V, Becker TC, Masopust D, Kaech SM, Antia R, von Andrian UH, Ahmed R. Nat Immunol. 2003;4:225–234. doi: 10.1038/ni889. [DOI] [PubMed] [Google Scholar]
- 31.Rizzardi GP, Tambussi G, Bart PA, Chapuis AG, Lazzarin A, Pantaleo G. AIDS. 2000;14:2257–2263. doi: 10.1097/00002030-200010200-00006. [DOI] [PubMed] [Google Scholar]
- 32.Schockmel GA, Yerly S, Perrin L. J Acquired Immune Defic Syndr. 1997;14:179–183. doi: 10.1097/00042560-199702010-00013. [DOI] [PubMed] [Google Scholar]
- 33.Kondo E, Akatsuka Y, Kuzushima K, Tsujimura K, Asakura S, Tajima K, Kagami Y, Kodera Y, Tanimoto M, Morishima Y, Takahashi T. Blood. 2004;103:630–638. doi: 10.1182/blood-2003-03-0824. [DOI] [PubMed] [Google Scholar]
- 34.Currier JR, Kuta EG, Turk E, Earhart LB, Loomis-Price L, Janetzki S, Ferrari G, Birx DL, Cox JH. J Immunol Methods. 2002;260:157–172. doi: 10.1016/s0022-1759(01)00535-x. [DOI] [PubMed] [Google Scholar]
- 35.Sayer D, Whidborne R, Brestovac B, Trimboli F, Witt C, Christiansen F. Tissue Antigens. 2001;57:46–54. doi: 10.1034/j.1399-0039.2001.057001046.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.