Abstract
rab6 is a ubiquitous ras-like GTPase involved in intra-Golgi transport. We have studied at both morphological and biochemical levels the behavior of Golgi resident proteins in HeLa cells overexpressing wild-type rab6 and GTP- and GDP-bound mutants of rab6 (rab6 Q72L and rab6 T27N, respectively). We show that wild-type rab6 and rab6 Q72L overexpression induces the redistribution of the trans-Golgi protein β-1,4-galactosyltransferase into the endoplasmic reticulum (ER) and allows the addition of sialylated O-glycans on an ER-retained protein, the major histocompatibility complex class II-associated invariant chain. Remarkably, rab6 Q72L effects, which require the integrity of microtubules, were almost indistinguishable from those induced by brefeldin A, a fungic metabolite that causes a mixing of Golgi and ER membranes. In contrast, overexpression of rab6 T27N does not cause the redistribution of Golgi proteins, but inhibits basal O-glycosylation of the major histocompatibility complex class II-associated invariant chain.
Small GTPases of the rab family are now recognized as key players of the protein machinery involved in vesicular transport within eukaryotic cells. Many of them have been localized to various compartments of both the biosynthetic/secretory and endocytic pathways. In vivo and in vitro studies performed with rab proteins locked in their GDP- or GTP-bound conformations have led to the hypothesis that rab proteins are involved in docking/fusion of transport vesicles with their target membranes. Good evidence also exists that rab proteins fulfill their function through a cycle between a GDP-bound cytosolic and a GTP-bound membrane form (for reviews see refs. 1–3). Nonetheless, their exact function is still poorly understood. The vesicle/target soluble N-ethylmaleimide-sensitive factor acceptor receptor (v/t SNARE) hypothesis has recently challenged the idea that rab proteins can, by themselves, specify docking/fusion of transport vesicles with their acceptor membrane. More likely, rab proteins participate in, or modulate the formation of, the v/t SNARE complexes (4–7). However, strong evidence for such a role is still lacking.
Consistent with their crucial role in transport, the alteration of rab function has profound effects on the morphology of organelles. For instance, overexpression of a GTPase mutant of rab5, a protein involved in early steps of endocytosis (8), greatly increases the size of early endosomes, whereas a GDP-bound mutant of rab5 has the opposite effect (9). Similarly, overexpression of the GDP-bound mutant of rab1a, a protein involved in endoplasmic reticulum (ER) to Golgi transport (10), affects the morphology of the Golgi apparatus (11). These experiments suggest that rab proteins either play a direct role in homotypic fusion between organelles, as demonstrated for rab5 (12), or modify the morphology of intracellular organelles by altering the entry and exit of transport vesicles from these organelles.
We have previously shown that rab6, a ubiquitous rab associated with the membranes of the Golgi apparatus as well as that of the trans-Golgi network (13, 14) functions in intra-Golgi transport (15). We also noticed that overexpression of the GTPase mutant of rab6 (rab6 Q72L) and to a lesser extent that of wild-type (wt) rab6 progressively alters the morphology of the Golgi apparatus (15). Here, we have studied in further detail this phenomenon and have investigated at both morphological and biochemical levels the behavior of Golgi resident proteins in cells overexpressing wt rab6 or rab6 mutants. We show that GTP-bound forms of rab6, but not GDP-bound forms, induce a brefeldin A (BFA)-like effect and redistribute Golgi resident proteins into the ER.
MATERIALS AND METHODS
Cells.
HeLa cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) containing 4.5 g/liter glucose (GIBCO) supplemented with 10% fetal calf serum (GIBCO) and penicillin/streptomycin.
Plasmids.
Construction of pGEM-1 vectors encoding for wt rab6, rab6 Q72L, and rab6 T27N has been previously described (15). pGEM vector containing human invariant chain cDNA (pGEM-Ii) was a generous gift of Vincent Lotteau (INSERM U391, Rennes, France).
Antibodies.
The following antibodies were used: affinity-purified rabbit antibody raised against human β-1,4-galactosyltransferase (gal-T)–β-galactosidase fusion protein expressed in Escherichia coli (for indirect immunofluorescence; ref. 16) and O14 affinity-purified rabbit antibody raised against purified soluble human gal-T (for immuno-electron microscopy; ref. 17); PIN.1.1 monoclonal antibody (kindly provided by A. Sant, University of Chicago), recognizing a luminal epitope of the human invariant chains Iip31 and Iip33 (18); rabbit antiserum (RαhIi) raised against the peptide PKESLELEDPSSGLGVTKQDLG [corresponding to amino acids 191–212 of major histocompatibility complex class II-associated invariant chain (Ii)] coupled to keyhole limpet hemocyanin; monoclonal antibody against protein disulfide isomerase (kindly provided by S. Fuller, EMBL, Heidelberg, Germany); and rabbit antiserum against mannosidase II (a gift from M. Farquhar, University of California, La Jolla). Texas red-labeled donkey anti-mouse antibody and fluorescein-labeled donkey anti-rabbit antibody were purchased from Amersham.
Infection with Vaccinia Virus and Transfection Procedure.
HeLa cells were plated on tissue culture dishes 18–24 h before the experiments to obtain 80% confluency at the time of infection. They were then infected with the vT7 recombinant vaccinia virus (19) and cotransfected using DOTAP (Boehringer Mannheim) with pGEM-Ii and either pGEM-1 (control cells) or pGEM vectors encoding for rab6 constructs, as previously described (15).
Immunofluorescence.
HeLa cells grown on 12-mm round glass coverslips were cotransfected with pGEM-Ii and control or rab6-encoding plasmids. Cells were processed 6 h after transfection for immunofluorescence as previously described (20). Confocal laser scanning microscopy and immunofluorescence analysis were performed using a TCS4D confocal microscope based on a DM microscope interfaced with an argon/krypton laser. Simultaneous double fluorescence acquisitions were performed using the 488-nm and the 568-nm laser lines to excite fluorescein isothiocyanate (FITC) and Texas red dyes using a 63× oil immersion Planapo 100 objective. The fluorescence was selected with appropriate double fluorescence dichroic mirror and band pass filters and measured with blue-green sensitive and red side sensitive-one photomultipliers.
Immuno-Electron Microscopy.
HeLa cells grown on 50-cm2 round tissue culture plates were infected and transfected as described above. Cells were processed 6 h after transfection for cryosectioning according to Slot et al. (21). The cryosections were made at −120°C using a cryoultramicrotome (Leica–Reichert) and retrieved with a 6:4 solution of 2.3 M sucrose/2% methyl cellulose. Immunogold double labeling was performed by introducing an intermediate fixation step (1% glutaraldehyde for 5 min) between the two successive incubations with antibodies (21). For monoclonal antibodies, a rabbit anti-mouse antibody linker (Dako) was used before protein A-gold labeling (purchased from J. W. Slot, Utrecht University, Utrecht, the Netherlands).
Pulse–Chase Experiments.
Cells were incubated for 15 min in medium without methionine and cysteine (ICN) and metabolically labeled for 10 min with 150 μCi/ml (1 Ci = 37 GBq) of [35S]methionine and [35S]cysteine (Amersham), 4 h 15 min after transfection. After being washed in serum-free medium, cells were either directly lysed or chased in complete medium supplemented with 2.5 mM methionine and cysteine. At the end of the chase, cells were washed once with ice-cold PBS and lysed at 4°C in buffer I (20 mM Tris·HCl, pH 8/150 mM NaCl/5 mM EDTA/1% Triton X-100/0.2% BSA) supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF) and a mixture of protease inhibitors (consisting of 0.1 μg/ml each of leupeptine, chymostatin, pepstatin, antipain, and aprotinin). Ii present in cell lysates was then immunoprecipitated using RαhIi and protein A-Sepharose. Immune precipitates were washed four times in buffer I and once in 50 mM Tris·HCl (pH 8) and then boiled for 5 min in sample buffer. Proteins were separated on 10–15% polyacrylamide/SDS gradient gels, as previously described (15). Bands corresponding to Ii were quantified with a PhosphorImager (Molecular Dynamics) equipped with the image quant software. In some experiments, quantification was directly carried out by counting the samples in a β scintillation counter.
N-Glycosylation and Sialylation Analysis.
Cells infected and transfected were pulsed and chased as described above. Ii immunoprecipitates were then treated with endoglycosidase H (endo H), N-glycanase, or neuraminidase as previously described (15).
Jacalin Binding Assay.
Ii immune precipitates were boiled two times for 3 min in 30 μl of 50 mM Tris·HCl, pH 6.8/1% SDS/5 mM DTT. Eluates were pooled, and one-tenth of each sample was collected. These aliquots were either diluted in sample buffer for SDS/PAGE analysis or directly used for scintillation counting. The rest of each sample was diluted with 600 μl of buffer I (without BSA) containing PMSF and protease inhibitors. Biotinylated jacalin (50 μg) and streptavidin-agarose (Pierce) were added, and samples were incubated for 4 h at 4°C. Pellets were washed two times in buffer II (20 mM Tris·HCl, pH 8/150 mM NaCl/0.2% Triton X-100). O-Glycosylated proteins bound to jacalin were specifically eluted with buffer II containing 0.8 M galactose, PMSF, and protease inhibitors. Eluates were either trichloroacetic acid-precipitated for SDS/PAGE analysis or directly used for scintillation counting.
RESULTS
Overexpression of the GTPase Mutant of rab6 Induces the Redistribution of the Golgi Enzyme β-1,4-Galactosyltransferase into the ER.
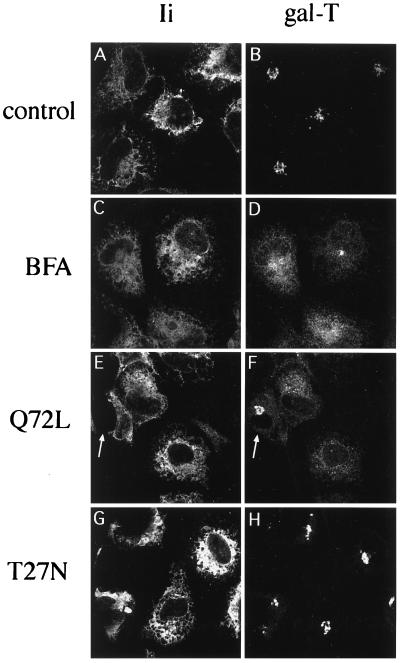
We first compared by immunofluorescence and confocal microscopy the localization of a Golgi resident protein (gal-T) to the distribution of an ER marker in cells overexpressing rab6 constructs. As an ER marker, we used the invariant chains Iip31/Iip33 of the human major histocompatibility complex class II antigens. When expressed in cells without major histocompatibility complex II α and β chains, Ii chains are retained in the ER, due to the presence of a specific signal (RRXX) on the cytoplasmic N terminus of Iip33 (22). Iip31, generated through a second initiation codon (23), does not contain the RRXX motif but is retained in the ER, because it forms multimeric complexes with Iip33 (mainly trimers; ref. 18). In the experiment shown in Fig. 1, HeLa cells were cotransfected with a plasmid encoding Ii chains and a plasmid encoding either rab6 Q72L or rab6 T27N. The staining pattern obtained after labeling of cells with anti-Ii antibody indicates that the bulk of Ii was associated with the ER (Fig. 1 A, C, E, and G). In control cells (cotransfected with Ii and empty pGEM-1 plasmids), anti-gal-T antibody decorated, as expected, typical Golgi structures (Fig. 1B). In contrast, most of the cells transfected with rab6 Q72L displayed a dispersed gal-T staining resembling the one obtained for Ii (Fig. 1 E and F). In many cells, gal-T was detected in the perinuclear membrane (this is more easily observed at higher magnification; see Fig. 7). In addition, the few cells in the rab6 Q72L transfected cell population that did not express Ii (Fig. 1E, arrow) displayed normal Golgi staining (Fig. 1F, arrow). Since nearly 100% of cells are cotransfected (i.e., with Ii- and rab6 Q72L-encoding plasmids) under our experimental conditions, this indicates that the dispersion of gal-T staining was due to overexpression of rab6 Q72L. Remarkably, gal-T staining observed in cells overexpressing rab6 Q72L was similar to the one seen in cells incubated with BFA (Fig. 1 C and D). We noticed, however, that gal-T staining was not completely redistributed in the ER of BFA-treated cells (Fig. 1 C and D), as previously observed (24).
Figure 1.
Overexpression of rab6 Q72L promotes the redistribution of Golgi gal-T into the ER. HeLa cells cotransfected with Ii and either pGEM-1 (A–D), rab6 Q72L (E and F), or rab6 T27N (G and H) plasmids were fixed 6 h after transfection with 4% paraformaldehyde and processed for immunofluorescence. BFA (5 μg/ml) was added 2 h before fixation (C and D). After permeabilization with saponin, cells were double labeled with a monoclonal antibody against Ii (A, C, E, and G) and a polyclonal antibody against gal-T (B, D, F, and H). The arrows in E and F point to a nontransfected cell that displays normal gal-T staining. All pictures shown here represent stacks of four medial optical slices obtained by confocal microscopy and separated from each other by 0.5 μm.
Figure 7.
rab6 Q72L-induced redistribution of gal-T is microtubule-dependent. HeLa cells were cotransfected with Ii and either pGEM-1 (A–D) or rab6 Q72L (E and F) plasmids. Nocodazole (10 μM; B, D, and F) and BFA (5 μg/ml; C and D) were added to the cells 3 and 4 h posttransfection, respectively. Cells were fixed 6 h posttransfection with 4% paraformaldehyde and double labeled with anti-Ii and anti-gal-T antibodies. We only show in this figure gal-T staining. All of these cells were also stained with anti-Ii antibody.
Overexpression of the GDP-bound mutant of rab6 (rab6 T27N) did not induce a redistribution of gal-T staining, as illustrated in Fig. 1 G and H. On the other hand, overexpression of wt rab6 also led to an ER-like distribution of gal-T staining, although this effect was not as pronounced as in rab6 Q72L-expressing cells (data not shown).
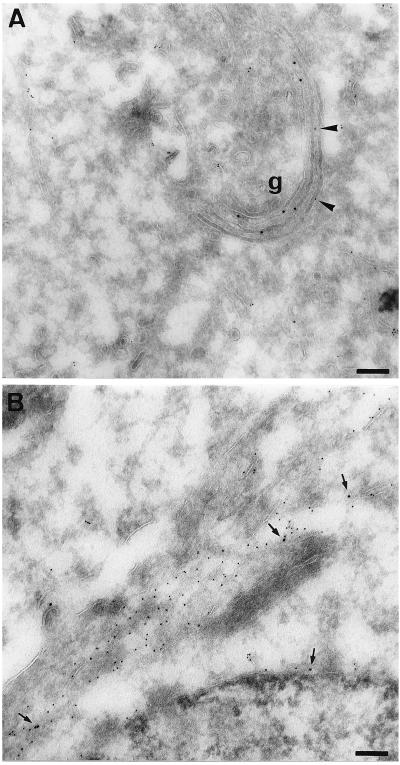
The observation that rab6 Q72L induces the redistribution of gal-T into the ER was confirmed by immuno-electron microscopy on ultrathin cryosections. In control cells, the gal-T antigen was found in cisternae of the Golgi apparatus and Ii was associated with ER membranes (Fig. 2A). However, Ii was often detected close to Golgi membranes and sometimes within the cisternae (Fig. 2A, arrowheads). In contrast, in cells transfected with rab6 Q72L, gal-T displayed a scattered distribution, part of which was found in membrane compartments strongly labeled by antibodies against Ii (Fig. 2B, arrows). In addition, gal-T was detected in the ER-connected nuclear envelope (Fig. 2B, arrows at bottom). In cells transfected with rab6 Q72L alone, a part of gal-T colocalized with protein disulfide isomerase, an endogenous resident ER enzyme (data not shown).
Figure 2.
Colocalization of gal-T and Ii in rab6 Q72L transfected cells. HeLa cells were cotransfected with Ii and either pGEM-1 (A) or rab6 Q72L (B) plasmids. Cells were fixed 6 h after transfection and processed for cryosectioning and immuno-electron microscopy. Cells were double labeled with antibody against gal-T and anti-Ii antibody. Primary antibodies were visualized with protein A-gold conjugates (gal-T, 10 nm; Ii, 5 nm). In control cells (A), anti-gal-T antibody labels typical Golgi stacks (g). Arrowheads in A point to Ii molecules localized in Golgi stacks in control cells. Arrows in B point to gal-T molecules localized within ER cisternae (Ii-positive) in rab6 Q72L transfected cells. (Bars = 0.1 μm.)
Similar results were obtained with the cis/medial marker α-mannosidase II that was shown to be completely redistributed in rab6 Q72L-expressing cells (data not shown). It was also interesting to monitor the behavior of rab6 itself in overexpressing cells. Since most of the overexpressed rab6 proteins were cytosolic (15), cells were permeabilized with saponin before fixation. Both endogenous rab6 and rab6 Q72L (and to a lesser extent overexpressed wt rab6) were found to be redistributed to the ER (data not shown).
Overexpression of wt rab6 and rab6 Q72L Causes the Addition of Sialylated O-Glycans on Ii.
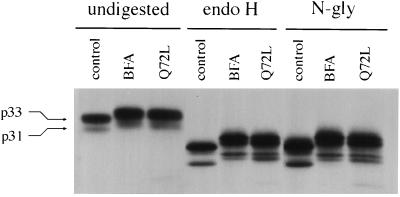
To further establish that Golgi proteins are redistributed into the ER in cells overexpressing rab6 Q72L, we determined whether Golgi-specific posttranslational modifications such as glycosylations can be detected on Ii. Experiments performed in BFA-treated cells have already indicated that Golgi glycosidases and glycosyltransferases can be still active after redistribution into the ER (25). We performed a series of pulse–chase experiments in which Ii was immunoprecipitated from control cells, BFA-treated cells, or cells transfected with rab6 Q72L. As shown in Fig. 3, Ii present in rab6 Q72L-overexpressing cells after a 10-min pulse and a 3-h chase migrated with a slower mobility on SDS/PAGE than Ii taken from control cells. Interestingly, Ii displayed exactly the same shift in mobility in cells incubated with BFA (Fig. 3, undigested). On the other hand, no difference in electrophoretic mobility of Ii was found between control and rab6 Q72L transfected cells after a 10-min pulse, indicating that posttranslational modifications were indeed responsible for the shift in Ii mobility (data not shown).
Figure 3.
rab6 Q72L and BFA induce posttranslational modifications of Ii that do not correspond to a maturation of N-linked sugars. HeLa cells cotransfected with Ii and either pGEM-1 (control, BFA) or rab6 Q72L (Q72L) plasmids were metabolically labeled for 10 min with [35S]methionine and [35S]cysteine and chased in medium alone (control, Q72L) or medium containing 5 μg/ml BFA. Ii present in cells was immunoprecipitated after 3 h. Immune precipitates were then divided into three equal parts and treated with or without endo H or N-glycanase (N-gly). Human Ii cDNA contains two ATGs, which generate two forms of Ii, Iip31 and Iip33 (arrows). Ii present in rab6 Q72L overexpressing cells and in BFA-treated cells displayed the same shift in mobility as compared with control cells (tracks undigested). In all cell lysates, Ii was endo H-sensitive (tracks endo H). The treatment with N-glycanase did not abolish the difference in mobility seen between Ii from control cells and Ii from BFA-treated or rab6 Q72L transfected cells.
Fig. 3 shows that the electrophoretic shift of Ii induced by rab6 Q72L expression or BFA treatment was not due to maturation of N-linked sugars. Ii immunoprecipitated from all cell lysates after a 3 h chase was still sensitive to endo H (Fig. 3, endo H). N-glycanase treatment, which removes all N-linked sugars from glycoproteins, did not also abolish the difference in mobility seen between Ii from control cells and Ii from BFA-treated or rab6 Q72L transfected cells (Fig. 3, N-gly). A reason why no maturation of N-linked sugars on Ii was detected could be that N-glycans of Ii are poor substrates for Golgi enzymes. Alternatively, although it is known that mature forms of Ii contain galactose and sialic acids residues (26), the fact that they are O-linked, N-linked, or both has not been firmly established.
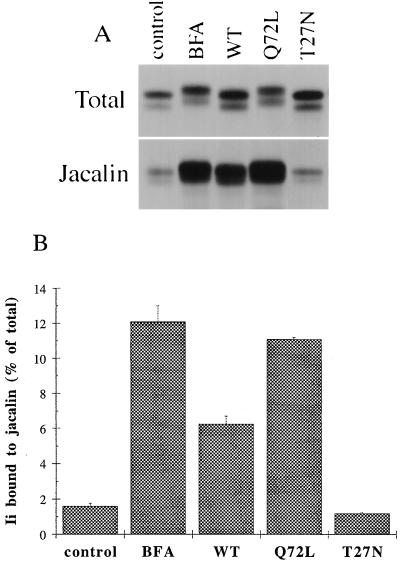
We therefore tested the possibility that the electrophoretic shift in Ii mobility was due to the addition of O-linked sugars. O-glycosylation of proteins following the biosynthetic/secretory pathway has been shown to start in the cis Golgi and/or the intermediate compartment, by the addition of N-acetylgalactosamine (GalNAc) on serine and threonine residues (27–30). Galactose, fucose, sialic acid, and terminal GalNAc residues are then added in more distal parts of the Golgi apparatus (31). Ii immune precipitates were solubilized and incubated with jacalin, a lectin that specifically binds to the central motif of O-glycans: Ser/Thr-GalNAc-Gal(β1–3) (for a review on jacalin, see ref. 32). It should be pointed out that substitutions of the central motif of O-glycosylation, such as addition of sialic acids, do not alter the binding capacity of jacalin (32). The experiments presented in Fig. 4 A and B show that only a small amount of Ii can bind to jacalin in control cells. In contrast, overexpression of rab6 Q72L greatly increased (≈7 times, Fig. 4B) the amount of Ii eluted from the jacalin beads. Interestingly, almost the same amount was found in BFA-treated cells. Moreover, overexpression of wt rab6 also increased the amount of O-glycosylated Ii (≈4 times more over control levels, Fig. 4B). On the other hand, overexpression of rab6 T27N did not increase, but rather decreased the binding of Ii to jacalin, as we will discuss later (see Figs. 4 A and B and 6).
Figure 4.
rab6 Q72L, wt rab6, and BFA increase O-glycosylation of Ii. HeLa cells cotransfected with Ii and either empty plasmid (control, BFA) or the indicated rab6 constructs (WT, Q72L, T27N) were metabolically labeled for 10 min and chased for 3 h. In BFA-treated cells, 5 μg/ml BFA was added to the chase medium. Ii immune precipitates were solubilized with SDS and nine-tenths of each eluate was diluted in Triton X-100 containing buffer and incubated with biotinylated jacalin and streptavidin-agarose. O-glycosylated Ii bound to jacalin was specifically eluted with galactose. (A) Visualization by autoradiography of Ii present in the different cell lysates before (Total) or after binding to jacalin (Jacalin). Total represents one-tenth of the whole immunoprecipitate for each sample. (B) Ii bands were quantified by scanning with the PhosphorImager. This graph represents the means ± SD of two independent experiments.
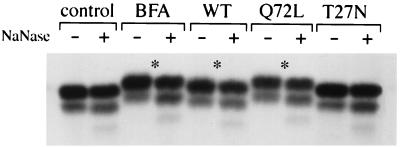
We then examined the stage of maturation of the O-linked sugars present on Ii. Ii immune precipitates were treated with neuraminidase, an exoglycosidase that removes terminal sialic acids present on both O- and N-linked glycans. As shown in Fig. 5, Ii present in rab6 Q72L and wt rab6 transfected cells as well as in BFA-treated cells was sialylated, as demonstrated by the shift in mobility on SDS/PAGE obtained after treatment with neuraminidase (Fig. 5, asterisks). In contrast, Ii present in control cells or in rab6 T27N transfected cells was not sialylated. It is noteworthy that, when Ii is sialylated, the whole band (and not just a fraction of it) was shifted down upon neuraminidase treatment. This result, and the observation that N-linked sugars present on Ii molecules remain endo H-sensitive (and thus unsialylated), demonstrate that sialylation occurred on O-linked sugars. In addition, one can conclude that Ii was uniformly O-glycosylated in BFA-treated cells, rab6 Q72L, and wt rab6 transfected cells.
Figure 5.
Ii is sialylated in BFA-treated cells and in cells overexpressing wt rab6 and rab6 Q72L. HeLa cells were transfected, pulsed, and chased as described in the legend to Fig. 4. After immunoprecipitation, Ii immunoprecipitates were treated with (+) or without (−) neuraminidase. Asterisks (∗) point to the shift in mobility of Ii caused by the removal of sialic acids. Note that both p31 and p33 are shifted down upon neuraminidase treatment.
The GDP-Bound Form of rab6 (rab6 T27N) Inhibits O-Glycosylation of Ii.
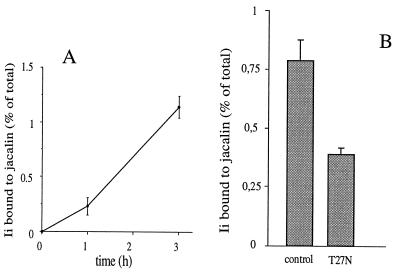
The amount of O-glycosylated Ii is very low in control cells (Fig. 4), consistent with morphological data showing that the bulk of Ii is associated with the ER (Figs. 1 and 2). However, as shown in the pulse–chase experiment described in Fig. 6A, the amount of Ii eluted from jacalin beads in control cells significantly increased over the chase period. Two nonmutually exclusive hypotheses may account for this result. Although the significance of this process is unclear, a recycling of Golgi resident enzymes through early Golgi compartments (33, 34) and the ER (35) has been recently documented. Such a process may progressively increase the amount of Ii retained on jacalin beads under control conditions. Alternatively, it has been proposed that the RRXX motif present in the N-terminal region of Iip33 functions as an ER retrieval signal, like the KKXX motif of ER-retained type I membrane proteins (22). Ii could then be progressively O-glycosylated in Golgi compartments before being retrieved to the ER. It should be pointed out that some Ii molecules can be detected in Golgi compartments by immuno-electron microscopy (see Fig. 2A, arrowheads).
Figure 6.
(A) Ii progressively acquires O-linked sugars in control cells. HeLa cells cotransfected with Ii and empty pGEM-1 plasmids were metabolically labeled for 10 min and chased for 0, 1, or 3 h. Ii immune precipitates were used in the jacalin binding assay as described in the legend to Fig. 4. Radioactivity in each sample (total and jacalin-bound) was directly measured with the β counter. Error bars represent the range in three independent experiments. (B) rab6 T27N inhibits O-glycosylation of Ii. HeLa cells cotransfected with Ii and either pGEM-1 (control) or rab6 T27N (T27N) plasmids were metabolically labeled for 10 min and chased for 3 h. Ii bound to jacalin was calculated as described above. Error bars represent the range in three independent experiments. The data in A and B are shown after subtraction of background (counts obtained after 10-min pulse).
This observation led us to investigate in further detail a possible effect of the GDP-bound mutant rab6 T27N on Ii O-glycosylation. Cells expressing both Ii and this mutant were pulse-labeled for 3 h before Ii being immunoprecipitated and bound to jacalin. As shown in Fig. 6B, overexpression of rab6 T27N inhibited by 50% the amount of O-glycosylated Ii eluted from the jacalin beads.
The Effects of rab6 Q72L Require the Integrity of Microtubules.
The above results show that overexpression of rab6 Q72L induces a BFA-like phenotype. We were then curious to see whether the effect of rab6 Q72L was also microtubule dependent, as it is well documented for BFA-induced redistribution of Golgi proteins (36). The immunofluorescence shown in Fig. 7 indicates that, as expected, incubation of cells with nocodazole, a drug that disrupts the microtubular network, before the addition of BFA prevents the redistribution of gal-T into the ER (Fig. 7 C and D). Interestingly, nocodazole also antagonizes the effect of the GTPase mutant of rab6. In cells overexpressing rab6 Q72L, but incubated with nocodazole before this protein starts to be expressed (3 h posttransfection), gal-T staining was found associated with typical dispersed Golgi structures, indistinguishable from the ones seen in nocodazole-treated control cells (Fig. 7 E and F). The disruption of microtubules also prevents O-glycosylation of Ii in rab6 Q72L-overexpressing cells (data not shown).
DISCUSSION
rab6 localizes to the Golgi (mainly medial and trans) and trans-Golgi network membranes (13, 14). Consistent with this localization, we have previously shown that in HeLa cells overexpression of GTP-bound forms of rab6 (wt rab6 and rab6 Q72L) affects transport of both membrane and secretory proteins between an α-mannosidase II-positive and a sialyl-transferase-positive Golgi compartment (15). In these cells, no effect was found on transport between trans-Golgi network and plasma membrane (15). The main finding of the present study is that overexpression of GTP-bound forms of rab6 induces the redistribution of Golgi resident proteins into the ER. Altogether, the above results suggest that increased levels of rab6:GTP affect the dynamics and homeostasis of medial/late Golgi-trans-Golgi network compartments. As a consequence, resident proteins of these compartments, such as gal-T and sialyltransferases, may be relocalized into early Golgi compartments. From there, Golgi proteins and membranes might be included in the active retrograde flow existing between early Golgi and ER. Such an interpretation does not suppose a direct role of rab6 in Golgi to ER transport.
The Golgi apparatus is an organelle whose structure is likely maintained by a controlled anterograde and retrograde membrane flow. The anterograde flow allows efficient transport of secretory proteins and growing evidence indicates that proteins, including ER escaped proteins, Golgi resident proteins, and toxins, can be transported in a retrograde way between Golgi compartments (37–42). Interestingly, overexpression of the human KDEL receptor ELP-1 has been shown to induce a BFA-like phenotype (43), suggesting that hyperactive retrograde traffic may cause Golgi disassembly. The redistribution of Golgi resident proteins seen in cells overexpressing GTP-bound forms of rab6 may be a consequence of a block of anterograde transport that would unbalance anterograde and retrograde flow in favor of retrograde transport. Alternatively, rab6 may be directly involved in the retrograde pathway. This latter hypothesis would be in agreement with the fact that overexpression of GTP-bound forms of rab proteins generally results in a gain of function (2, 3).
The results that we obtained with rab6 T27N are also more in favor of a direct involvment of rab6 in an intra-Golgi retrograde pathway. GDP-bound forms of rab proteins have been shown to be potent inhibitors of transport events (44, 45). Nevertheless, we previously found that overexpression of rab6 T27N has no effect on the kinetics of intra-Golgi transport of secretory markers (15). Here, we show that rab6 T27N does not induce the redistribution of Golgi proteins into the ER but inhibits basal O-glycosylation of Ii. Such an effect could be explained by a rab6 T27N induced inhibition of the recycling of Golgi enzymes through early Golgi compartments (assuming that Ii cycle between ER and these compartments) or through the ER. The reciprocal effect of increased rab6 activity (overexpression of GTP-bound forms) would be the forced recycling of Golgi enzymes. The consequence could be a dramatic alteration of Golgi morphology, since Golgi enzymes have been proposed as being directly involved in the maintenance of the structure of Golgi stacks [kin recognition hypothesis, (46)].
A striking result of this study is that overexpression of rab6 Q72L causes almost the same phenotypic effects as does the addition of BFA. It is unlikely, however, that these effects arise via analogous mechanisms. BFA impairs the cycle of association and dissociation of coat proteins, including COP I, and the formation of transport vesicles in vitro (47). The removal of coat proteins and/or inhibition of the formation of vesicles probably explains why BFA induces mixing of Golgi and ER membranes (48, 49). Most of the data accumulated so far on rab proteins do not support a role of these proteins in the assembly of coats, or the formation of transport vesicles. It should be also pointed out that the typical necklaces and tubular structures emanating from the Golgi of BFA-treated cells (36) were never observed in cells overexpressing wt rab6 or rab6 Q72L, even at early times of overexpression.
In conclusion, our data demonstrate that alteration of rab6 function has a dramatic effect on the architecture of the Golgi apparatus. Although the presence of rab6 on putative post-Golgi vesicles in some specialized cell types may suggest an additional role for this protein (50, 51), the body of evidence points to a role for rab6 in a microtubule dependent retrograde membrane traffic within the Golgi.
Acknowledgments
We thank Drs. Gordon Langsley and Catherine Desaymard for critical reading of this manuscript and Daniel Meur and Dominique Morineau for expert photographic work. This work was supported by grants from the Association de la Recherche contre le Cancer (1405), from the Fondation pour la Recherche Medicale, from the European Community (ERB CHR XCT 940 592), and from the Swiss National Science Foundation (to E.G.B., 3100-40749.94).
ABBREVIATIONS
- ER
endoplasmic reticulum
- wt
wild-type
- BFA
brefeldin A
- Ii
major histocompatibility complex class II-associated invariant chain
- endo H
endoglycosidase H
- gal-T
β-1,4-galactosyltransferase
References
- 1.Goud B, McCaffrey M. Curr Opin Cell Biol. 1991;3:626–633. doi: 10.1016/0955-0674(91)90033-u. [DOI] [PubMed] [Google Scholar]
- 2.Zerial M, Stenmark H. Curr Opin Cell Biol. 1993;5:613–620. doi: 10.1016/0955-0674(93)90130-i. [DOI] [PubMed] [Google Scholar]
- 3.Pfeffer S R. Curr Opin Cell Biol. 1994;6:522–526. doi: 10.1016/0955-0674(94)90071-x. [DOI] [PubMed] [Google Scholar]
- 4.Brennwald P, Kearns B, Champion K, Keranen S, Bankaitis V, Novick P. Cell. 1994;79:245–258. doi: 10.1016/0092-8674(94)90194-5. [DOI] [PubMed] [Google Scholar]
- 5.Lian J P, Stone S, Jiang Y, Lyons P, Ferro-Novick S. Nature (London) 1994;372:698–701. doi: 10.1038/372698a0. [DOI] [PubMed] [Google Scholar]
- 6.Sogaard M, Tani K, Ye R R, Geromanos S, Tempst P, Kirchhausen T, Rothman J E, Sollner T. Cell. 1994;78:937–948. doi: 10.1016/0092-8674(94)90270-4. [DOI] [PubMed] [Google Scholar]
- 7.Johannes L, Doussau F, Clabecq A, Henry J P, Darchen F, Poulain B. J Cell Sci. 1996;109:2875–2884. doi: 10.1242/jcs.109.12.2875. [DOI] [PubMed] [Google Scholar]
- 8.Bucci C, Parton R G, Mather I H, Stunnenberg H, Simons K, Hoflack B, Zerial M. Cell. 1992;70:715–728. doi: 10.1016/0092-8674(92)90306-w. [DOI] [PubMed] [Google Scholar]
- 9.Stenmark H, Parton R G, Steele-Mortimer O, Lutcke A, Gruenberg J, Zerial M. EMBO J. 1994;13:1287–1296. doi: 10.1002/j.1460-2075.1994.tb06381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tisdale E J, Bourne J R, Khosravi-Far R, Der C J, Balch W E. J Cell Biol. 1992;119:749–761. doi: 10.1083/jcb.119.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilson B S, Nuoffer C, Meinkoth J L, McCaffery M, Feramisco J R, Balch W E, Farquhar M G. J Cell Biol. 1994;125:557–571. doi: 10.1083/jcb.125.3.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorvel J P, Chavrier P, Zerial M, Gruenberg J. Cell. 1991;64:915–925. doi: 10.1016/0092-8674(91)90316-q. [DOI] [PubMed] [Google Scholar]
- 13.Goud B, Zahraoui A, Tavitian A, Saraste J. Nature (London) 1990;345:553–556. doi: 10.1038/345553a0. [DOI] [PubMed] [Google Scholar]
- 14.Antony C, Cibert C, Geraud G, Santa Maria A, Maro B, Mayau V, Goud B. J Cell Sci. 1992;103:785–796. doi: 10.1242/jcs.103.3.785. [DOI] [PubMed] [Google Scholar]
- 15.Martinez O, Schmidt A, Salamero J, Hoflack B, Roa M, Goud B. J Cell Biol. 1994;127:1575–1588. doi: 10.1083/jcb.127.6.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watzele G, Bachofner R, Berger E G. Eur J Cell Biol. 1991;56:451–458. [PubMed] [Google Scholar]
- 17.Roth J, Berger E G. J Cell Biol. 1982;93:223–229. doi: 10.1083/jcb.93.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roche P A, Marks M S, Cresswell P. Nature (London) 1991;354:392–394. doi: 10.1038/354392a0. [DOI] [PubMed] [Google Scholar]
- 19.Fuerst T R, Niles E G, Studier F W, Moss B. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roa M, Cornet V, Yang C Z, Goud B. J Cell Sci. 1993;106:789–802. doi: 10.1242/jcs.106.3.789. [DOI] [PubMed] [Google Scholar]
- 21.Slot J W, Geuze H J, Gigengack S, Lienhard G E, James D E. J Cell Biol. 1991;113:123–135. doi: 10.1083/jcb.113.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schutze M P, Peterson P A, Jackson M R. EMBO J. 1994;13:1696–1705. doi: 10.1002/j.1460-2075.1994.tb06434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strubin M, Long E O, Mach B. Cell. 1986;47:619–625. doi: 10.1016/0092-8674(86)90626-4. [DOI] [PubMed] [Google Scholar]
- 24.Strous G J, Berger E G, van Kerkhof P, Bosshart H, Berger B, Geuze H J. Biol Cell. 1991;71:25–31. doi: 10.1016/0248-4900(91)90048-r. [DOI] [PubMed] [Google Scholar]
- 25.Lippincott-Schwartz J, Yuan L C, Bonifacino J S, Klausner R D. Cell. 1989;56:801–813. doi: 10.1016/0092-8674(89)90685-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cresswell P. Annu Rev Immunol. 1994;12:259–293. doi: 10.1146/annurev.iy.12.040194.001355. [DOI] [PubMed] [Google Scholar]
- 27.Tooze S A, Tooze J, Warren G. J Cell Biol. 1988;106:1475–1487. doi: 10.1083/jcb.106.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krijnse-Locker J, Ericsson M, Rottier P J, Griffiths G. J Cell Biol. 1994;124:55–70. doi: 10.1083/jcb.124.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deschuyteneer M, Eckhardt A E, Roth J, Hill R L. J Biol Chem. 1988;263:2452–2459. [PubMed] [Google Scholar]
- 30.Roth J, Wang Y, Eckhardt A E, Hill R L. Proc Natl Acad Sci USA. 1994;91:8935–8939. doi: 10.1073/pnas.91.19.8935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roth J. Biochim Biophys Acta. 1987;906:405–436. doi: 10.1016/0304-4157(87)90018-9. [DOI] [PubMed] [Google Scholar]
- 32.Kabir S, Daar A S. Immunol Invest. 1994;23:167–188. doi: 10.3109/08820139409087798. [DOI] [PubMed] [Google Scholar]
- 33.Hoe M H, Slusarewicz P, Misteli T, Watson R, Warren G. J Biol Chem. 1995;270:25057–25063. doi: 10.1074/jbc.270.42.25057. [DOI] [PubMed] [Google Scholar]
- 34.Harris S L, Waters M G. J Cell Biol. 1996;132:985–998. doi: 10.1083/jcb.132.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cole N, Ellenberg J, Song J, Lippincott-Schwartz J. Mol Biol Cell. 1995;6:1913. [Google Scholar]
- 36.Lippincott-Schwartz J, Donaldson J G, Schweizer A, Berger E G, Hauri H P, Yuan L C, Klausner R D. Cell. 1990;60:821–836. doi: 10.1016/0092-8674(90)90096-w. [DOI] [PubMed] [Google Scholar]
- 37.Jackson M R, Nilsson T, Peterson P A. J Cell Biol. 1993;121:317–333. doi: 10.1083/jcb.121.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Griffiths G, Ericsson M, Krijnse-Locker J, Nilsson T, Goud B, Soling H D, Tang B L, Wong S H, Hong W. J Cell Biol. 1994;127:1557–1574. doi: 10.1083/jcb.127.6.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moremen K W, Touster O. J Biol Chem. 1985;260:6654–6662. [PubMed] [Google Scholar]
- 40.Johnston P A, Stieber A, Gonatas N K. J Cell Sci. 1994;107:529–537. doi: 10.1242/jcs.107.3.529. [DOI] [PubMed] [Google Scholar]
- 41.Sandvig K, Garred O, Prydz K, Kozlov J V, Hansen S H, van Deurs B. Nature (London) 1992;358:510–512. doi: 10.1038/358510a0. [DOI] [PubMed] [Google Scholar]
- 42.Majoul I V, Bastiaens P I H, Söling H D. J Cell Biol. 1996;133:777–789. doi: 10.1083/jcb.133.4.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hsu V W, Shah N, Klausner R D. Cell. 1992;69:625–635. doi: 10.1016/0092-8674(92)90226-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li G, Stahl P D. J Biol Chem. 1993;268:24475–24480. [PubMed] [Google Scholar]
- 45.Nuoffer C, Davidson H W, Matteson J, Meinkoth J, Balch W E. J Cell Biol. 1994;125:225–237. doi: 10.1083/jcb.125.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nilsson T, Hoe M H, Slusarewicz P, Rabouille C, Watson R, Hunte F, Watzele G, Berger E G, Warren G. EMBO J. 1994;13:562–574. doi: 10.1002/j.1460-2075.1994.tb06294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Donaldson J G, Finazzi D, Klausner R D. Nature (London) 1992;360:350–352. doi: 10.1038/360350a0. [DOI] [PubMed] [Google Scholar]
- 48.Pelham H R. Cell. 1991;67:449–451. doi: 10.1016/0092-8674(91)90517-3. [DOI] [PubMed] [Google Scholar]
- 49.Elazar Z, Orci L, Ostermann J, Amherdt M, Tanigawa G, Rothman J E. J Cell Biol. 1994;124:415–424. doi: 10.1083/jcb.124.4.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jasmin B J, Goud B, Camus G, Cartaud J. Neuroscience. 1992;49:849–855. doi: 10.1016/0306-4522(92)90361-5. [DOI] [PubMed] [Google Scholar]
- 51.Tixier-Vidal A, Barret A, Picart R, Mayau V, Vogt D, Wiedenmann B, Goud B. J Cell Sci. 1993;105:935–947. doi: 10.1242/jcs.105.4.935. [DOI] [PubMed] [Google Scholar]