Abstract
The product of the dek oncogene is the 43-kDa DEK nuclear protein. DEK was first identified in a fusion with the CAN nucleoporin protein in a specific subtype of acute myelogenous leukemia. DEK has also been shown to be an autoantigen in patients with pauciarticular onset juvenile rheumatoid arthritis. Further, the last 65 amino acids of DEK can partially reverse the mutation-prone phenotype of cells from patients with ataxia-telangiectasia. However, in spite of these significant disease associations, the function of DEK has remained unclear. The HIV-2 peri-ets (pets) site is a TG-rich element found between the two Elf-1 binding sites in the HIV-2 enhancer. The pets element mediates transcriptional activation whether the enhancer is stimulated by phorbol 12-myristate 13-acetate (PMA) alone, phytohemagluttinin (PHA) alone, PMA plus PHA, soluble antibodies to the T cell receptor, immobilized antibodies to the T cell receptor, or by antigen. Previously, we purified and characterized the pets factor, demonstrating that it is a 43-kDa nuclear protein. We now describe the identification of DEK as this 43-kDa pets factor. Using a modified Southwestern screening procedure, we find that DEK can recognize the pets element. We demonstrate the ability of recombinant DEK to bind specifically to the pets site using the electrophoretic mobility shift assay (EMSA) and DNase I footprinting. “Supershift” EMSA further confirms that DEK is the dominant protein binding to the pets site in T cell extracts. Our findings show that DEK is a site-specific DNA binding protein that is likely involved in transcriptional regulation and signal transduction. This has implications for multiple pathogenic processes, including hematologic malignancies, arthritis, ataxia-telangiectasia, and AIDS caused by HIV-2.
A (6;9) chromosomal translocation is associated with a specific subtype of acute myelogenous leukemia (AML) (1–3). This translocation results in the fusion of two genes, dek and can, and the expression of a leukemia-specific, chimeric dek-can mRNA and fusion protein (1). How this 165-kDa chimeric protein might lead to leukemia has been unclear, however. It has recently been shown that CAN is a nucleoporin, and it has therefore also been termed nup214 (nucleoporin of 214 kDa; ref. 4). The 3′ portion of CAN can form a fusion protein not only with DEK, leading to AML, but also with the SET protein, leading to undifferentiated leukemia (2). One group has shown that CAN localizes exclusively to the cytoplasmic side of the nuclear pore complex (4). A second group has demonstrated that, when CAN is overexpressed, it is found not only on the cytoplasmic side of the nuclear membrane, but also on the nucleoplasmic side (5). Like their normal counterparts DEK and SET, DEK–CAN, and SET–CAN were localized exclusively to the nucleus. It was concluded that the relocation of the carboxyl-terminal portion of CAN from the nuclear envelope to the nucleoplasm may reinforce a nuclear function of CAN (5), implying that this relocation plays a role in leukemogenesis.
In the DEK–CAN chimera, the N-terminal two-thirds of the 43-kDa DEK protein is fused to the C-terminal two-thirds of CAN (1). Unlike CAN, DEK has not been demonstrated to be a member of any known family of proteins. In fact, comparison of the predicted amino acid sequence for DEK with the European Molecular Biology Laboratory database did not shown any substantial similarity to known sequences (1). While the protein is predicted to have a continuous stretch of acidic residues at the N terminus, three acidic regions interspersed with serines, and a very high percentage (42%) of charged amino acids, no specific structures could be recognized. However, while DEK has been a protein of unknown function and mechanism of action, it has been associated with several diseases and implicated in an important cellular pathway. When DEK is linked to CAN in a chimera, the fusion protein appears to be oncogenic. However, on its own, DEK may have “anti-oncogenic” effects, as it can partially reverse the transformation-prone phenotype of the cells of patients with ataxia-telangiectasia (A-T), a disease associated with a greatly increased likelihood of hematologic malignancy. When introduced into these cells, the last 65 amino acids of DEK can partially complement the phenotype of mutagen sensitivity, high spontaneous recombination rate, and radio-resistant DNA synthesis (6). Further, DEK appears to be an autoantigen in certain disease states, as antibodies to DEK are found in 77% of patients with pauciarticular onset juvenile rheumatoid arthritis (7, 8). Therefore, DEK is an important cellular protein having significant associations with cancer and other diseases, but whose mechanism of action has previously been completely unknown.
HIV-2 is found in West Africa and, increasingly, in other parts of the world. Like HIV-1, HIV-2 can cause AIDS, but typically the asymptomatic period following HIV-2 infection is much longer than that following HIV-1 infection (9). Unlike HIV-1, in which the the two κB sites play the dominant role in regulating inducible enhancer function in activated T cells, with some contribution from the 5′ Sp1 site, HIV-2 enhancer activation in T cells is regulated by at least four distinct cis-acting elements: two purine-rich sites (PuB1 and PuB2), which bind the ets protooncogene family member Elf-1; the peri-ets (pets) site, which binds a 43-kDa nuclear factor; and a single κB site, which binds the well described components of NF-κB (10–19). Mutation of any of these elements markedly diminishes the response of the enhancer to cellular activation at the RNA level but does not affect the response to Tat or to other viral transactivating proteins. Therefore, these elements specifically mediate enhancer stimulation in activated cells.
The pets site is a TG-rich element (TTGGTCAGGG) found between the two Elf-1 binding sites in the HIV-2 enhancer. The pets element mediates activation whether the enhancer is stimulated by phorbol 12-myristate 13-acetate (PMA) alone, phytohemagluttinin (PHA) alone, PMA plus PHA, soluble antibodies to the T cell receptor, immobilized antibodies to the T cell receptor, or by antigen (12, 15). We and others (20, 21) have also demonstrated that a similar TG-rich pets-like site, again adjacent to an Elf-1 binding site, plays a significant role in mediating activation of the human T cell leukemia virus type I enhancer in T cells. TG-rich sites adjacent to ets binding sites are also found in murine retroviruses, and alteration of these pets-like sites can change the type of malignancy seen in mice following infection (22–24).
The crucial role played by the pets site in the response of the HIV-2 enhancer to T cell stimulation suggested that the cellular protein interacting with the pets site would be of importance to the biology of HIV-2 and also was likely to be involved in signal transduction in T cells. Further, by analogy with other proteins known to bind to retroviral enhancer elements, we reasoned that the pets factor was likely to be an oncoprotein and, as the pets site sequence did not correspond to other known DNA enhancer elements, we suspected that this would be a cellular protein not previously shown to bind DNA in a site-specific manner. Our previous studies employing the electrophoretic mobility shift assay (EMSA) and DNase footprinting showed that the pets site contained the sequence TTGGTCAGGG (12, 15). Using multiple biochemical techniques, we previously demonstrated that the size of the pets binding factor is 43 kDa (19). In this report, we employ modified Southwestern screening of a T cell cDNA library to identify DEK as the 43-kDa pets site binding factor. Using EMSA and DNase I footprinting, we demonstrate recombinant DEK binding to the HIV-2 pets site in vitro. Furthermore, antibody targeted against DEK “supershifts” the protein–DNA complex seen in EMSA when the pets site probe is incubated with Jurkat nuclear extract. Based on these observations, we conclude that DEK is the nuclear factor that binds to the HIV-2 pets site. Because DEK has little similarity to other known cellular factors and no recognizable DNA binding motifs (1, 2), our results indicate that DEK, an autoantigen that has been linked to cancer and now to signal transduction and regulation of HIV-2 transcription, is the first member of what is likely to be a new family of DNA binding/transcription factors implicated in oncogenesis.
MATERIALS AND METHODS
Materials.
The poly-histidine tagging proEx-1 vector was obtained from GIBCO/BRL. Ni-NTA resin was purchased from Qiagen (Chatsworth, CA), and pGEX vector and glutathione-Sepharose 4B were purchased from Pharmacia. Poly-dIdC was obtained from Sigma. Jurkat T cell nuclear extract was prepared as described (12). All deoxyribonucleotides were synthesized by the University of Michigan DNA Core Facility with an Applied Biosystems model 300B synthesizer. Rabbit polyclonal antibody to DEK was prepared as described (5). Human antisera to DEK was obtained from W. Szer (8). DNase I was purchased from Boehringer Mannheim, and Sequenase was obtained from Amersham. PCR reagents were purchased from GIBCO/BRL, and all other cloning and labeling enzymes were purchased from New England Biolabs. The Jurkat λgt11 cDNA library was a gift of J. Leiden (University of Chicago).
Southwestern Screening of Phage Library and Isolation of the cDNA Clone.
Southwestern screening of a Jurkat λgt11 library was performed as described by Singh et al. (25, 26) with the following modifications: PCR generation of a 32P-labeled 8× multimeric pets probe (with all sites in the same orientation) was as described (19). The library was amplified according to standard procedures except that 1 mM isopropy β-d-thiogalactoside was added to the top agar layer. The amplified library, ≈1 × 1012 plaque forming units in 10 ml SM buffer (50 mM Tris·HCl, pH 7.5/10 mM MgSO4/100 mM NaCl/0.01% gelatin), was cleared of cellular debris by centrifugation and mixed with 0.5 ml DNA-affinity Sepharose CL-2B resin (27) containing the concatemerized pets oligonucleotide at a concentration of 20 μg/ml. The resin was gently mixed on a rotating platform for 20 min at room temperature before the supernatant was removed following centrifugation at 5000 × g for 1 min. The resin was then washed four times with 50 ml buffer A (20 mM Tris·HCl, pH 7.9/100 mM KCl/10 μM ZnSO4/10% glyerol/0.01% Nonidet P-40) for 10 min on a rotating platform at room temperature. Buffer A (2 ml) containing 1 M KCl was added to the resin, and after 10 min of gentle mixing the resin was spun down and the supernatant collected. This eluate was diluted to 100 mM KCl, and the enrichment process was repeated with 0.2 ml fresh DNA-Sepharose resin. The final eluate (0.5 ml) from this second enrichment round was used to infect Escherichia coli Y1090. Two candidates were identified from ≈1 × 105 clones screened. These two clones were plaque purified and screened with either the pets probe, or for a control, a nonrelated probe generated from an 8× HIV-2 κB construct. Lysogen extracts were made in E. coli Y1089 as described (25), and 2 μl per reaction was used in EMSA to test for specific binding to the pets probe. The phage DNA from the positive clones was extracted and purified using the cetyldimethylethyl-ammonium bromide (CTAB) method (28). Primers flanking the EcoRI site of the λgt11 DNA were synthesized and used to PCR amplify the phage insert. The PCR amplification product was cut with EcoRI and cloned into pGem 7F− (Promega) and sequenced with Sequenase (United States Biochemical).
Subcloning, Expression, and Purification of Recombinant DEK.
For the full-length DEK cDNA, primers (with added EcoRI ends) flanking the published DEK sequence (1) were synthesized and used to reverse transcribe (RT)-PCR amplify a 1200-bp cDNA fragment using Elongase (GIBCO/BRL). The PCR product was cut with EcoRI and cloned into pGEX (Pharmacia) and subsequently subcloned into ProEx-1 in frame with the poly-histidine tag. Full-length DEK was produced as a glutathione S-transferase (GST) fusion protein or as a poly-histidine-tagged fusion protein in E. coli strain BL21 (Novagen) and purified with glutathione-Sepharose (Pharmacia) or Ni-NTA (Qiagen) according to manufactures’ guidelines, except that 0.1 M KCl was added to all the buffers. The purified recombinant proteins were verified by Coomassie staining following SDS/PAGE and by Western blot analysis.
EMSA.
EMSA was performed as described (12). Binding reactions were performed in the presence of 100 mM KCl. For supershift assays, 10 min before loading the samples onto the gel, 1 μl of immune serum was added to the binding reaction following an initial incubation of the protein sample with the probe for 10 min at room temperature. The sequence of the HIV-2 pets oligonucleotide used was 5′-GATCCAGCTATACTTGGTCAGGGCGAATTCTAACTA. The mutant version of the pets site oligonucleotide used was 5′-GATCCAGCTATACTAGATCTGGGCGAATTCTAACTA. For EMSA employing purified recombinant DEK, a 2× multimerized binding site probe was used.
DNase I Footprinting.
A 32P-labeled 190-bp fragment of the HIV-2 enhancer encompassing the pets site was generated using PCR with an end-labeled primer. The PCR product was purified using PAGE and sequenced before use. Footprinting was performed as described (14), using recombinant GST–DEK.
SDS/PAGE and Western Analysis.
SDS/PAGE was performed as described (29). Western blot analysis was performed using either a horseradish peroxidase detection kit (Amersham) or an alkaline phosphatase detection kit (GIBCO/BRL), depending on the secondary antibody used.
Computer Homology Searches.
Dek was identified by using the blast search at the National Center for Biotechnology Information. Potential phosphorylation sites for DEK were identified with prosite at expasy (University of Geneva).
RESULTS
We previously purified the pets factor from bovine spleen, demonstrating that it is 43 kDa in size (19). Biochemical characterization demonstrated that the human pets factor was also 43 kDa in size (19). Unfortunately, sequencing of the 43-kDa polypeptide that recognizes the pets site was hindered by several factors. We had previously attempted to clone the pets factor using routine Southwestern screening of a Jurkat T cell library, using a concatemerized, single site probe, but failed to obtain a candidate for the pets factor. Therefore, we made two improvements in the technique. (i) We noted that a probe with multiple copies of the DNA recognition site in the proper orientation (i.e., not randomly concatemerized) increased the sensitivity from our Southwestern screening. Whereas previous authors (25, 26) have generally suggested four copies of the DNA site, we prepared a probe that had eight copies of the pets site, all in the same orientation. This probe appeared to function much better than the randomly concatemerized single copy probe in Southwestern blotting experiments (19). (ii) In considering Southwestern screening of phage cDNA libraries, we reasoned that, due to the high percentage of the recombinant polypeptide present in the phage following induction, at least a small amount of that polypeptide might be associated with the capsid. Therefore, we enriched phage from the Jurkat T cell library using DNA-Sepharose containing the pets site. Phage retained on the the beads were then eluted, plated, and screened using the Southwestern technique. This technique appeared to enrich ≈10-fold for phage encoding DNA-binding factors, although not specificially for the pets factor (G.K.F. and D.M.M., unpublished observations).
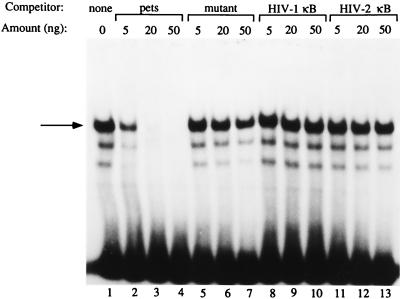
Before beginning cloning experiments, we confirmed and extended our previous work (12, 15), showing that a factor(s) from Jurkat nuclear extracts binds specifically to the HIV-2 pets site (Fig. 1, lane 1). Using EMSA, we demonstrated the specificity of this binding using cold, unlabeled oligonucleotide competitors. Addition of unlabeled pets site oligonucleotide competed away the binding (lanes 3 and 4), while addition of an excess amount of a pets oligonucleotide with the binding site mutated in a manner known to eliminate function (12, 15) (lanes 5–7) or oligonucleotides with other unrelated HIV enhancer elements (lanes 8–13) did not compete away the band.
Figure 1.
A nuclear factor(s) binds specifically to the HIV-2 pets site. EMSA using Jurkat nuclear extracts with the HIV-2 pets site probe. The shifted band seen in lane 1 (indicated by the arrow on the left) can be competed away by the addition of unlabeled self-competitor (lanes 3 and 4), but not by a mutant version of the pets site oligonucleotide (lanes 6 and 7) or by unrelated HIV enhancer elements (lanes 7–13).
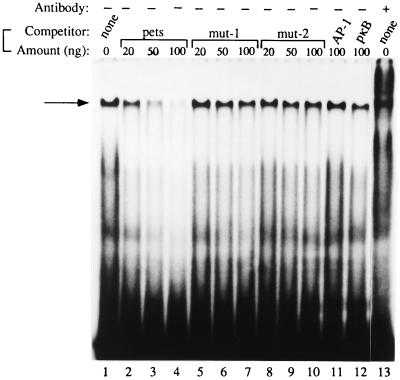
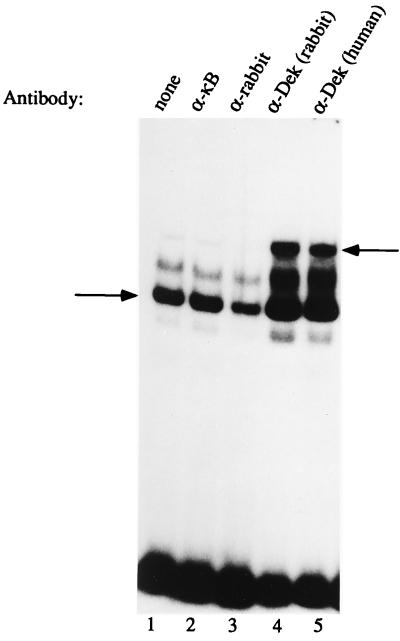
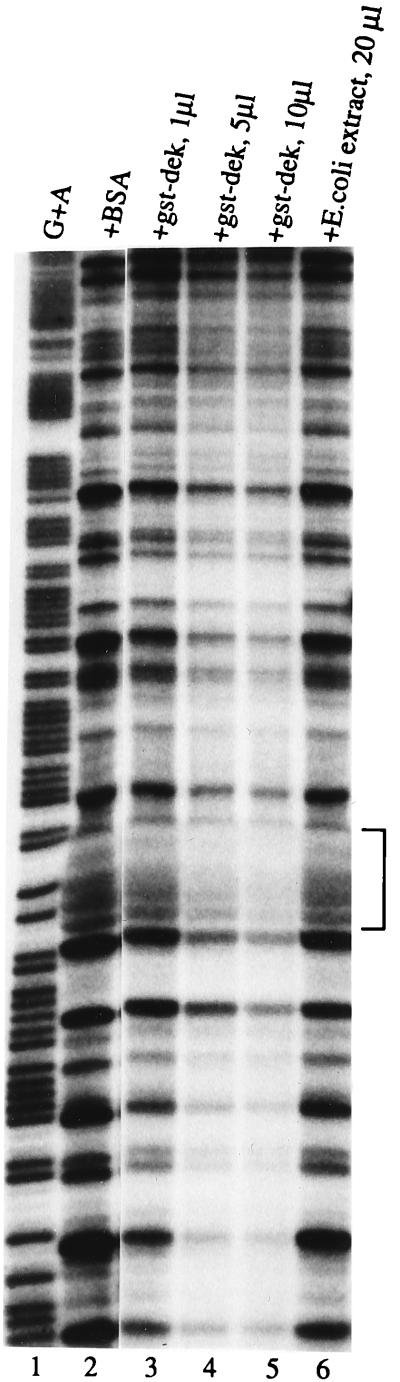
The human T cell factor that binds to the HIV-2 pets site, identified using our enriched Southwestern screening technique, is the 43-kDa DEK protein which, as discussed above, has not previously been known to be a site-specific DNA binding protein and has no obvious DNA binding motif. The β-galactosidase–DEK fusion protein that we cloned from the T cell library bound in a site-specific manner to the pets site (not shown). This clone was lacking the first 200 bp at the 5′ end of the known DEK sequence. To further investigate whether or not DEK is capable of site-specific DNA binding, we prepared a full-length DEK–(HIS)6 fusion protein and used it to show sequence-specific binding to the HIV-2 pets site (Fig. 2). Addition of increasing amounts of unlabeled pets site oligonucleotide competed away the band (Fig. 2, lanes 2–4) but not when mutant versions of the pets site or unrelated binding site oligonucleotides were used (lanes 5 to 12). Addition of human anti-DEK antibody to the reaction caused a “supershift,” further supporting the finding of DEK-specific binding to the pets site (lane 13). Having shown that recombinant DEK is able to bind to the pets site in vitro, we wished to assess whether it is the dominant protein interacting with the pets site, as our biochemical studies would suggest (19). Therefore, we used both human and rabbit antibody known to strongly react against DEK in EMSA with the pets probe and T cell nuclear extract. We observed a supershift when the DEK antibody was added to the reaction (Fig. 3, lanes 4 and 5), but not when control antisera were used (lanes 2 and 3). Incubation of the pets site probe with anti-DEK antibody in the absence of cellular extract did not cause a shift to occur (not shown). In addition to the supershift results, the 43-kDa protein found in our highly purified protein fractions with enriched binding activity for the pets site (19) is recognized by antibodies to DEK (not shown). Therefore DEK is the dominant protein recognizing the pets site in nuclear extracts. We also produced a full-length GST–DEK fusion protein and used it in a DNase I footprinting assay to show that DEK protects the 5′ end (TTGG) and five bases upstream (TATAC) of the pets site of the HIV-2 enhancer from DNase digestion (Fig. 4, lanes 4 and 5). Taken together these results indicate that DEK binds specifically to the HIV-2 pets site.
Figure 2.
Purified recombinant DEK–(HIS)6 binds to the HIV-2 pets site in EMSA (lane 1). Addition of cold unlabeled pets oligonucleotide competed away the shifted band indicated by the arrow (lanes 3 and 4) but not when mutant or unrelated binding site oligonucleotides were added (lanes 5–12). The mutant oligonucleotide (mut-2) added in lanes 8–10, unlike mut-1 (lanes 5–7), did not have the downstream Elf-1 binding site mutated. In lane 13, human anti-DEK serum has been added to the reaction, leading to a supershift.
Figure 3.
Identification of the nuclear factor binding to the HIV-2 pets site using supershift EMSA. The arrow on the left indicates a band shift seen in EMSA using the pets probe with Jurkat nuclear extract (lane 1). Addition of antibody recognizing DEK supershifted this complex (lanes 4 and 5) as indicated by the arrow to the right. Addition of control antibodies to NF-κB p50 or to rabbit immunoglobulin does not supershift the complex (lanes 2 and 3).
Figure 4.
Binding of recombinant DEK to the HIV-2 pets site can be demonstrated using DNase I footprinting. No protection was seen when bovine serum albumin (BSA) was added to the probe (lane 2) or when control E. coli extract was used (lane 6). Addition of increasing amounts of E. coli extract containing GST–DEK (lanes 3–5) protects the TATACTTGG bases of the HIV-2 enhancer as indicated by the bracket on the right. A G+A ladder (lane 1) identifies the position of the footprint as the pets site.
DISCUSSION
We have demonstrated that DEK is a site-specific DNA binding protein that recognizes the pets site, an element in the HIV-2 enhancer that responds to T cell signaling. The pets site was originally defined by DNase I footprinting, using crude Jurkat T cell nuclear extracts, as consisting of the sequence TTGGTCAGGG (15). Mutation of the second, fourth, and seventh bases in this sequence (TAGATCTGGG) markedly reduces the ability of the pets site to mediate HIV-2 enhancer activation following T cell or monocyte stimulation (11, 12, 15). DNase I footprinting studies using highly purified pets binding factor (which contains primarily DEK) show two areas of protection (19), which include most of the previously defined pets site and several bases upstream (GCTATA is the 5′ footprint, the CT which follows is not protected, and TGGTCAGG is then protected in the 3′ footprint). In the the studies using recombinant DEK presented in this paper, the more 5′ sequence is protected from DNase I digestion (TATACTTGG). Preliminary data using supershift EMSA indicate that DEK in nuclear extracts can bind to pets-like sites in which the more 3′ elements are better conserved than are the 5′ elements (G.K.F. and D.M.M., unpublished results). Therefore, it would appear that both the 5′ and 3′ portions of the pets site play a role in DEK binding within the nucleus of a cell.
In view of the role of DEK–CAN in leukemogenesis, we sought pets-like sites in promoters for genes expressed in neutrophils and used as markers in neutrophilic leukemias. Interestingly, reasonable matches were found in the promoters for myeloperoxidase and neutrophil elastase, genes that match this description (30, 31). Preliminary data indicate that DEK does bind to these somewhat divergent pets-like sites in the neutrophil elastase and myeloperoxidase promoters (not shown). Therefore, DEK would appear to have one or more important cellular targets that potentially link it to gene expression in immature neutrophils.
The gene mutated in A-T has recently been identified (32) and is not similar to the DEK protein. Therefore, it is unclear why DEK is able to reverse some of the phenotypic changes (including radiation sensitivity) seen in cells from A-T patients (6). It has been speculated that DEK may be acting upon the same protein as does the A-T gene product or might regulate the A-T gene itself, and this theory fits with our observations that DEK binds to specific functional promoter elements. This is of particular interest in view of the fact that A-T patients are much more susceptible to developing breast and hematologic malignancies, again linking DEK to an oncogenic (or anti-oncogenic) phenotype.
Very recently, it has been shown that a translocation which produces a chimeric protein involving the HOXA9 transcription factor and the nucleoporin protein nup98 has been linked to certain cases of AML (33, 34). Our finding that DEK is a site-specific DNA binding protein which is able to recognize a transcriptionally active element in the HIV-2 promoter would suggest that the DEK–CAN fusion protein is another example of a chimeric protein involving a transcription factor and a nucleoporin protein in AML. Therefore, this type of chimera is a new class of oncoproteins now implicated in two subtypes of AML. While CAN is found at the nuclear membrane, both DEK and DEK–CAN are found in the nucleus (5). Therefore, unless it is a rather subtle effect, it would not appear that the oncogenic mechanism of this fusion protein involves altering the location of DEK, although the change in the subcellular localization of CAN may be a factor (5). Whether the fusion with CAN, which involves the N-terminal two-thirds of DEK, alters the DNA binding or transciptional properties of DEK will be investigated in future experiments.
The finding that DEK can bind specifically to an inducible element in the HIV-2 promoter suggests that this protein, which is not similar to any other protein in the computer database, is a site-specific DNA binding protein that is involved in transcriptional regulation. DEK contains no recognizable DNA binding motifs and, therefore, it would appear that its nucleic acid binding region might also consist of a previously unknown motif. DEK would thus appear to be the first member described of a new family of site-specific DNA binding factors likely to be involved in signal transduction and transcriptional regulation. As noted above, this quality of DEK might have implications for multiple pathogenic processes, including hematologic malignancies, arthritis, A-T, and AIDS caused by HIV-2.
Acknowledgments
We thank Dr. W. Szer for antibodies, Dr. J. Leiden for the Jurkat cDNA library, Nilesh Shah for reagent preparation, and Shekelia Taylor for preparation of the manuscript. This work was supported by grants to D.M.M. from the National Institutes of Health (AI30924 and AI36685). G.K.F. was supported in part by the Cancer Biology Training Grant of the University of Michigan (T32 CA09676) and a University of Michigan Rackham Dissertation Grant.
ABBREVIATIONS
- AML
acute myelogenous leukemia
- A-T
ataxia-telangiectasia
- pets
peri-ets
- EMSA
electrophoretic mobility shift assay
- GST
glutathione S-transferase
References
- 1.von Lindern M, Fornerod M, van Baal S, Jaegle M, de Wit T, Buijs A, Grosveld G. Mol Cell Biol. 1992;12:1687–1697. doi: 10.1128/mcb.12.4.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.von Lindern M, Breems D, van Baal S, Adriaansen H, Grosveld G. Genes Chromosomes Cancer. 1992;5:227–234. doi: 10.1002/gcc.2870050309. [DOI] [PubMed] [Google Scholar]
- 3.Soekarman D, von Lindern M, van der Plas D C, Selleri L, Bartram C R I, Martiat P, Culligan D, Padua R A, Hasper-Voogt K P, Hagemeijer A, Grosveld G. Leukemia. 1992;6:489–494. [PubMed] [Google Scholar]
- 4.Kraemer D, Wozniak R W, Blobel G, Radu A. Proc Natl Acad Sci USA. 1994;91:1519–1523. doi: 10.1073/pnas.91.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fornerod M, Boer J, van Baal S, Jaegle M, von Lindern M, Murti K G, Davis D, Bonten J, Buijs A, Grosveld G. Oncogene. 1995;10:1739–1748. [PubMed] [Google Scholar]
- 6.Meyn M S, Lu-Kuo J M, Herzing L B K. Am J Hum Genet. 1993;53:1206–1216. [PMC free article] [PubMed] [Google Scholar]
- 7.Sierakowska H, Williams K R, Szer I S, Szer W. Clin Exp Immunol. 1993;94:435–439. doi: 10.1111/j.1365-2249.1993.tb08214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szer I S, Sierakowska H, Szer W. J Rheum. 1994;21:2136–2142. [PubMed] [Google Scholar]
- 9.Markovitz D M. Ann Intern Med. 1993;118:211–218. doi: 10.7326/0003-4819-118-3-199302010-00010. [DOI] [PubMed] [Google Scholar]
- 10.Hannibal M, Markovitz D M, Clark N, Nabel G J. J Virol. 1993;67:5035–5040. doi: 10.1128/jvi.67.8.5035-5040.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hannibal M C, Markovitz D M, Nabel G J. Blood. 1994;83:1839–1846. [PubMed] [Google Scholar]
- 12.Hilfinger J, Clark N, Robinson K, Smith M, Markovitz D M. J Virol. 1993;67:4448–4453. doi: 10.1128/jvi.67.7.4448-4453.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leiden J M, Wang C-W, Petryniak B, Markovitz D M, Nabel G J, Thompson C B. J Virol. 1992;66:5890–5897. doi: 10.1128/jvi.66.10.5890-5897.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Markovitz D M, Hannibal M, Perez V L, Gauntt C, Folks T M, Nabel G J. Proc Natl Acad Sci USA. 1990;87:9098–9102. doi: 10.1073/pnas.87.23.9098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Markovitz D M, Smith M, Hilfinger J, Hannibal M C, Petryniak B, Nabel G J. J Virol. 1992;66:5479–5484. doi: 10.1128/jvi.66.9.5479-5484.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Markovitz D M, Hannibal M C, Smith M, Cossman R, Nabel G J. J Virol. 1992;66:3961–3965. doi: 10.1128/jvi.66.6.3961-3965.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nabel G, Baltimore D. Nature (London) 1987;326:711–713. doi: 10.1038/326711a0. [DOI] [PubMed] [Google Scholar]
- 18.Perkins N D, Edwards N L, Duckett C S, Agranoff A B, Schmid R M, Nabel G J. EMBO J. 1993;12:3551–3558. doi: 10.1002/j.1460-2075.1993.tb06029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu G K, Markovitz D M. J Biol Chem. 1996;271:19599–19605. doi: 10.1074/jbc.271.32.19599. [DOI] [PubMed] [Google Scholar]
- 20.Gitlin S D, Bosselut R, Gegonne A, Ghysdael J, Brady J N. J Virol. 1991;65:5513–5523. doi: 10.1128/jvi.65.10.5513-5523.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clark N, Smith M, Hilfinger J, Markovitz D M. J Virol. 1993;67:5522–5528. doi: 10.1128/jvi.67.9.5522-5528.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Golemis E A, Speck N A, Hopkins N. J Virol. 1990;64:534–542. doi: 10.1128/jvi.64.2.534-542.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Speck N A, Renjifo B, Golemis E, Fredrickson N, Hartley J W, Hopkins N. Genes Dev. 1990;4:233–242. doi: 10.1101/gad.4.2.233. [DOI] [PubMed] [Google Scholar]
- 24.Speck N A, Renjifo B, Hopkins N. J Virol. 1990;64:543–550. doi: 10.1128/jvi.64.2.543-550.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh H, Clerc R G, Lebowitz J H. BioTechniques. 1989;7:252–261. [PubMed] [Google Scholar]
- 26.Singh H, Sen R, Baltimore D, Sharp P. Nature (London) 1986;319:154–158. doi: 10.1038/319154a0. [DOI] [PubMed] [Google Scholar]
- 27.Kadonaga J T, Tjian R. Proc Natl Acad Sci USA. 1986;83:5889–5893. doi: 10.1073/pnas.83.16.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manfioletti G, Schneider C. Nucleic Acids Res. 1988;16:2873–2884. doi: 10.1093/nar/16.7.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 30.Austin G E, Lam L, Zaki S R, Chan W C, Hodge T, Hou J, Swan D, Zhang W, Racine M, Whitsett C, Brown T. Leukemia. 1993;7:1445–1450. [PubMed] [Google Scholar]
- 31.Han J, Unlap T, Rado T A. Biochem Biophys Res Commun. 1991;181:1462–1468. doi: 10.1016/0006-291x(91)92104-r. [DOI] [PubMed] [Google Scholar]
- 32.Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, et al. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 33.Borrow J, Shearman A M, Stanton V P, Becher R, Collins T, Williams A J, Dube I, Katz F, Kwong Y L, Morris C, Ohyashiki K, Toyama K, Rowley J, Housman D E. Nat Genet. 1996;12:159–167. doi: 10.1038/ng0296-159. [DOI] [PubMed] [Google Scholar]
- 34.Nakamura T, Largaespada D A, Lee M P, Johnson L A, Ohyashiki K, Toyama K, Chen S J, Willman C L, Chen I-M, Feinberg A P, Jenkins N A, Copeland N G, Shaughnessy J D. Nat Genet. 1996;12:154–158. doi: 10.1038/ng0296-154. [DOI] [PubMed] [Google Scholar]